Abstract
Growing well-diffracting crystals constitutes a serious bottleneck in structural biology. A recently proposed crystallization methodology for “stubborn crystallizers” is to engineer surface sequence variants designed to form intermolecular contacts that could support a crystal lattice. This approach relies on the concept of surface entropy reduction (SER), i.e., the replacement of clusters of flexible, solvent-exposed residues with residues with lower conformational entropy. This strategy minimizes the loss of conformational entropy upon crystallization and renders crystallization thermodynamically favorable. The method has been successfully used to crystallize more than 15 novel proteins, all stubborn crystallizers. But the choice of suitable sites for mutagenesis is not trivial. Herein, we announce a Web server, the surface entropy reduction prediction server (SERp server), designed to identify mutations that may facilitate crystallization. Suggested mutations are predicted based on an algorithm incorporating a conformational entropy profile, a secondary structure prediction, and sequence conservation. Minor considerations include the nature of flanking residues and gaps between mutation candidates. While designed to be used with default values, the server has many user-controlled parameters allowing for considerable flexibility. Within, we discuss (1) the methodology of the server, (2) how to interpret the results, and (3) factors that must be considered when selecting mutations. We also attempt to benchmark the server by comparing the server's predictions with successful SER structures. In most cases, the structure yielding mutations were easily identified by the SERp server. The server can be accessed at http://www.doe-mbi.ucla.edu/Services/SER.
Keywords: crystallography, SER variants, sequence replacements, surface entropy reduction (SER), X-ray methods
Crystallization remains the rate-limiting step in macromolecular X-ray diffraction analysis. In spite of dramatic progress in the design of extensive screens, the advent of sophisticated nanovolume robots and, microfluidic technology, the process relies on screening because to date it has not been possible to assess the solubility of a protein, or to predict its behavior in crystallization screens, based on its amino acid sequence alone. High-throughput structural genomics laboratories utilize crystallization robots that are able to generate over 100,000 samples per day while minimizing the sample volumes down to 50 nL or less, yet the number of conditions tested is not directly correlated to the likelihood of success of crystallizing a given protein, which even for a subset of relatively small, single-domain prokaryotic proteins does not typically exceed ∼30% (Stevens 2000).
Although prediction of a protein's macroscopic physical properties such as solubility and crystallizability constitutes at present an insurmountable challenge, it is well established that even small changes in the amino acid sequence can dramatically change a protein's behavior. The canonical example is the β6 Glu → Val mutation in hemoglobin, which critically reduces the solubility of the protein leading to sickle cell anemia. Thus, in principle one should be able to generate relatively minor modifications to proteins by site-directed mutagenesis that would result in molecules that are more amenable to crystallization than the wild-type sequence. The question is, however, if we can identify relatively simple rules that would allow for rational design of mutants with enhanced crystallizability.
Crystallization involves the formation of intermolecular contacts which facilitate the assembly of the protein molecules or multimers into a crystalline lattice. While without this step a macromolecular crystal would never form, the phenomenon historically attracted limited attention with only a handful of papers written on the subject. The chemistry of crystal contacts is, however, both interesting and important. Immobilization of side chains with high conformational entropy at the point of crystal contacts is energetically unfavorable and is expected to impede crystallization. It follows that—all other things being equal—specific solvent-exposed amino acid sequence motifs with lower conformational entropy may permit thermodynamically favorable crystal contacts in some proteins. This notion was explicitly used by one of us to formulate the surface entropy reduction (SER) approach to protein crystallization (Derewenda 2004b) in which clusters of two to three amino acids with high conformational entropy, such as Lys, Glu, and Gln, are replaced with Ala.
The SER concept was tested with very encouraging results in a model system of human RhoGDI (guanine-nucleotide dissociation inhibitor) and was subsequently used to obtain crystallizable variants of a number of proteins, or to generate new crystal forms with significantly improved diffraction properties (Table 1). Moreover, in those cases where the method was successful, typically very few mutants (no more than three) were necessary to identify a crystallizable variant. Heretofore, the SER approach required a subjective analysis of the amino acid sequence of the target protein and a manual selection of the mutants by the investigator, without the aid of tools that might objectively gauge the probability of success. The analysis of protein sequences by the server described in this paper aims to automate the tasks of identifying sites most suitable for mutation designed to confer enhanced crystallizability based on a range of predictions and conditions, stemming from both objective criteria as well as the database of structures crystallized by the SER strategy.
