Abstract
Transcarbamylases catalyze the transfer of the carbamyl group from carbamyl phosphate (CP) to an amino group of a second substrate such as aspartate, ornithine, or putrescine. Previously, structural determination of a transcarbamylase from Xanthomonas campestris led to the discovery of a novel N-acetylornithine transcarbamylase (AOTCase) that catalyzes the carbamylation of N-acetylornithine. Recently, a novel N-succinylornithine transcarbamylase (SOTCase) from Bacteroides fragilis was identified. Structural comparisons of AOTCase from X. campestris and SOTCase from B. fragilis revealed that residue Glu92 (X. campestris numbering) plays a critical role in distinguishing AOTCase from SOTCase. Enzymatic assays of E92P, E92S, E92V, and E92A mutants of AOTCase demonstrate that each of these mutations converts the AOTCase to an SOTCase. Similarly, the P90E mutation in B. fragilis SOTCase (equivalent to E92 in X. campestris AOTCase) converts the SOTCase to AOTCase. Hence, a single amino acid substitution is sufficient to swap the substrate specificities of AOTCase and SOTCase. X-ray crystal structures of these mutants in complexes with CP and N-acetyl-L-norvaline (an analog of N-acetyl-L-ornithine) or N-succinyl-L-norvaline (an analog of N-succinyl-L-ornithine) substantiate this conversion. In addition to Glu92 (X. campestris numbering), other residues such as Asn185 and Lys30 in AOTCase, which are involved in binding substrates through bridging water molecules, help to define the substrate specificity of AOTCase. These results provide the correct annotation (AOTCase or SOTCase) for a set of the transcarbamylase-like proteins that have been erroneously annotated as ornithine transcarbamylase (OTCase, EC 2.1.3.3).
Keywords: argF gene, transcarbamylase, N-succinylornithine, N-acetylornithine, arginine biosynthesis, protein engineering, crystal structure
The transcarbamylase family of enzymes catalyzes the transfer of a carbamyl group from carbamyl phosphate (CP) to an amino group of the second substrate. Aspartate transcarbamylase (ATCase) and ornithine transcarbamylase (OTCase) are the two best-known members of this family. Phylogenetic analysis for nearly 450 transcarbamylase sequences demonstrated that most of them are either ATCases or OTCases, forming two major branches in the phylogenetic tree (Naumoff et al. 2004). OTCases are essential enzymes that catalyze the carbamylation of L-ornithine in the arginine biosynthetic pathway in lower organisms or in the urea cycle in mammals. Along the OTCase branch, several small clades exist between the root and OTCase branch. The work reported herein demonstrates that most of the transcarbamylase sequences in these clades are incorrectly annotated and belong to novel classes of transcarbamylase.
The canonical biosynthesis of arginine in microbes and plants proceeds from glutamic acid through eight enzymatic steps using acetylated intermediates in the first five steps (Cunin et al. 1986; Davis 1986; Slocum 2005). The recent discovery of N-acetylornithine transcarbamylase (AOTCase) indicates that some plant pathogens in the Xanthomonadaceae genus use a modified biosynthetic pathway to produce arginine (Shi et al. 2005a, 2006b; Morizono et al. 2006). In these organisms, two enzymes, N-acetylornithine deacetylase (AODase) and ornithine transcarbamylase (OTCase), are missing; instead, two novel enzymes, AOTCase and N-acetylcitrulline deacetylase (ACDase), replace them (Shi et al. 2005b, 2007). Thus, the order of deacetylation and transcarbamylation is reversed relative to the canonical pathway. When we compared the structure of AOTCase from Xanthomonas campestris with the structure of a transcarbamylase-like protein essential for arginine biosynthesis in Bacteroides fragilis (Shi et al. 2002), an additional novel enzyme, N-succinyl-transcarbamylase (SOTCase), was uncovered (Shi et al. 2006a). This discovery implies that B. fragilis, a clinically important human pathogen, and other bacteria with homologous enzymes may use succinyl intermediates rather than acetyl intermediates for arginine biosynthesis (Shi et al. 2006a). This finding provides interesting clues about the evolutionary relationships among enzymes in the arginine biosynthesis pathway and allows a new potential strategy for inhibiting human and plant pathogens using these unique enzymes as targets for antibacterial agents. The previous work also indicated that E92 of X. campestris AOTCase (the corresponding residue P90 in B. fragilis SOTCase) is critical for conferring specificity toward the second substrate.
