Abstract
The burial of nonpolar surface area is known to enhance markedly the conformational stability of proteins. The contribution from the burial of polar surface area is less clear. Here, we report on the tolerance to substitution of Ser75 of bovine pancreatic ribonuclease (RNase A), a residue that has the unusual attributes of being buried, conserved, and polar. To identify variants that retain biological function, we used a genetic selection based on the intrinsic cytotoxicity of ribonucleolytic activity. Cell growth at 30°C, 37°C, and 44°C correlated with residue size, indicating that the primary attribute of Ser75 is its small size. The side-chain hydroxyl group of Ser75 forms a hydrogen bond with a main-chain nitrogen. The conformational stability of the S75A variant, which lacks this hydrogen bond, was diminished by ΔΔG = 2.5 kcal/mol. Threonine, which can reinstate this hydrogen bond, provided a catalytically active RNase A variant at higher temperatures than did some smaller residues (including aspartate), indicating that a secondary attribute of Ser75 is the ability of its uncharged side chain to accept a hydrogen bond. These results provide insight on the imperatives for the conservation of a buried polar residue.
Keywords: conformational stability, genetic selection, hydrogen bond, hydrophobic core, molecular evolution, ribonucleases
The conformational stability of a protein derives from the energetic difference between its unfolded and folded states. For the majority of proteins, this energetic difference (ΔG°conf) falls within the narrow range from –5 to –15 kcal/mol (Kumar et al. 2006; Rose et al. 2006). The small energetic distinction between the unfolded and folded states is maintained through the burial of nonpolar surface area, the formation of hydrogen bonds, and other forces (Dill 1990; Myers and Pace 1996; Fleming and Rose 2005; Rose et al. 2006).
Conformational stability is only one attribute that leads to the existence of a residue at a particular position in an amino acid sequence. Proteins often concede stability for function (Shoichet et al. 1995; Beadle and Shoichet 2002; DePristo et al. 2005; Pál et al. 2006). For example, altering residues in the active site of AmpC β-lactamase increases the stability of that enzyme, but greatly decreases its catalytic activity (Beadle and Shoichet 2002).
The selective pressures that determine the amino acid composition at specific positions have been elucidated for nonpolar residues buried in the interior of a protein (Lim and Sauer 1989, 1991; Guo et al. 2004; Smith and Raines 2006). These residues are relatively invariant to substitution, as they are required to maintain the structural integrity of the protein and they have lower substitution rates than expected by neutral drift (Choi et al. 2006). Still, it is unclear whether these arguments also apply to buried polar residues.
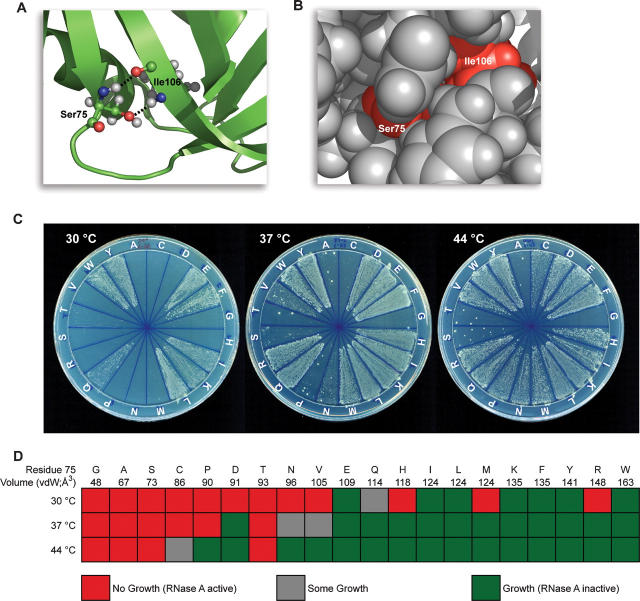
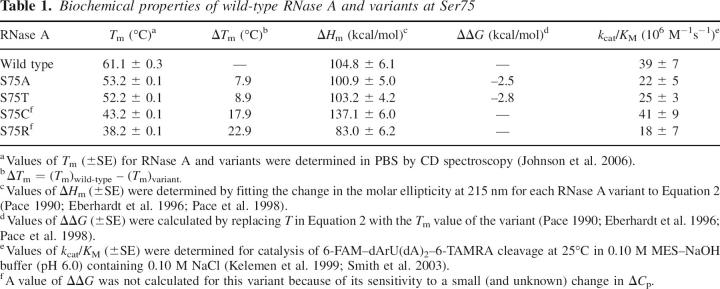
Recently, the residues critical for the function of bovine pancreatic ribonuclease (RNase A) were revealed by using a novel genetic selection system (Smith and Raines 2006). Residues found to be essential included the canonical active-site residues (His12, Lys41, and His119), half-cystines, and residues buried in the hydrophobic core (Smith and Raines 2006). Although serine is the most abundant residue in RNase A, only one of its 15 serine residues was found to be critical (Smith and Raines 2006). That unique residue is Ser75, which is the most buried of the serine residues (ASA = 1.9 Å2) (Fig. 1A,B) and is highly conserved among homologous ribonucleases (Beintema 1987; Beintema et al. 1988). In the hydrophobic core of RNase A, Ser75 forms two hydrogen bonds with Ile106, another essential residue that expedites protein folding (Coll et al. 1999). Predictably, replacing Ile106 with an alanine residue lowers the value of T m by 14°C (Coll et al. 1999). No variant at position 75 has been studied previously.