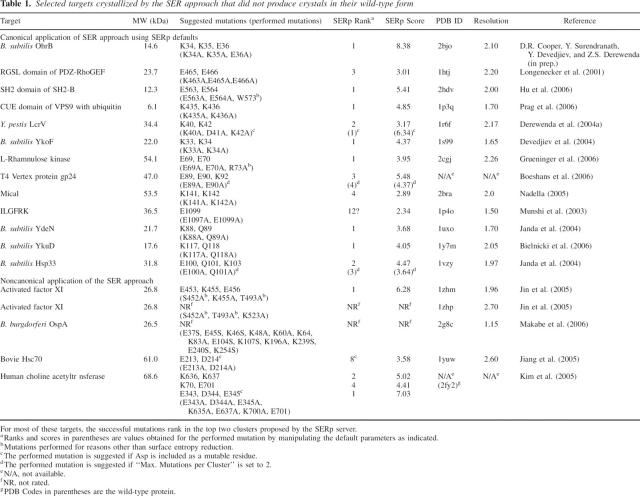
Table 1.
Selected targets crystallized by the SER approach that did not produce crystals in their wild-type form
Description of the SER server
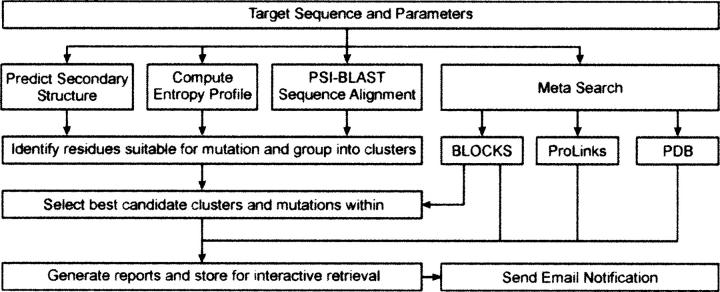
The submitted amino acid or DNA sequence undergoes three principal analyses, whose combined results determine sites most suitable for mutation (Scheme 1). These three analyses are (1) prediction of the surface conformational entropy, (2) prediction of secondary structure, and (3) analysis of patterns of evolutionary sequence conservation. The overall objective is to identify a site containing solvent-exposed residues with high conformational entropy that may impede the formation of crystal contacts and which are poorly conserved by evolution, suggesting that they are not involved in functionally critical active sites or other functional epitopes. Most default processing parameters can be easily adjusted and are reflected in almost instantaneously updated results. Intermediate scores from each principal analysis as well as composite scores for selected residues are provided in the server's results.
Scheme 1.
Flowchart summarizing the SER prediction process. The submitted gene or peptide sequence undergoes three principal analyses: secondary structure prediction, computation of its side chain entropy profile, and a search for homologous sequences. Potential residue candidates are grouped into clusters and scored using several key principles (see text). Additional meta searches identify functionally important regions, structural homologs, and linkages to potential interacting protein partners.
Conformational entropy profile
A side-chain conformational entropy profile is computed using the values for individual amino acids proposed by Pickett and Sternberg (1993) as a basis. These values are then multiplied by the solvent exposure index (i.e., the frequency a particular residue type is solvent-exposed in known structures, as described by Baud and Karlin (1999) and normalized with respect to lysine, the amino acid with the highest conformational surface entropy. This produces a profile that contains high scores for residues with both high conformational entropy and high probability of significant solvent exposure, using a relative scale with the range of 0.0 (Ala) and 1.0 (Lys). To overcome local noise in this profile, a smoothing filter with a sliding window of three residues is applied, such that the side chain entropy for each residue is equal to the average for the three residues, centered on the target residue. This average is the per-residue entropy score returned by this analysis.
Secondary structure prediction
The secondary structure prediction is obtained with PSIPRED (Jones 1999) to identify loops between distinct secondary structure elements such as α-helices and β-strands. Such loops are given priority because they have a high probability of harboring solvent-exposed residues and have been shown to be effective in SER mutants designed to date to enhance crystallizability (D.R. Cooper, Y. Surendranath, and Z.S. Devedjiev, in prep.). Conversely, SER was found to be less effective for sites that lie on the solvent-exposed face of a helix, probably because the main chain groups in helices are not available for intermolecular H-bonding, thus impeding formation of crystal contacts. Residues that are predicted to be in an α-helix and β-strand strand with a PSIPRED confidence above the cutoff threshold (default value 0.2) are penalized and score lower. Confidences below this threshold are truncated to reduce some of the noise intrinsic to PSIPRED predictions. Additionally, loops shorter than three residues are ignored to reduce false positives in the secondary structure prediction. The per-residue secondary structure score from this analysis is in the range from 0.0 (not in a loop) to 1.0 (high confidence to be in a surface-exposed loop).
