Abstract
Recent studies indicate that the matrix domain (MA) of the HIV-1 Gag polyprotein directs Gag to the plasma membrane for virus assembly via a phosphatidylinositol-4,5-bisphosphate (PIP2)–dependent myristyl switch mechanism. MA also has been reported to direct nuclear trafficking via nuclear import and export functions, and some studies suggest that nuclear targeting may be regulated by MA phosphorylation (although this proposal remains controversial). We have prepared and studied a series of HIV-1 MA mutants containing Ser-to-Asp substitutions designed to mimic phosphorylation, including substitutions in regions of the protein involved in protein–protein interactions and known to influence the myristyl switch (S6D, S9D, S67D, S72D, S6D/S9D, and S67D/S72D). We were particularly interested in substitutions at residue 6, since conservative mutations adjacent to this site strongly perturb the myristyl switch equilibrium, and this site had not been genetically tested due to its involvement in post-translational myristylation. Our studies reveal that none of these mutations, including S6D, influences the PIP2- or concentration-dependent myristyl switch equilibrium. In addition, all of the mutants bind liposomes with affinities that are only slightly reduced in comparison with the native protein. In contrast, the myristylated mutants bind liposomes with substantially greater affinity than that of the native, unmyristylated protein. These findings support the hypothesis that phosphorylation is unlikely to significantly influence membrane-mediated intracellular trafficking.
Keywords: human immunodeficiency virus type-1 (HIV-1); myristyl (myr); matrix (MA); Gag; phosphatidylinositol-4,5-bisphosphate (PIP2); liposome; nuclear magnetic resonance (NMR)
The major structural constituent of all retroviruses is a protein called Gag, a multidomain polypeptide that is capable of assembling into virus-like particles (VLP) when expressed in various cell types in the absence of other viral constituents. Viral infectivity is critically dependent on the ability of Gag to selectively and tightly bind to proper cellular membranes for assembly. Gag's matrix (MA) domain, which is post-translationally myristylated on the N terminus in most retroviruses, plays several important roles during the replication cycle (for reviews, see Freed 1998; Demirov and Freed 2004). For most retroviruses, tight membrane binding requires a myristyl group (myr) and a conserved patch of basic residues on the surface of the MA protein (Yuan et al. 1993; Zhou et al. 1994). NMR-based structural studies revealed that the HIV-1 MA myristyl group can adopt sequestered (myr[s]) and exposed (myr[e]) conformations, and that myristate exposure can be promoted by factors that increase protein self-association (Tang et al. 2004). More recent studies have shown that myristate exposure can also be triggered by the binding of phosphatidyl-4,5-bisphosphate (PIP2) (Saad et al. 2006), a cellular factor located in the inner leaflet of the plasma membrane (PM) that targets Gag to the PM for virus assembly (Ono et al. 2004).
Other studies have suggested that MA may play a role in directing Gag in and out of the nucleus prior to assembly. Mutagenesis studies indicate that MA contains nuclear localization (NLSs) and nuclear export (NESs) signals, and that disruption of the NES can lead to accumulation of Gag in the nucleus and to the production of RNA-deficient virions (Dupont et al. 1999; Scheifele et al. 2002). Mutations in Rous sarcoma virus (RSV) MA that block nuclear localization of Gag can also lead to the production of particles deficient in genomic RNA (Scheifele et al. 2002; Callahan and Wills 2003). MA also has been reported to help direct the preintegration complex to the nucleus during the early phase of replication (Bukrinsky et al. 1992, 1993a,b; Dupont et al. 1999; Haffar et al. 2000). A fraction of MA molecules appears to be phosphorylated in infected cells, and it has been suggested that phosphorylation may play a role in retargeting MA from the PM to the nucleus (Dupont et al. 1999). However, in vivo mutagenesis studies were unable to identify a specific Ser, Thr, or Tyr residue that is essential for nuclear targeting. Mutation of individual serine residues to alanine had little effect on virus production and infectivity, and the simultaneous substitution at positions 9, 67, 72, and 77 was required to significantly impair viral infectivity without affecting virus assembly (Kaushik and Ratner 2004). Based on these results, it was suggested that phosphorylation at multiple sites changes the overall charge balance on the surface of MA protein, which might disrupt the electrostatic interaction between MA and the PM (Kaushik and Ratner 2004). It is noteworthy, however, that genetic studies were not performed for Ser-6 due to its role as a consensus recognition element for N-terminal myristylation (Freed and Martin 1994; Kaushik and Ratner 2004).