Figure 1.
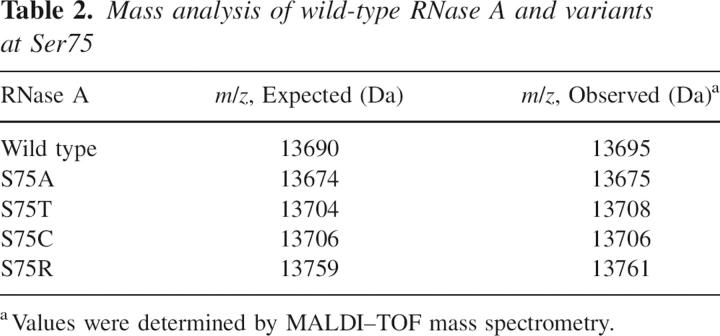
Genetic selection to reveal the influence of substitutions at position 75 to the activity and stability of RNase A. (A) Ribbon diagram of RNase A (green) in which the atoms of Ser75 and Ile106 are depicted explicitly and putative hydrogen bonds are shown with black dashed lines. (B) Space-filling model of Ser75 and Ile106 in the same orientation as in A. Ser75 and Ile106 are in red, and all other RNase A residues are in gray. Images in A and B were created with PDB entry 7RSA (Wlodawer et al. 1988) and the program PyMOL (DeLano Scientific). (C) Growth of Origami E. coli cells that produce an RNase A variant containing the indicated amino acid residue at position 75. Plates were incubated at the indicated temperature for 48 h. Cells producing active and thus cytotoxic RNase A variants will be inviable; those producing inactive variants will be viable. (D) Graphical representation of the results from C. Amino acids are listed in order of increasing van der Waals volume (Richards 1974).
Here, we report on the biochemical attributes that lead to the conservation of a buried polar residue. To do so, we create RNase A variants with each of the 20 proteinogenic amino acids at position 75. We then subject these 20 variants to the extant genetic selection (Smith and Raines 2006), but at different temperatures so as to glean additional information. Finally, we characterize the five most interesting variants in detail. The results provide a comprehensive picture of the effect of a buried polar residue on the conformational stability and catalytic activity of a protein, and serve to reveal the physicochemical attributes that lead to its evolutionary conservation.
Results
Genetic selection at Ser75
RNase A produced in traditional Escherichia coli strains aggregates to form enzymatically inactive inclusion bodies, as its requisite four disulfide bonds cannot form in the reducing environment of the cytosol (delCardayré et al. 1995). Engineered E. coli strains, such as the Origami strain (Novagen), provide a more oxidizing cytosol, wherein RNase A can fold (Smith and Raines 2006). Folded RNase A catalyzes the degradation of bacterial RNA, resulting in cell death. Even leaky expression of wild-type RNase A from the uninduced P tac promoter is adequate to cause cell death (Smith and Raines 2006).
We used the ability of properly folded, active RNase A to elicit cell death to explore the role of Ser75. Twenty plasmids encoding variants in which position 75 was substituted with all 20 natural amino acids were created by site-directed mutagenesis, and these plasmids were transformed individually into Origami cells. The transformed Origami cells were grown at four different temperatures (23°C, 30°C, 37°C, and 44°C) and selected for growth. At 23°C, no variant reduced the ribonucleolytic activity enough to allow for the growth of E. coli cells (data not shown). Results at the other three temperatures are shown in Figure 1C. At 44°C, which is 17°C lower than the T m value of wild-type RNase A (Table 1), only serine (wild type) or its substitution with glycine, alanine, or threonine significantly inhibited cell growth. At 30°C, 12 variants were identified (glycine, alanine, serine, cysteine, proline, aspartate, threonine, aspargine, valine, histidine, methionine, and arginine) that inhibited cell growth.
Table 1.
Biochemical properties of wild-type RNase A and variants at Ser75
The amino acid substitutions that diminished ribonucleolytic activity sufficiently to allow for the growth of E. coli cells are depicted in Figure 1D, in which the amino acids are listed in the order of their van der Waals volume (Creighton 1993). The inhibition of growth correlates with the van der Waals volume of the amino acid substitution. At 44°C, the amino acid substitutions that retain enzymatic activity are the smallest amino acids with the exception of threonine, which has a van der Waals volume that is larger than cysteine, proline, or aspartate. The ability of the threonine substitution at position 75 to maintain the catalytic activity and stability of RNase A at 44°C, even with its larger van der Waals volume, suggests that it still forms a hydrogen bond with Ile106.
Data from 37°C and 30°C follow a similar pattern, where growth of the Origami cells is related to the van der Waals volume of the substituted amino acid (Fig. 1D). At 30°C, there are also outliers to this trend. For example, the van der Waals volume of arginine is 24 Å2 greater than that of methionine, which is the amino acid with the next closest volume that inhibited growth (Fig. 1D). To better understand the relationship between the van der Waals volume and hydrogen-bonding capability of amino acid substitutions at position 75 and the resulting ribonucleolytic activity and conformational stability of the RNase A variants, five variants (serine, alanine, threonine, cysteine, and arginine) were isolated (Table 2) and subjected to detailed analysis. These variants were chosen based on their small size (alanine), ability to form a hydrogen bond with Ile106 (serine and threonine), and unexpected retention of ribonucleolytic activity in the genetic selection (cysteine and arginine).
Table 2.