Evolutionary conservation
The third analysis estimates the evolutionary conservation of each residue in the submitted sequence from PSI-BLAST alignments. The conservation level is equal to the number of aligned sequences containing the identical residue at a specific position normalized by the total number of aligned sequences. The residue replacement level is equal to the number of sequences containing an alanine or another target residue at a specific position in any of the homologs, and is also normalized by the total number of aligned sequences. Conserved residues are avoided as targets for mutations while sites already containing one of the proposed target residues in homologs are preferred. The per-residue conservation score from this analysis is equal to (replacement level—conservation level) * sequence count weight, and thus is in the range from −1.0 (highly conserved) to +1.0 (changed to a target residue in all aligned sequences). The sequence count weight gives more significance to the conservation score if there are more aligned sequences. Lastly, residues that fall within a conserved block (as determined by the Blocks meta search, if enabled) are penalized by adding a negative value (default −0.5) to the conservation score.
Final evaluation, scoring, and additional features
Results from the three principal analyses are combined to establish the potential for mutation of each residue. Ideal candidates are nonconserved, high entropy residues that lie in surface-exposed, entropy-rich regions of the protein. The final per-residue score is a weighted sum of the contributions from each principal analysis, which are included in the detailed server results. The weights can be customized and by default are 1.0 for the entropy and secondary structure analyses, and 0.5 for the evolutionary conservation analysis.
Once scores are assigned to residues, the server looks for several suitable residues in close proximity, which are then grouped into a cluster. A cluster starts and ends with either a low entropy residue (e.g., Ala) or a potentially mutable high entropy residue (Lys, Glu, or Gln) and contains a continuous segment of only such residues. Disruptions of cluster continuity by a single or two consecutive other residues are allowed. The overall cluster score (SERp score) indicative of the predicted success is computed from scores of residues selected for mutation within said cluster. This calculation involves the use of specific weights, which currently are adjusted based on the database of proteins crystallized using the SER concept, but—like most other parameters—they can be modified by the user.
The following principles are taken into account while ranking mutable residues and selecting the exact mutations in a cluster. Removal of long, polar side chains may have a negative impact on protein solubility, thus the number of required mutations per cluster is minimized. Typically no more than three mutations per target are allowed, with a maximum gap of one residue between mutations. The length of the low entropy patch post-mutation is maximized to favor the creation of sufficiently large low entropy patches, which can form new crystal contacts. The number and length of interruptions of the low entropy patch by higher entropy residues is minimized, as continuous, uniform patches were found to work best. Finally, the change in entropy is maximized to favor the greatest reduction of entropy.
In the final output, the server suggests mutations for each candidate cluster in a simple summary form, along with detailed results from all analyses used to select the mutations. To achieve a sufficient change in surface entropy, all proposed mutations within a chosen cluster must be introduced into the target concurrently. Such a mutated cluster is expected to form a low entropy patch capable of mediating an intermolecular contact leading to successful crystallization. The server proposes several alternative candidate clusters ordered by their SERp scores indicative of predicted success.
Additional meta searches are performed on the submitted sequence to screen for other potential sources of failure of crystallization and to identify sequence regions that may be important for function. These include a search of the Prolinks, Blocks, and the PDB databases. The Prolinks database is a collection of inference methods used to predict functional linkages between proteins (Bowers et al. 2004). It has been suggested that crystallization may require or be improved by co-expression with an interacting partner (Strong et al. 2006). The Prolinks search identifies potentially interacting partners that could be co-expressed to improve crystallizability. The Blocks database contains multiply aligned, ungapped segments corresponding to the most highly conserved regions of proteins (Henikoff et al. 1999). This search identifies highly conserved motifs such as metal or ligand binding sites. Oftentimes the addition of the corresponding ligand may be required for or promote crystallization. Additionally, this search identifies functional regions of the protein, which are disfavored for mutation. Residues in such conserved regions receive a slight scoring penalty during selection of mutation candidates. A search of the PDB aims to identify known homologous structures, whose secondary structure is analyzed with DSSP (Kabsch and Sander 1983) to identify solvent accessible residues. The solvent accessibility is shown on the results graph (Fig. 1B) to provide additional indication of regions which are likely on the protein surface. Only results from the Blocks search have an impact on the selection and scoring of proposed mutations; results from all other meta searches are provided to assist in the final selection of proposed clusters. We are currently exploring new ways to incorporate these additional results into the cluster scoring and mutation selection process.