We recently demonstrated that mutations of residues adjacent to Ser-6 and Ser-9 (V7R, L8A, and L8I) can dramatically alter the myristyl switch equilibrium (Saad et al. 2007), leading us to speculate that phosphorylation of Ser-6 might similarly influence the myristyl switch. We therefore conducted NMR, thermodynamic, and membrane binding studies on a series of serine-to-aspartate mutants designed to mimic phosphorylated states of the protein.
Results
A variety of studies have shown that mutation of serine to aspartate/glutamate and serine to alanine mimics phosphorylation and dephosphorylation, respectively (Lu and Ou 2002; Kaushik and Ratner 2004; Bivona et al. 2006). By using native chemical ligation methods (Wu et al. 2004), we obtained, for the first time, highly pure myristylated S6D and S6D/S9D protein samples. For NMR studies, we selectively labeled seven residues in the N terminus of MA (Gly-2, Ala-3, Ala-5, Val-7, Leu-8, Gly-10, and Gly-11).
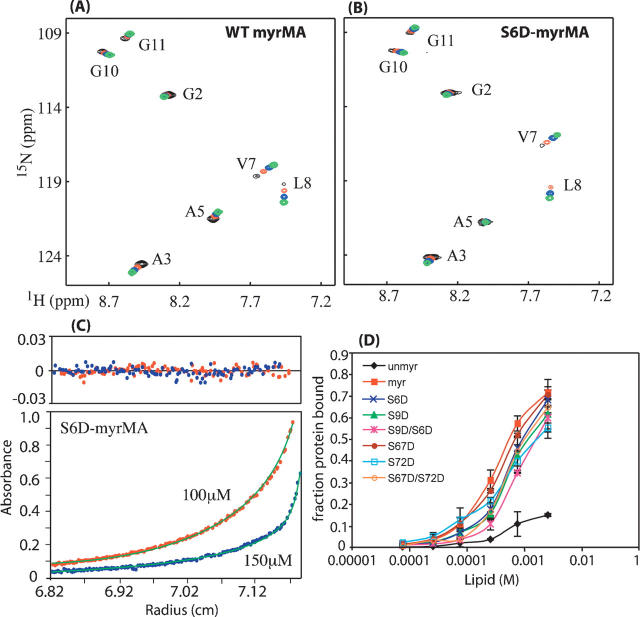
Two-dimensional [1H–15N] HSQC spectra obtained for S6D, S9D, and S6D/S9D myrMA proteins at concentrations of 50–800 μM display a chemical shift pattern that is essentially identical to the wild-type (WT) myrMA spectrum (Fig. 1 and Supplemental material). These results demonstrate that phosphorylation of serine residues in N terminus of MA does not perturb the myristyl switch mechanism and does not induce any significant conformational changes in the N-terminal domain of HIV-1 MA. These findings are in sharp contrast to those obtained for V7R, L8A, and L8I mutants, in which a single-point mutation nearby Ser-6 and Ser-9 induced only minor conformational changes that favor the myristyl-sequestered form (Saad et al. 2007). Representative sedimentation equilibrium (SE) data obtained for WT and S6D myrMA are shown in Figure 1. Analysis of the SE data afforded association constant values (Kassoc) = 2.2 ± 0.5 × 108 and 4.7 ± 1.1 × 108 M−2 (20°C) for S6D and S9D myrMA, respectively. These values are very similar to that obtained for WT myrMA (1.2 ± 0.4 × 108 M−2) and best fit a monomer-trimer equilibrium. Equilibrium data demonstrate that substitution of Ser-6 and Ser-9 has no effect on the equilibrium constant and oligomerization properties of myrMA.
Figure 1.