Mass analysis of wild-type RNase A and variants at Ser75
Physiochemical properties of RNase A variants
To insure that all five RNase A variants were folded properly at 25°C, far-UV circular dichroism (CD) spectroscopy was used to discern any changes in secondary structure (Kelly et al. 2005). Each variant was found to have a similar CD spectrum (Fig. 2A), suggesting that substitution at position 75 does not have a large effect on the secondary structure of RNase A. This result is consistent with studies in other proteins, which show that single substitutions tend to have small effects on global protein structure (Matthews 1987; Stites et al. 1991; Chen et al. 1993). RNase A variants with lower conformational stability (Table 1) (arginine and cysteine) did, however, have lower mean residue-weight ellipticity, consistent with minor changes to local secondary structure.
Figure 2.
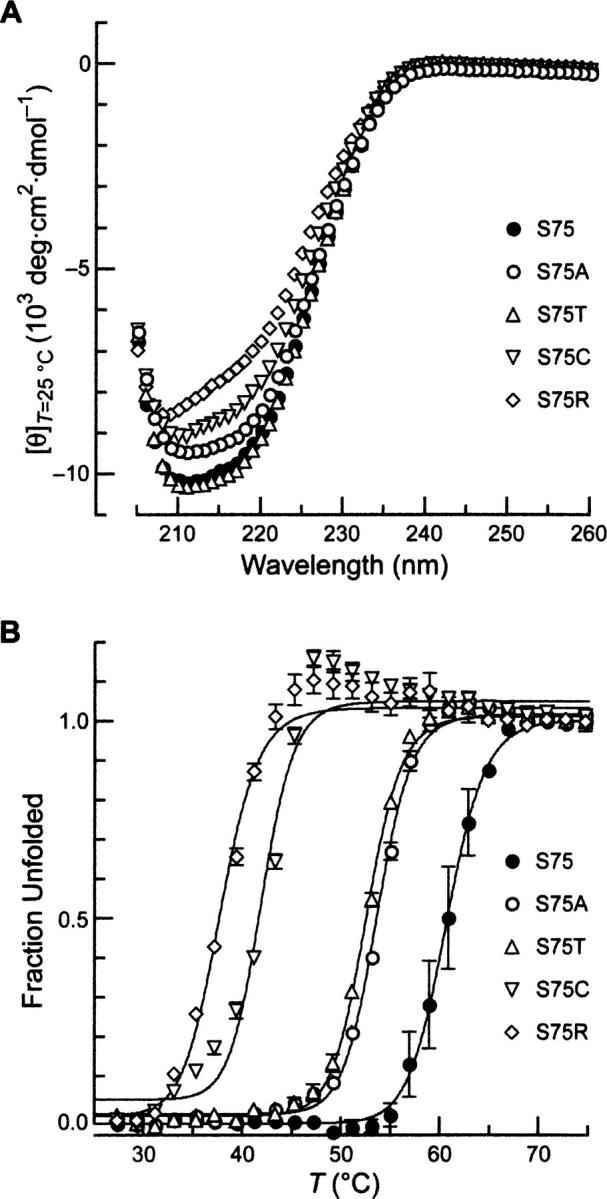
Conformational stability of five variants of RNase A. (A) Far-UV CD spectra of the indicated position 75 variants of RNase A (0.5 mg/mL in PBS). (B) Thermal denaturation of the indicated position 75 variants of RNase A (0.5 mg/mL in PBS). Molar ellipticity at 215 nm was monitored after a 2-min equilibration at each temperature. Data points are mean values (±SE) from ≥2 experiments, and were fitted to a two-state model to determine values of T m (Table 1).
To characterize these RNase A variants further, their conformational stabilities were determined by monitoring their CD spectrum at 215 nm as a function of temperature (Fig. 2B; Johnson et al. 2006). The temperature at the midpoint of the transition curve (T m) and associated change in enthalpy (ΔH m) are listed in Table 1. A large range of conformational stabilities was generated upon substitution at position 75. Alanine and threonine residues decrease the value of T m by 8°C, whereas arginine decreases the T m by 23°C. A similar trend is observed for the values of ΔH m, except for the S75C variant, which has a ΔH m value that is 32 kcal/mol greater than that of wild-type RNase A. The S75C variant exhibits a steady increase in ellipticity following denaturation (Fig. 2B), suggesting that a two-state model for its unfolding is invalid. This anomalous behavior could be caused by the free cysteine residue in S75C RNase A eliciting dimerization via disulfide bond formation, leading to aggregation. Otherwise, the T m values for the purified RNase A variants closely follow the growth inhibition results (Fig. 1C,D). The T m values of the S75T and S75A variants are >44°C, that of the S75C variant is >37°C, and that of the S75R is >30°C (Table 1), demonstrating the ability of the genetic selection to report accurately on conformational stability.
This large variation in other physicochemical attributes of the variants is not reflected in their catalytic activity (Table 1). At 25°C, all variants have ribonucleolytic activity within twofold of the wild-type enzyme, suggesting, as do the data in Figure 2A, that each variant assumes the structure of wild-type RNase A at 25°C. This negligible effect of substitution at position 75 on ribonucleolytic activity is most likely true for any amino acid substitution, as the growth of all transformed E. coli cells was inhibited at 23°C (data not shown). Thus, position 75 in RNase A strongly influences the conformational stability of the enzyme, but does not affect its ribonucleolytic activity.