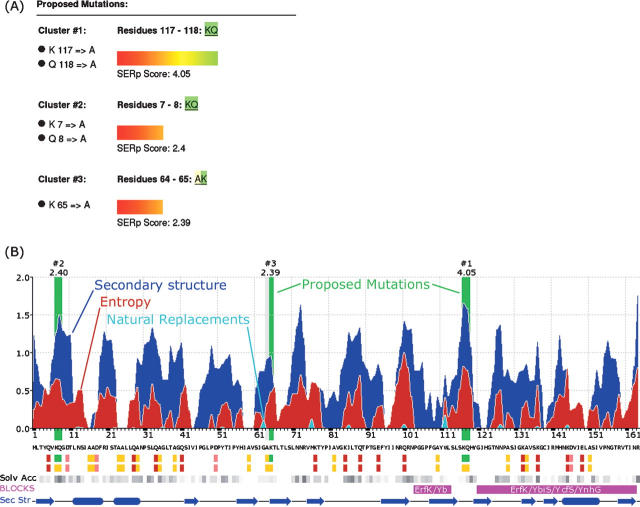
Figure 1.
(A) Summary of candidate clusters and proposed mutations therein for YkuD (PDB code 1Y7M). The SERp score (arbitrary units) is shown for each cluster. (B) A stacked summary graph showing the contribution from each principal analysis. The blue peaks (top) indicate residues predicted to be in loops. Red peaks correspond to high entropy regions (middle). Cyan peaks (bottom) indicate residues that in homologous structures are changed to one of the target residues. Residues at peaks of this stacked graph are preferred for mutation. Proposed mutations are highlighted in green. Residue types are shown below the protein sequence: high entropy residues in pink, mutable residues in red, low entropy target residues in yellow, and high entropy residues proposed for mutation in green. Selected results from performed meta searches are also shown. The solvent accessibility estimated from homologous structures (Kabsch and Sander 1983) is shown as shades of gray (darker shades indicate higher solvent accessibility). Highly conserved sequence regions identified with Blocks (Henikoff et al. 1999) are marked in magenta. Predicted secondary structure is shown schematically at the very bottom in blue.
The final selection of mutable sites is naturally dependent on the weights assigned to each of the parameters. Optimal weights can be defined only in an empirical fashion, so that mutations that led to readily crystallizable mutants are accurately predicted. We carried out such optimization when selecting the current default values for all parameters of the SERp server.
Results and Discussion
Overview of output
Figure 1A shows a representative sample of the SERp server's summary for the YkuD protein from Bacillus subtilis. This protein was originally selected as a target for the Midwest Center for Structural Genomics (MCSG). After it failed to crystallize in the high-throughput pipeline, Bielnicki et al. (2006) used the SER approach to determine the structure of this 18-kDa protein to 2.0 Å resolution, revealing a novel, ubiquitous family of bacterial enzymes. The scoring details of each suggested cluster can be viewed by selecting the “Score Details” tab in the top of the results page. Several graphical representations of the calculations are presented in the “Graphs” tab, including one graph that depicts the overall contribution of the entropy profile, secondary structure prediction, and sequence conservation (Fig. 1B). The secondary structure prediction is shown schematically below the main graph. In this main graph, suggested clusters are indicated by green vertical bars labeled with the cluster number and the SERp score for that cluster. The sequence is shown below the graph with a color-coded representation of high and low entropy residues, mutable residues, and proposed mutations. The graphical representation also includes some of the meta search results, including the calculated solvent accessibility of homologous proteins (darker shades of gray indicate higher solvent accessibility) and the location of conserved sequence motifs identified with Blocks.