Overlay of two-dimensional 1H-15N HSQC spectra for WT (A) and S6D-myrMA (B) proteins collected at different concentrations (800–900 μM [black], 300–400 μM [red], 150 μM [blue], 50 μM [green]). Signals indicative of myristyl switch from sequestered form at low concentration to exposed form at high concentration shift similarly for WT and S6D-myrMA proteins. (C) Representative sedimentation profiles obtained for S6D-myrMA (100 μM [red], 150 μM [blue]). (D) Effect of lipid concentration on MA–lipid interaction. The apparent binding constant is ∼10−3 M. Data show that mutant HIV-1 myrMA proteins bound to liposomes with ∼1–2 lower affinity than native myrMA for most mutants except S6D/S9D, which showed ∼3 times weaker than native. Unmyristylated MA bound very weakly to liposomes.
In the X-ray structure of HIV-1 MA, Thr-70 and Ser-72 of one monomer form salt bridges with Asn-47 and Ala-45 of another monomer, respectively. Thus, we predicted that phosphorylation of these residues may disrupt interactions between monomers and shift the monomer-trimer equilibrium toward the monomer form. Two-dimensional [1H–15N] HSQC spectra and SE data obtained for S67D, S72D, and S67D/S72D confirmed that proteins exist in a monomer-trimer equilibrium with equilibrium constants very similar to the WT myrMA (Supplemental material), suggesting that phosphorylation on these positions does not induce significant conformational changes of the MA protein.
PIP2 binding to mutants
Binding studies were conducted with di-C4-phosphatidylinositol phosphate (di-C4-PIP2), a soluble analog of PIP2 containing truncated acyl chains. Representative two-dimensional [1H-15N] HSQC NMR data obtained upon titration of S6D- and S9D-myrMA with di-C4-PIP2 are shown in the Supplemental material. Titration of di-C4-PIP2 led to changes in the backbone 1H and 15N NMR chemical shifts of residues Ala-3, Ala-5, Val-7, and Leu-8. These signals correspond to residues that are well removed from the PIP2 binding site, but their shifts are indicative of a progressive shift from myr(s) in the PIP2-free state to myr(e) in the PIP2-bound state. For S9D-myrMA, in addition to the shifts observed for the 1H-15N NMR signals of Ala-3 to Gly-11, titration of di-C4-PIP2 led to significant changes in the backbone 1H and 15N NMR chemical shifts of residues Arg-22, Lys-26, Lys-27, His-33, Glu-73, Glu-74, Leu-75, and Ser-77 (ΔδHN [Δδ1 H]2+[Δδ15 N]2)1/2 = 0.1 – 0.8 ppm; Supplemental material). These residues reside on the β-II-V cleft and were previously shown to contribute to the PIP2 binding site (Saad et al. 2006). Nonlinear least squares fits of the titration data afforded a dissociation constant (Kd) values 116 ± 17 and 113 ± 8 μM for S6D- and S9D-myrMA, respectively. In summary, our results demonstrate that Ser-to-Asp mutations near the N terminus of MA (Ser-6 and Ser-9) or at downstream sites (Ser-67 and Ser-72) do not alter the binding properties of PIP2 or its ability to trigger myristate exposure.
Effect of mutations on liposome binding
To study the effect of phosphorylation on MA binding to membranes, we conducted liposome binding studies with the WT and mutant proteins using a published in vitro liposome-binding assay (Dalton et al. 2005). All of the Ser-to-Asp mutations had a minor effect upon membrane binding (Fig. 1). The most dramatic effect was seen for the S6D/S9D double mutant, which bound liposomes 3.5-fold less tightly than the WT protein. However, none of the mutations reduced binding to the level of myr(−)MA. These data reveal that the Ser-to-Asp mutations do not dramatically reduce membrane binding of HIV-1 MA, as suggested by other in vivo studies (Gallay et al. 1995a,b; Bukrinskaya et al. 1996). The small decrease in binding affinities observed for the mutant proteins in the in vitro assay likely reflects a reduction in the electrostatic contribution to membrane binding.