Discussion
Role of a buried polar residue in conformational stability
The neutral theory of molecular evolution states that the vast majority of mutations do not affect the fitness of an organism (Kimura 1968, 1983). Still, amino acid substitutions often do have detrimental effects on the structure and function of a particular protein (DePristo et al. 2005; Pál et al. 2006). Understanding the tolerance of specific conserved residues to substitution and the evolutionary pressures that select for conservation could reveal the underlying evolutionary mechanisms and facilitate identification of disease-inducing residues (Benner 2001; Guo et al. 2004; DePristo et al. 2005; Smith and Raines 2006). We have combined data from a genetic selection with phylogenetic data to investigate these selective pressures as applied to a buried polar residue.
As our model system we chose RNase A, which is one of the best-characterized proteins (D'Alessio and Riordan 1997; Raines 1998). In seminal work, Anfinsen used RNase A to show that “the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment” (Anfinsen 1973). Subsequently, RNase A has been used to evaluate the factors involved in protein folding and stability, especially disulfide bond formation and prolyl peptide bond isomerization (Wedemeyer et al. 2000, 2002). The RNase A superfamily has also served as an archetype for studying many aspects of molecular evolution, including mammalian speciation (Beintema 1987; Beintema et al. 1988, 1997; Yu and Zhang 2006), adaptive parallel evolution (Schienman et al. 2006; Zhang 2006), the shuffling of disulfide bonds (Zhang 2007), and gene duplication (Sassi et al. 2007). Sequences of RNase A homologs have been reported from >200 organisms (Beintema 1987; Beintema et al. 1988, 1997; Nitto et al. 2006; Pizzo et al. 2006; Yu and Zhang 2006), making RNase A an exceptional protein with which to study the selective pressures that lead to amino acid conservation.
We have focused on the basis for the conservation of a particular buried polar residue in RNase A, Ser75. Within the 266 known members of the RNase A superfamily (Smith and Raines 2006), Ser75 is conserved in 238 (89.5%). This level of conservation is indicative of biological importance. Recently, we showed that Ser75 is indeed critical to ribonuclease function (Smith and Raines 2006). Still, whether Ser75 played a role in enzymatic catalysis or conformational stability (or both) had been unknown.
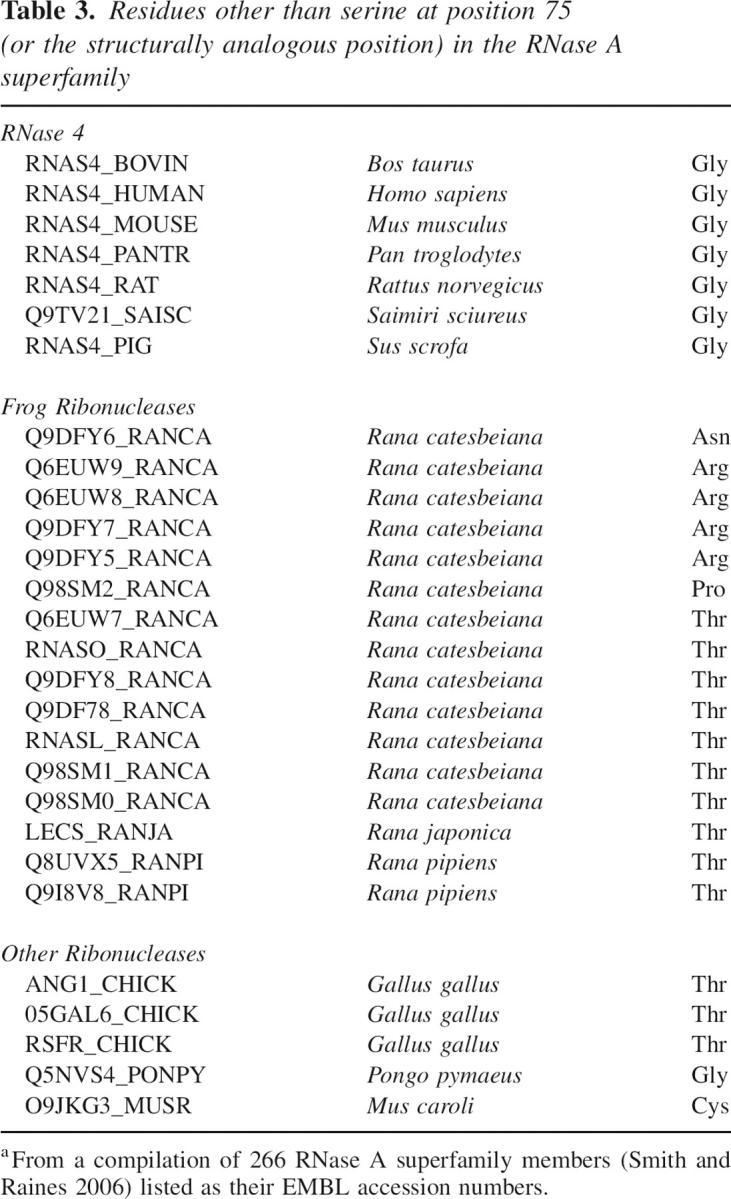
The 28 known ribonucleases that diverge from a serine residue at position 75 are listed in Table 3. Interestingly, each of these divergent ribonucleases has a residue that enables function at 30°C in our genetic selection (Fig. 1C,D). Moreover, most of the amino acids that replace serine at position 75 in these divergent ribonucleases fall into the subset of smaller amino acids that lead to cytotoxicity at 44°C (glycine, threonine, and cysteine). This direct correlation between the ribonuclease sequence data (Table 3) and our genetic selection data (Fig. 1C,D) indicates that a small amino acid at position 75 is necessary for ribonucleases to function properly (Fig. 1B).