For example, when the SERp server is run with all default parameters for a previously successful SER structure, YkuD, three clusters are suggested. The highest ranking cluster (SERp score 4.05) contains two adjacent residues, K117 and Q118, which should be mutated concurrently. Both residues are predicted to be in an entropy-rich region within a stretch of random coil (PSIPRED confidence 70%–85%), and their evolutionary conservation within the family is low (2%–13%). Together these three factors make this cluster a good candidate for SER. Indeed, these are mutations that were selected by hand to generate the structure (PDB ID 1Y7M). The second cluster contains the proposed mutation K65A but has a SERp score 40% lower than the top cluster (2.40). Single-residue substitutions are usually not the most effective in the SER approach, but in this case there is an adjacent glycine–alanine pair; thus, the K65A mutation would create a larger, low entropy patch that has a high probability of being on the surface. The third cluster has a score very close to the second cluster (2.39) and would require the mutation of two high entropy residues (K7, Q8) that are not well conserved but have a low confidence of being in a loop.
The SERp server also provides additional results that the user may find useful. Links to sequence alignments of homologs identified with PSI-BLAST and homolgous structured in the Protein Data Bank are provided. Highly conserved sequence regions identified with Blocks and potential protein–protein interactions identified with the ProLinks server are summarized. Complete results are available on the respective subtabs of the main “Results” tab.
Prediction evaluation
The reliability of the SERp server's predictions can be put to a stringent test only when the crystallizability of the mutants that it predicts is assessed by experiments. We are in the process of testing these predictions using a pool of targets from Structural Genomics Centers shown to be recalcitrant to crystallization in their wild-type form. However, we have gained useful information from the reassessment of amino acid sequences of proteins already crystallized by SER using manually designed variants and from the comparison of these mutants with those identified by the SERp server. Space limitations preclude full discussion of all results, but interested readers will find an extensive analysis on the SERp server's Web site.
Although the database of proteins crystallized by the SER approach is still modest (Table 1), the methodology has proven effective and is gaining popularity. A number of new structures were solved in this way by Derewenda and coworkers (2001), and other laboratories have reported a number of successful applications. In most cases, the change of two or three residues yielded well-diffracting crystals where none were available for the wild-type protein, or caused the protein to crystallize in a novel space group with diffraction superior to that of existing wild-type crystals. In two of these cases, the method was used to generate new, better-diffracting crystal forms for purpose of structure-based drug design.
We have submitted all of the reported successful SER structures to the SERp server and compared the default output with reported experimental data. In the vast majority of cases the manually chosen mutation sites of the crystallized variants were easily identified by the SERp server. In 11 out of a total 13 cases representing the canonical use of the SER approach, the SERp server identifies the crystal-yielding mutation as one of the top recommendations.
We note that within any protein ∼250–400 amino acids in length, there are typically relatively few sites identified with high SERp scores, suggesting suitability for surface engineering. This is encouraging, because it suggests that screening of a limited number of mutants may be sufficient to obtain high-quality crystals. On the other hand, it remains to be seen if these easily identifiable high conformational entropy sites routinely show critical impact on protein crystallizability and if the effective success rate of the SERp server's predictions can indeed be as high as Table 1 suggests.
Overall, the SERp server's predictions agree extremely well with the existing experimental data. In seven cases one of the suggested mutations was identical to the mutations that produced the structure. In the remaining cases, there are minor differences between the suggested and manually selected mutations that produced structures. For example, for Hsp33 the SERp server suggests a triple mutation, E100A, Q101A, K103A, in place of the double-mutant E100A, Q101A which yielded crystals (Janda et al. 2004). For comparison, we have included in Table 1 SERp scores generated using nondefault parameters that ensured the structure-producing mutations were suggested. For Hsp33 the SERp score for the double-mutant can be generated by limiting the “maximum mutations per cluster” to two. In this case, the SERp score for the triple and double-mutation are 4.47 and 3.64, respectively. We speculate that the triple mutant would work equally well, but it is important to note that, if the maximum number of allowed mutations per cluster is limited to two, the SERp server still assigns a top score to this site.
It should be noted that several of the proteins solved by the SER approach have mutated aspartate residues. The default values currently include only Lys, Glu, and Gln as mutable residues. At this time we do not suggest mutating aspartates as a first approach, simply because the propensity of aspartates to participate in protein–protein interactions, and therefore possible crystal contacts, is higher than that of the default mutable residues (Conte et al. 1999; Fernandez et al. 2003).