Discussion
In contrast to conservative point mutations of Val-7 and Leu-8, which dramatically alter the myristyl-switch equilibrium (Saad et al. 2007), substitution of the adjacent serine residues (Ser-6, Ser-9, or both) by Asp had no effect on the concentration- or PIP2-dependent myristyl switch equilibrium. It therefore seems unlikely that phosphorylation at these sites would significantly influence membrane targeting. Substitution of other surface Ser residues by Asp, including residues at the intermolecular interface of a trimeric myr(−)MA crystal structure, also had a minimal effect on binding to liposomes, suggesting that MA–MA interactions observed in the trimeric myr(−)MA X-ray crystal structure (Hill et al. 1996) may not exist when MA is bound to membranes. These findings are consistent with recent electron cryotomography studies of immature virions, which revealed an ordered hexagonal capsid shell but a MA shell without regular structure (Wright et al. 2007). Our findings are also consistent with studies by Spearman et al. (1997), which showed that the level of MA phosphorylation did not appear to differ between membrane-free and membrane-bound forms, suggesting that MA phosphorylation is not a major regulator of membrane association.
Materials and Methods
Sample preparation
S6D and S6D/S9D proteins were synthesized as described (Wu et al. 2004). Molecular weights were confirmed by electrospray ionization mass spectrometry. For proper solubilization and refolding, protein samples were first dissolved in H2O prior to dialysis in NMR buffer (50 mM sodium phosphate at pH 5.5, 100 mM NaCl, and 5 mM DTT). S9D, S67D, S72D, and S67D/S72D plasmids were made by site-directed mutagenesis of a coexpression vector harboring the yeast N-terminal myristyl transferase (yNMT) and HIV-1 MA gene (Tang et al. 2004). Unlabeled and 15N-enriched protein samples were prepared as described (Saad et al. 2007).
NMR spectroscopy
NMR data (35°C) were collected with a Bruker DMX (600 MHz 1H) spectrometer equipped with a cryoprobe, processed with NMRPIPE (Delaglio et al. 1995) and analyzed with NMRVIEW (Johnson and Blevins 1994). Binding isotherms from 1H-15N NMR HSQC titration experiments were calculated with ORIGIN 7.0 software (MicoCal).
Analytical ultracentrifugation
SE measurements were made as described elsewhere (Saad et al. 2007).
Liposome binding assay
Membrane flotation assays were performed as described (Dalton et al. 2005). See Supplemental material for more details.
Electronic supplemental material
Electronic supplemental material includes description of liposome binding assay and two-dimensional [1H-15N] HSQC spectra for mutant proteins as a function of protein- or PIP2-concentration.
Acknowledgments
This work was supported by NIH grants AI30917 (M.F.S.), CA20081 (V.M.V.), and AI058939 (W.L.).
Footnotes
Supplemental material: www.proteinscience.org
Reprint requests to: Michael F. Summers, Howard Hughes Medical Institute and Department of Chemistry and Biochemistry, University of Maryland, Baltimore County, 1000 Hilltop Circle, Baltimore, MD 21250, USA; e-mail: summers@hhmi.umbc.edu; fax: (410) 455-1174.
Abbreviations: MA, myristylated HIV-1 matrix protein; Gag, myristylated HIV-1 Gag polyprotein; myr(−), unmyristylated; myr(s), myristate-sequestered state; myr(e), myristate-exposed state; PIP2, phosphatidylinositol-(4,5)-bisphosphate; NMR, nuclear magnetic resonance; HSQC, heteronuclear single-quantum coherence; AU, analytical ultracentrifugation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072987607.