Table 3.
Residues other than serine at position 75 (or the structurally analogous position) in the RNase A superfamily
Another salient characteristic of this buried polar residue is its ability to interact with proximal residues in the folded protein (Fig. 1A,B). The hydrogen bond formed from the hydroxymethyl side chain of Ser75 adds an additional ΔΔG = 2.5 kcal/mol of energetic stability to RNase A over other small amino acids, but this hydrogen bond does not seem to be the only selective pressure. Substitution with threonine maintains ribonucleolytic activity at 44°C (Fig. 1C,D), presumably because it can form a hydrogen bond from its hydroxyethyl side chain to Ile106. S75T RNase A is, however, not as stable as the S75A variant (Table 1). Adding a methylene group to the side chain of Ser75 (as in the S75T variant) disrupts the energetic stability of wild-type RNase A more than does removing the oxygen atom from its side chain (as in the S75A variant) (Table 1). Thus, the amino acid at position 75 in ribonucleases, especially RNase A, appears to have been selected primarily because of its van der Waals volume. Among the amino acids with a small van der Waals volume, serine had been selected because of the additional energetic stability imparted by its ability to form a hydrogen bond with Ile106. Similarly, a conserved glutamate residue in pyrrolidone carboxyl peptidases from hyperthermophiles is completely buried and stabilizes that protein by forming hydrogen bonds with main-chain atoms (Kaushik et al. 2006).
Hydrogen bonds and conformational stability
The influence of hydrogen bonds on the conformational stability of proteins has generated much interest (Dill 1990; Myers and Pace 1996; Baldwin 2003). The recent observation by Rose and coworkers that virtually all of the nitrogens and oxygens in proteins participate in hydrogen bonds provides compelling evidence that these hydrogen bonds confer much stability (Fleming and Rose 2005; Rose et al. 2006). Pace and coworkers have demonstrated that intramolecular hydrogen bonds increase the packing density and van der Waals interactions of a protein (Schell et al. 2006). For example, the average volume occupied by a serine side chain decreases by 1 Å3 when the side chain is engaged in a hydrogen bond.
Ser75 of RNase A appears to function in this dual manner. The main-chain nitrogen of Ile106 needs a hydrogen-bonding partner, and other residues are not in a position to fulfill that role. On average, replacing a serine residue with alanine results in ΔΔG = –1.1 ± 1.5 kcal/mol for serine residues that are 77% ± 25% buried from solvent (Myers and Pace 1996). Ser75 of RNase A falls on the extreme end of this continuum, having ΔΔG = –2.5 kcal/mol (Table 1) and being 98.1% buried. These data underscore the importance of the Ser75–Ile106 hydrogen bond to conformational stability. It is noteworthy that the stability conferred by Ser75 is comparable to that measured for the buried hydrogen bond between the side chain of Tyr97 and main-chain oxygen of Lys41 of RNase A (Eberhardt et al. 1996), as well as buried hydrogen bonds in the microbial ribonucleases, RNase T1 and barnase (Shirley et al. 1992; Chen et al. 1993). A buried salt bridge in triosephosphate isomerase is likewise critical to the function of that enzyme (Silverman et al. 2001).
The importance of the Ser75–Ile106 hydrogen bond is apparent from its environment. The relative importance of a particular hydrogen bond to protein stability is related to its mobility (Pace et al. 2001). Ser75 is packed tightly in the interior of RNase A (Fig. 1B), being surrounded by seven residues that lie within 3.9 Å. Thus, the Ser75–Ile106 hydrogen bond could contribute to stability not only by its satisfying the hydrogen-bonding requirement of Ile106, but also by eliciting an increase in van der Waals contacts within the hydrophobic core of RNase A.
Conclusions
By combining genetic selection with phylogenetic data, we sought to reveal the characteristics that select for a buried polar residue and understand the tolerance to mutation of this conserved residue. A trend was observed in both genetic and evolutionary selection toward amino acids with small van der Waals volume, implying a requirement for a small amino acid at position 75 to maintain conformational stability. A serine substitution adds an additional 2.5 kcal/mol of energetic stability beyond an alanine residue due to its ability to form a hydrogen bond with Ile106, which could also increase the packing density of the protein interior. Ser75 functions largely to maintain the stability of RNase A, but has no direct impact on its catalytic activity at 25°C. Our genetic selection could be useful to probe the contribution of other ribonuclease residues to their biological function and the selective pressures responsible for their conservation.
Materials and Methods
General
E. coli BL21 (DE3) cells, E. coli Origami B (DE3) cells, and plasmid pET27b(+) were from Novagen. The fluorogenic ribonuclease substrate, 6-FAM–dArU(dA)2–6-TAMRA, and DNA oligonucleotides for mutagenesis were from Integrated DNA Technologies. Protein purification columns and the pGEX vector were from GE Healthcare. MES buffer (Sigma–Aldrich) was purified by anion-exchange chromatography to remove trace amounts of oligomeric vinylsulfonic acid (Smith et al. 2003). All other chemicals were of commercial grade or better, and were used without further purification.
Luria–Bertani (LB) agar was contained in 1.00 L tryptone (10 g), yeast extract (5 g), NaCl (10 g), and agar (15 g). PBS (pH 7.4), was contained in 1.00 L NaCl (8.0 g), KCl (2.0 g), Na2HPO4·7H2O (1.15 g), KH2PO4 (2.0 g), and NaN3 (0.10 g).