In L-rhamnulose kinase the successful site is predicted with the third highest score, but it should be noted that Geueninger et al. (2006) also mutated an adjacent Arg, so that the overall effect of the triple mutation may be higher than that predicted for the double-mutant. Similarly, gp24 was crystallized using a variant with a double-mutation that is identified with the third highest score. Finally, the double-mutant used in the study of the two-domain, 53.5-kDa protein MICAL, is predicted only as the fourth top score. It is quite possible that the top sites predicted by the SERp server would work at least as well.
In all of the above cases, the mutated epitope was later found to be in a crystal contact, in perfect agreement with the central premise of the SER approach. However, there are additional examples in the literature of noncanonical use of the method (Table 1), which warrant some discussion. Jin et al. (2005) obtained two novel crystal forms of Factor XI, using single-residue, Lys → Ala mutations. Although such mutations may occasionally work especially if the high entropy residue is flanked by a low entropy residue, we believe that mutating a minimum of two residues is far more effective. The structure of OspA is an extreme example of the SER approach, in which 13 lysines and glutamates were mutated to alanines or serines, yet without compromising the protein's solubility. Hsp70 is an interesting example of a two domain protein in which the successful double-mutation (E213A, D214A) is located at an interface between the two domains, rather than on the surface, and clearly effects the mutual disposition of the two modules, thus promoting crystallization in a way different from the canonical SER approach. Choline acetyltransferase was crystallized by combining three clusters, each of which is predicted by the SERp server with scores ranking 1, 2, and 4.
Future directions and developments
It is assumed that the users will have some insight into the structure of their target protein, at least with respect to its domain architecture, possible disorder in the absence of cofactors, etc. The surface engineering approach of the SER method has been demonstrated to enhance the protein's propensity to form crystals, but it may not overcome other impediments such as intrinsic flexibility of a multidomain system. The current version of the software does not check for disordered regions, and it does not provide three-dimensional structure prediction or domain analysis. These are all improvements that we envisage in the future. Another possible addition is to recommend replacement residues based on empirical residue counts at crystal contacts (Dasgupta et al. 1997). Future versions may automatically design primers if the DNA sequence is used as the input.
Access to the server
The Surface Entropy Reduction prediction server is available at http://www.doe-mbi.ucla.edu/Services/SER. A job submission requires only the amino acid or DNA sequence and a valid e-mail address. While the default processing parameters should suit most users, most parameters can be adjusted prior to the initial submission or at a later time. Prediction results can be delivered by e-mail, although the server was designed to present results interactively on the Web site offering many internal links to analysis details as well as cross references to external resources. Users who wish to process as many as 25 sequences per day can upload a FASTA file for the batch processing mode. The server version (v1.5) described in this paper has been made publicly available in January 2007, replacing an earlier version that has been available since January 2006.
Acknowledgments
We thank NIH PSI ISFI Center and HHMI for support. L.G. is supported by a Ralph and Shirley Shapiro Fellowship.
Footnotes
Reprint requests to: David Eisenberg, Howard Hughes Medical Institute, UCLA-DOE Institute of Genomics and Proteomics, 611 Charles E. Young Drive East, Boyer 105, Los Angeles, CA 90095-1570, USA; e-mail: david@mbi.ucla.edu; fax: (310) 206-3914.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072914007.