References
- Bivona T.G., Quatela, S.E., Bodemann, B.O., Ahearn, I.M., Soskis, M.J., Mor, A., Miura, J., Wiener, H.H., Wright, L., Saba, S.G., et al. 2006. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell 21: 481–493. [DOI] [PubMed] [Google Scholar]
- Bukrinskaya A.G., Ghorpade, A., Heinzinger, N.K., Smithgall, T.E., Lewis, R.E., and Stevenson, M. 1996. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc. Natl. Acad. Sci. 93: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I., Sharova, N., Dempsey, M.P., Stanwick, T.L., Bukrinskaya, A.G., Haggerty, S., and Stevenson, M. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. 89: 6580–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I., Haggerty, S., Dempsey, M.P., Sharova, N., Adzhubei, A., Spitz, L., Lewis, P., Goldfarb, D., Emerman, M., and Stevenson, M. 1993a. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365: 666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I., Sharova, N., McDonald, T.L., Pushkarskaya, T., Tarpley, W.G., and Stevenson, M. 1993b. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. 90: 6125–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan E.M. and Wills, J.W. 2003. Link between genome packaging and rate of budding for Rous Sarcoma Virus. Virology 77: 9388–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton A.K., Murray, P.S., Murray, D., and Vogt, V.M. 2005. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 79: 6227–6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Demirov D. and Freed, E.O. 2004. Retrovirus budding. Virus Res. 106: 87–102. [DOI] [PubMed] [Google Scholar]
- Dupont S., Sharova, N., DeHoratius, C., Virbasius, C.-M.A., Zhu, X., Bukrinskaya, A.G., Stevenson, M., and Green, M.R. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402: 681–685. [DOI] [PubMed] [Google Scholar]
- Freed E.O. 1998. HIV-1 Gag proteins: Diverse functions in the virus life cycle. Virology 251: 1–15. [DOI] [PubMed] [Google Scholar]
- Freed E.O. and Martin, M.A. 1994. HIV-1 infection of non-dividing cells. Nature 369: 107–108. [DOI] [PubMed] [Google Scholar]
- Gallay P., Swingler, S., Aiken, C., and Trono, D. 1995a. HIV-1 infection of non-dividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80: 379–388. [DOI] [PubMed] [Google Scholar]
- Gallay P., Swingler, S., Song, J., Bishman, F., and Trono, D. 1995b. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83: 569–576. [DOI] [PubMed] [Google Scholar]
- Haffar O.K., Popov, S., Dubrovsky, L., Agostini, I., Tang, H., Pushkarsky, T., Nadler, S.G., and Bukrinsky, M. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 2999: 359–368. [DOI] [PubMed] [Google Scholar]
- Hill C.P., Worthylake, D., Bancroft, D.P., Christensen, A.M., and Sundquist, W.I. 1996. Crystal structures of the trimeric HIV-1 matrix protein: Implications for membrane association. Proc. Natl. Acad. Sci. 93: 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A. and Blevins, R.A. 1994. NMRview: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Kaushik R. and Ratner, L. 2004. Role of human immunodeficiency virus type 1 matrix phosphorylation in an early post-entry step of virus replication. J. Virol. 78: 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W. and Ou, J.-H. 2002. Phosphorylation of hepatitis C virus core protein by protein kinase A and protein kinase C. Virology 300: 20–30. [DOI] [PubMed] [Google Scholar]
- Ono A., Ablan, S.D., Lockett, S.J., Nagashima, K., and Freed, E.O. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. 101: 14889–14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Miller, J., Tai, J., Kim, A., Ghanam, R.H., and Summers, M.F. 2006. Structural basis for targeting HIV-1 Gag to virus assembly sites on the plasma membrane. Proc. Natl. Acad. Sci. 103: 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Loeliger, E., Luncsford, P., Liriano, M., Tai, J., Kim, A., Miller, J., Joshi, A., Freed, E.O., and Summers, M.F. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele L.Z., Garbitt, R.A., Rhoads, J.D., and Parent, L.J. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. 99: 3944–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman P., Horton, R., Ratner, L., and Kuli-Zade, I. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71: 6582–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Loeliger, E., Luncsford, P., Kinde, I., Beckett, D., and Summers, M.F. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. 101: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E.R., Schooler, J.B., Ding, H.J., Kieffer, C., Fillmore, C., Sundquist, W.I., and Jensen, G.J. 2007. Electron crytomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26: 2218–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Alexandratos, J., Ericksen, B., Lubkowski, J., Gallo, R.C., and Lu, W. 2004. Total chemical synthesis of N-myristoylated HIV-1 matrix protein p17: Structural and mechanistic implications of p17 myristoylation. Proc. Natl. Acad. Sci. 101: 11587–11592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yu, X., Lee, T.-H., and Essex, M. 1993. Mutations in the N-terminal region of human immunodeficiency virus type I matrix protein block intracellular transport of the Gag precursor. J. Virol. 67: 6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Parent, L.J., Wills, J.W., and Resh, M.D. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag Protein which interacts with acidic phospholipids. J. Virol. 68: 2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]