Fluorescence spectroscopy was performed with a QuantaMaster1 photon-counting fluorimeter equipped with sample stirring (Photon Technology International). CD data were collected with a model 62A DS CD spectrometer (Aviv) equipped with a temperature controller. Matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectrometry was performed with a Voyager-DE-PRO Biospectrometry Workstation (Applied Biosystems). CD and MALDI–TOF mass spectrometry experiments were performed at the campus Biophysics Instrumentation Facility.
Genetic selection
Variants of RNase A with all 20 amino acids at position 75 were created by using plasmid pGEX-RNase A, which was derived from pGEX-4T3 (Smith and Raines 2006), and the Quikchange site-directed mutagenesis kit (Stratagene). Codons for position 75 substitution were chosen for optimized protein expression in E. coli, based on known codon usage (Aota et al. 1988). The presence of Met(–1) has been shown to have no discernable effect on the ribonucleolytic activity or conformational stability of RNase A (Arnold et al. 2002). The identity of codons at position 75 and elsewhere was confirmed by DNA sequence analysis. Purified plasmid DNA from all 20 RNase A variants was transformed into Origami B (DE3) cells. Transformed Origami B (DE3) cells were spread onto LB agar containing ampicillin (100 μg/mL), kanamycin (20 μg/mL), and tetracycline (12.5 μg/mL), and allowed to grow at 23°C, 30°C, 37°C, or 44°C for 48 h. Results were confirmed by repeating the selection (i.e., transformation, spreading, and growth).
Protein purification
RNase A and variants of RNase A were purified from inclusion bodies by using the same high-yielding procedure described previously (delCardayré et al. 1995; Rutkoski et al. 2005). A pET27b(+) plasmid containing DNA that encodes wild-type RNase A was used to produce Ser75 variants of RNase A (Leland et al. 1998; Rutkoski et al. 2005) using the Quikchange site-directed mutagenesis kit (Stratagene). The molecular mass of each purified RNase A variant was determined with MALDI–TOF mass spectrometry.
Enzymatic activity
The ribonucleolytic activity of wild-type RNase A and its Ser75 variants was determined by using 6-FAM–dArU(dA)2–6-TAMRA as the substrate (Kelemen et al. 1999). Assays were carried out at 23(±2)°C in 2.0 mL of 0.10 M MES–NaOH buffer (pH 6.0) containing NaCl (0.10 M). Fluorescence data were fitted to the equation:
In Equation 1, ΔI/Δt represents the initial reaction velocity, I 0 is the fluorescence intensity before the addition of a ribonuclease, I f corresponds to the final fluorescence intensity after complete substrate hydrolysis, and [E] is the concentration of added ribonuclease.
Secondary structure and conformational stability
CD spectroscopy was used to assess the secondary structure of the RNase A variants (Johnson et al. 2006). A solution of PBS containing an RNase A variant (0.5 mg/mL) was incubated for 5 min at 25°C, and CD spectra were then acquired from 260 to 205 nm in 1-nm increments. CD spectra were collected at least twice for each RNase A variant, and data points are shown as the mean residue-weight ellipticity at each wavelength.
CD spectroscopy was also used to evaluate the conformational stability of the variants of RNase A (Johnson et al. 2006). PBS containing each of the RNase A variants (0.5 mg/mL) was heated from 25°C to 75°C in 2°C increments. The change in molar ellipticity at 215 nm was measured after a 2-min equilibration at each temperature, and the entire experiment was repeated at least once. Raw CD data at 215 nm were used to determine the values of T m and ΔH m using Igor Pro 5.04B (Wavemetrics, Inc.) and the Gibbs–Helmholtz equation (Pace 1990; Eberhardt et al. 1996; Pace et al. 1998):
Values of ΔC p were held constant (ΔC p = 0), as the ΔC p values calculated by fitting the raw CD data to Equation 2 were small compared with ΔH m. Also, curve-fitting of the raw CD data with and without ΔC p had no affect on the T m values and only slightly changed the ΔH m value for wild-type RNase A (<5%). The values of ΔΔG for the S75A and S75T variants were calculated at the T m of wild-type RNase A by replacing T in Equation 2 with the T m values for S75A and S75T, respectively. Analogous values for S75C and S75R were not calculated because their large ΔT m values impose a high error in the calculation.
Acknowledgments
We are grateful to D.R. McCaslin for assistance with CD spectroscopy and thermodynamic calculations. We thank B.D. Smith and M.T. Borra for contributive discussions. This work was supported by grant CA73808 (NIH). R.J.J. was supported by Biotechnology Training grant 08349 (NIH). S.R.L. was supported by a Wisconsin/Hilldale Undergraduate/Faculty Research Award. The Biophysics Instrumentation Facility was established with grants BIR-9512577 (NSF) and RR13790 (NIH).
Footnotes
Reprint requests to: Ronald T. Raines, Department of Biochemistry, University of Wisconsin-Madison, 433 Babcock Drive, Madison, WI 53706-1544, USA; e-mail: raines@biochem.wisc.edu; fax: (608) 262-3453.
Abbreviations: ASA, accessible surface area; CD, circular dichroism; 6-FAM, 6-carboxyfluorescein; MALDI–TOF, matrix-assisted laser desorption/ionization–time-of-flight; MES, 2-(N-morpholino)ethanesulfonic acid; PBS, phosphate-buffered saline; PDB, protein data bank; RNase A, bovine pancreatic ribonuclease; 6-TAMRA, 6-carboxytetramethylrhodamine; vdW, van der Waals.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072938907.