References
- Baud F. and Karlin, S. 1999. Measures of residue density in protein structures. Proc. Natl. Acad. Sci. 96: 12494–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeshans K.M., Liu, F., Peng, G., Idler, W., Jang, S.I., Marekov, L., Black, L., and Ahvazi, B. 2006. Purification, crystallization and preliminary X-ray diffraction analysis of the phage T4 vertex protein gp24 and its mutant forms. Protein Expr. Purif. 49: 2: 235–243. [DOI] [PubMed] [Google Scholar]
- Bielnicki J., Devedjiev, Y., Derewenda, U., Dauter, Z., Joachimiak, A., and Derewenda, Z.S. 2006. B. subtilis ykuD protein at 2.0 Å resolution: Insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins 62: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers P.M., Pellegrini, M., Fierro, J., and Eisenberg, D. 2004. Prolinks: A database of protein functional linkages derived from coevolution. Genome Biol. http://genomebiology.com/2004/5/5/R35. [DOI] [PMC free article] [PubMed]
- Conte L.L., Chothia, C., and Janin, J. 1999. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285: 2177–2198. [DOI] [PubMed] [Google Scholar]
- Dasgupta S., Iyer, G.H., Bryant, S.H., Lawrence, C.E., and Jeffrey, B.A. 1997. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins 28: 494–514. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S. 2004a. The use of recombinant methods and molecular engineering in protein crystallization. Methods 34: 354–363. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S. 2004b. Rational protein crystallization by mutational surface engineering. Structure 12: 529–535. [DOI] [PubMed] [Google Scholar]
- Devedjiev Y., Surendranath, Y., Derewenda, U., Gabrys, A., Cooper, D.R., Zhang, R., Lezondra, L., Joachimiak, A., and Derewenda, Z.S. 2004. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J. Mol. Biol. 343: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Scott, L.R., and Scheraga, H.A. 2003. Amino acid residues at protein-protein interfaces: Why is propensity so different from relative abundance? J. Phys. Chem. B 107: 9929–9932. [Google Scholar]
- Grueninger D. and Schulz, G.E. 2006. Structure and reaction mechanism of L-Rhamnulose kinase from Escherichia coli. J. Mol. Biol. 359: 787–797. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff, J.G., and Pietrokovski, S. 1999. Blocks: A non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics 15: 471–479. [DOI] [PubMed] [Google Scholar]
- Hu J. and Hubbard, S.R. 2006. Structural basis for phosphotyrosine recognition by the Src homology-2 domains of the adapter proteins SH2-B and APS. J. Mol. Biol. 361: 69–79. [DOI] [PubMed] [Google Scholar]
- Janda I., Devedjiev, Y., Cooper, D., Chruszcz, M., Derewenda, U., Gabrys, A., Minor, W., Joachimiak, A., and Derewenda, Z.S. 2004. Harvesting the high-hanging fruit: The structure of the YdeN gene product from Bacillus subtilis at 1.8 Å resolution. Acta Crystallogr. D Biol. Crystallogr. D60: 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Prasad, K., Lafer, E.M., and Sousa, R. 2005. Structural basis of inter-domain communication in the Hsc70 chaperone. Mol. Cell 20: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Pandey, P., Babine, R.E., Weaver, D.T., Abdel-Meguid, S.S., and Strickler, J.E. 2005. Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: An essential step for the iterative structure-based design of factor XI inhibitors. Acta Crystallogr. D61: 1418–1425. [DOI] [PubMed] [Google Scholar]
- Jones D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kim R., Obransky, T., Rylett, R., and Shilton, B. 2005. Surface-entropy reduction used in the crystallization of human choline acetyltransferase. Acta Crystallogr. D61: 1306–1310. [DOI] [PubMed] [Google Scholar]
- Longenecker K.L., Lewis, M.E., Chikumi, H., Gutkind, J.S., and Derewenda, Z.S. 2001. Structure of the RGS-like domain from PDZ-RhoGEF: Linking heterotrimeric g protein-coupled signaling to Rho GTPases. Structure 9: 559–569. [DOI] [PubMed] [Google Scholar]
- Makabe K., Tereshko, V., Gawlak, G., Yan, S., and Koide, S. 2006. Atomic-resolution crystal structure of Borrelia burgdorferi outer surface protein A via surface engineering. Protein Sci. 15: 1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi S., Hall, D.L., Kornienko, M., Darke, P.L., and Kuo, L.C. 2003. Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. D59: 1725–1730. [DOI] [PubMed] [Google Scholar]
- Nadella M., Bianchet, M.A., Gabelli, S.B., Barrila, J., and Amzel, L.M. 2005. Structure and activity of the axon guidance protein mical. Proc. Natl. Acad. Sci. 102: 16830–16835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett S.D. and Sternberg, M.J. 1993. Empirical scale of side-chain conformational entropy in protein folding. J. Mol. Biol. 231: 825–839. [DOI] [PubMed] [Google Scholar]
- Prag G., Misra, S., Jones, E.A., Ghirlando, R., Davies, B.A., Horazdovsky, B.F., and Hurley, J.H. 2003. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113: 609–620. [DOI] [PubMed] [Google Scholar]
- Stevens R.C. 2000. High-throughput protein crystallization. Curr. Opin. Struct. Biol. 10: 558–563. [DOI] [PubMed] [Google Scholar]
- Strong M., Sawaya, M.R., Wang, S., Phillips, M., Cascio, D., and Eisenberg, D. 2006. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. 103: 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]