References
- Anfinsen C.B. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. [DOI] [PubMed] [Google Scholar]
- Aota S., Gojobori, T., Ishibashi, F., Maruyama, T., and Ikemura, T. 1988. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. Suppl. 16: r315–r402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold U., Hinderaker, M.P., and Raines, R.T. 2002. Semisynthesis of ribonuclease A using intein-mediated protein ligation. ScientificWorldJournal 2: 1823–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R.L. 2003. In search of the energetic role of peptide hydrogen bonds. J. Biol. Chem. 278: 17581–17588. [DOI] [PubMed] [Google Scholar]
- Beadle B.M. and Shoichet, B.K. 2002. Structural bases of stability–function tradeoffs in enzymes. J. Mol. Biol. 321: 285–296. [DOI] [PubMed] [Google Scholar]
- Beintema J.J. 1987. Structure, properties and molecular evolution of pancreatic-type ribonucleases. Life Chem. Rep. 4: 333–389. [Google Scholar]
- Beintema J.J., Schüller, C., Irie, M., and Carsana, A. 1988. Molecular evolution of the ribonuclease superfamily. Prog. Biophys. Mol. Biol. 51: 165–192. [DOI] [PubMed] [Google Scholar]
- Beintema J.J., Breukelman, H.J., Carsana, A., and Furia, A. 1997. Evolution of vertebrate ribonucleases: Ribonuclease A superfamily. In Ribonucleases: Structures and functions (eds. G. D'Alessio and J.F. Riordan), pp. 245–269. Academic Press, New York.
- Benner S.A. 2001. Natural progression. Nature 409: 459. doi: 10.1038/35054149. [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Fersht, A.R., and Henrick, K. 1993. Contribution of buried hydrogen bonds to protein stability. The crystal structures of two barnase mutants. J. Mol. Biol. 234: 1158–1170. [DOI] [PubMed] [Google Scholar]
- Choi S.S., Vallender, E.J., and Lahn, B.T. 2006. Systematically assessing the influence of 3-dimensional structural context on the molecular evolution of mammalian proteomes. Mol. Biol. Evol. 23: 2131–2133. [DOI] [PubMed] [Google Scholar]
- Coll M.G., Protasevich, I.I., Torrent, J., Ribó, M., Lobachov, V.M., Makarov, A.A., and Vilanova, M. 1999. Valine 108, a chain-folding initiation site-belonging residue, crucial for the ribonuclease A stability. Biochem. Biophys. Res. Commun. 265: 356–360. [DOI] [PubMed] [Google Scholar]
- Creighton T.E. 1993. Proteins: Structures and molecular propertie, 2nd ed. W.H. Freeman, New York.
- D'Alessio G. and Riordan, J.F. 1997. Ribonucleases: Structures and functions. Academic Press, New York.
- delCardayré S.B., Ribó, M., Yokel, E.M., Quirk, D.J., Rutter, W.J., and Raines, R.T. 1995. Engineering ribonuclease A: Production, purification, and characterization of wild-type enzyme and mutants at Gln11. Protein Eng. 8: 261–273. [DOI] [PubMed] [Google Scholar]
- DePristo M.A., Weinreich, D.M., and Hartl, D.L. 2005. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat. Rev. Genet. 6: 678–687. [DOI] [PubMed] [Google Scholar]
- Dill K.A. 1990. Dominant forces in protein folding. Biochemistry 29: 7133–7155. [DOI] [PubMed] [Google Scholar]
- Eberhardt E.S., Wittmayer, P.K., Templer, B.M., and Raines, R.T. 1996. Contribution of a tyrosine side chain to ribonuclease A catalysis and stability. Protein Sci. 5: 1697–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming P.J. and Rose, G.D. 2005. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 14: 1911–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.H., Choe, J., and Loeb, L.A. 2004. Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. 101: 9205–9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.J., Lin, S.R., and Raines, R.T. 2006. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 273: 5457–5465. [DOI] [PubMed] [Google Scholar]
- Kaushik J.K., Iimura, S., Ogasahara, K., Yamagata, Y., Segawa, S., and Yutani, K. 2006. Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxylpeptidases from hyperthermophiles. Biochemistry 45: 7100–7112. [DOI] [PubMed] [Google Scholar]
- Kelemen B.R., Klink, T.A., Behlke, M.A., Eubanks, S.R., Leland, P.A., and Raines, R.T. 1999. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 27: 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S.M., Jess, T.J., and Price, N.C. 2005. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751: 119–139. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- Kumar M.D., Bava, K.A., Gromiha, M.M., Prabakaran, P., Kitajima, K., Uedaira, H., and Sarai, A. 2006. ProTherm and ProNIT: Thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 34: D204–D206, doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland P.A., Schultz, L.W., Kim, B.-M., and Raines, R.T. 1998. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. 98: 10407–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W.A. and Sauer, R.T. 1989. Alternative packing arrangements in the hydrophobic core of λ repressor. Nature 339: 31–36. [DOI] [PubMed] [Google Scholar]
- Lim W.A. and Sauer, R.T. 1991. The role of internal packing interactions in determining the structure and stability of a protein. J. Mol. Biol. 219: 359–376. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. 1987. Genetic and structural analysis of the protein stability problem. Biochemistry 26: 6885–6888. [DOI] [PubMed] [Google Scholar]
- Myers J.K. and Pace, C.N. 1996. Hydrogen bonding stabilizes globular proteins. Biophys. J. 71: 2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitto T., Dyer, K.D., Czapiga, M., and Rosenberg, H.F. 2006. Evolution and function of leukocyte RNase A ribonucleases of the avian species, Gallus gallus . J. Biol. Chem. 281: 25622–25634. [DOI] [PubMed] [Google Scholar]
- Pace C.N. 1990. Measuring and increasing protein stability. Trends Biotechnol. 8: 93–98. [DOI] [PubMed] [Google Scholar]
- Pace C.N., Hebert, E.J., Shaw, K.L., Schell, D., Both, V., Krajcikova, D., Sevcik, J., Wilson, K.S., Dauter, Z., Hartley, R.W., et al. 1998. Conformational stabilty and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J. Mol. Biol. 279: 271–286. [DOI] [PubMed] [Google Scholar]
- Pace C.N., Horn, G., Hebert, E.J., Bechert, J., Shaw, K., Urbanikova, L., Scholtz, J.M., and Sevcik, J. 2001. Tyrosine hydrogen bonds make a large contribution to protein stability. J. Mol. Biol. 312: 393–404. [DOI] [PubMed] [Google Scholar]
- Pál C., Papp, B., and Lercher, M.J. 2006. An integrated view of protein evolution. Nat. Rev. Genet. 7: 337–348. [DOI] [PubMed] [Google Scholar]
- Pizzo E., Buonanno, P., Di Maro, A., Ponticelli, S., De Falco, S., Quarto, N., Cubellis, M.V., and D'Alessio, G. 2006. Ribonucleases and angiogenins from fish. J. Biol. Chem. 281: 27454–27460. [DOI] [PubMed] [Google Scholar]
- Raines R.T. 1998. Ribonuclease A. Chem. Rev. 98: 1045–1065. [DOI] [PubMed] [Google Scholar]
- Richards F.M. 1974. The interpretation of protein structures: Total volume, group volume distributions and packing density. J. Mol. Biol. 82: 1–14. [DOI] [PubMed] [Google Scholar]
- Rose G.D., Fleming, P.J., Banavar, J.R., and Maritan, A. 2006. A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. 103: 16623–16633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkoski T.J., Kurten, E.L., Mitchell, J.C., and Raines, R.T. 2005. Disruption of shape-complentarity markers to create cytotoxic variants of ribonuclease A. J. Mol. Biol. 354: 41–54. [DOI] [PubMed] [Google Scholar]
- Sassi S.O., Braun, E.L., and Benner, S.A. 2007. The evolution of seminal ribonuclease: Pseudogene reactivation or multiple gene inactivation events? Mol. Biol. Evol. 24: 1012–1024. [DOI] [PubMed] [Google Scholar]
- Schell D., Tsai, J., Scholtz, J.M., and Pace, C.N. 2006. Hydrogen bonding increases packing density in the protein interior. Proteins 63: 278–282. [DOI] [PubMed] [Google Scholar]
- Schienman J.E., Holt, R.A., Auerbach, M.R., and Stewart, C.B. 2006. Duplication and divergence of 2 distinct pancreatic ribonuclease genes in leaf-eating African and Asian Colobine monkeys. Mol. Biol. Evol. 23: 1465–1479. [DOI] [PubMed] [Google Scholar]
- Shirley B.A., Stanssens, P., Hahn, U., and Pace, C.N. 1992. Contribution of hydrogen bonding to the conformational stability of ribonuclease T1. Biochemistry 31: 725–732. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K., Baase, W.A., Kuroki, R., and Matthews, B.W. 1995. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. 92: 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J.A., Balakrishnan, R., and Harbury, P.B. 2001. Reverse engineering the (β/α)8 barrel fold. Proc. Natl. Acad. Sci. 98: 2958–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.D. and Raines, R.T. 2006. Genetic selection for critical residues in ribonucleases. J. Mol. Biol. 362: 459–478. [DOI] [PubMed] [Google Scholar]
- Smith B.D., Soellner, M.B., and Raines, R.T. 2003. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid). J. Biol. Chem. 278: 20934–20938. [DOI] [PubMed] [Google Scholar]
- Stites W.E., Gittis, A.G., Lattman, E.E., and Shortle, D. 1991. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J. Mol. Biol. 221: 7–14. [DOI] [PubMed] [Google Scholar]
- Wedemeyer W.J., Welker, E., Narayan, M., and Scheraga, H.A. 2000. Disulfide bonds and protein folding. Biochemistry 39: 4207–4216. [DOI] [PubMed] [Google Scholar]
- Wedemeyer W.J., Welker, E., and Scheraga, H.A. 2002. Proline cis–trans isomerization and protein folding. Biochemistry 41: 14637–14644. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Svensson, L.A., Sjölin, L., and Gilliland, G.L. 1988. Structure of phosphate-free ribonuclease A refined at 1.26 Å. Biochemistry 27: 2705–2717. [DOI] [PubMed] [Google Scholar]
- Yu L. and Zhang, Y.P. 2006. The unusual adaptive expansion of pancreatic ribonuclease gene in carnivora. Mol. Biol. Evol. 23: 2326–2335. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2006. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 38: 819–823. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2007. Disulfide-bond reshuffling in the evolution of an ape placental ribonuclease. Mol. Biol. Evol. 24: 505–512. [DOI] [PubMed] [Google Scholar]