In this paper, the activity and crystal structures of E92A, E92P, E92S, and E92V mutants of AOTCase from X. campestris and the P90E/T242L (T242L is a cloning artifact) mutant of SOTCase from B. fragilis have been investigated. The activity assays demonstrate that a single mutation in the active site interconverts the substrate specificity between AOTCase and SOTCase. The crystal structures of X. campestris AOTCase mutants complexed with CP and N-succinylnorvaline (SNOR) and a B. fragilis SOTCase mutant complexed with CP and N-acetylnovaline (ANOR) confirm that E92 (X. campestris numbering) is the key residue in determining substrate specificity.
Results and Discussion
Specificity of transcarbamylation activity
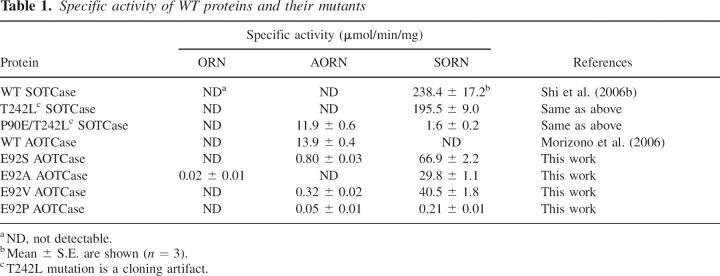
Specific activities of wild-type SOTCase from B. fragilis, wild-type AOTCase from X. campestris, and of the various mutants are listed in Table 1. These enzymatic assays clearly indicate that all four mutants of AOTCase preferentially catalyze the carbamylation of N-succinylornithine relative to two other potential substrates, N-acetylornithine and ornithine. Enzymatic activities of the mutants in the order from most to least active are: E92S > E92V > E92A > E92P. The shape and polarity of the side chains of the mutated residues also appear to significantly impact the catalytic activity since the E92P mutant shows 319-fold less activity than the E92S mutant. It is also interesting to note that the E92P mutant has 1135-fold less activity than the B. fragilis wild-type (WT) SOTCase in which the equivalent residue is also Pro. However, the specific activity of the P90E/T242L mutant of SOTCase, which preferentially catalyzes the carbamylation of N-acetylornithine, is only slightly less than the X. campestris wild-type AOTCase. While activity assays confirmed that Glu92 (X. campestris numbering) is a key residue involved in distinguishing AOTCase from SOTCase, the significantly lower catalytic efficiency of these mutants compared to the WT proteins implies that other structural differences also play a significant role in the catalytic reaction. To clarify these differences, structural information for these mutants complexed with bound substrates or substrate analogs is essential.
Table 1.
Specific activity of WT proteins and their mutants
Structural model
The structures of all mutant proteins complexed with CP and their second substrate analog, ANOR or SNOR, have been determined. Refinement statistics of the final structural models are listed in Table 2. The stereochemistry, assessed using program PROCHECK (Laskowski et al. 1993), indicates that all structures have acceptable geometry. As in the WT protein structures, three residues, Glu144, Thr145, and Leu 295 (X. campestris numbering), are in energetically unfavorable conformations and there is one cis peptide bond between Leu 295 and Pro 296. The mutant structures are very similar to the WT structure with an RMS difference <0.25 Å when all equivalent Cα atoms are superimposed.
Table 2.
Data collection and refinement statistics
The electron density maps show clear and interpretable electron density in the active site corresponding to the bound ligands, CP and SNOR or CP and ANOR (Fig. 1A–E), even in the early stages of refinement. The conformation of the 80's loop where the mutation is located is well defined. The electron density around Glu92 of AOTCase and Pro90 of SOTCase verifies the presence of the engineered mutations.
Figure 1.
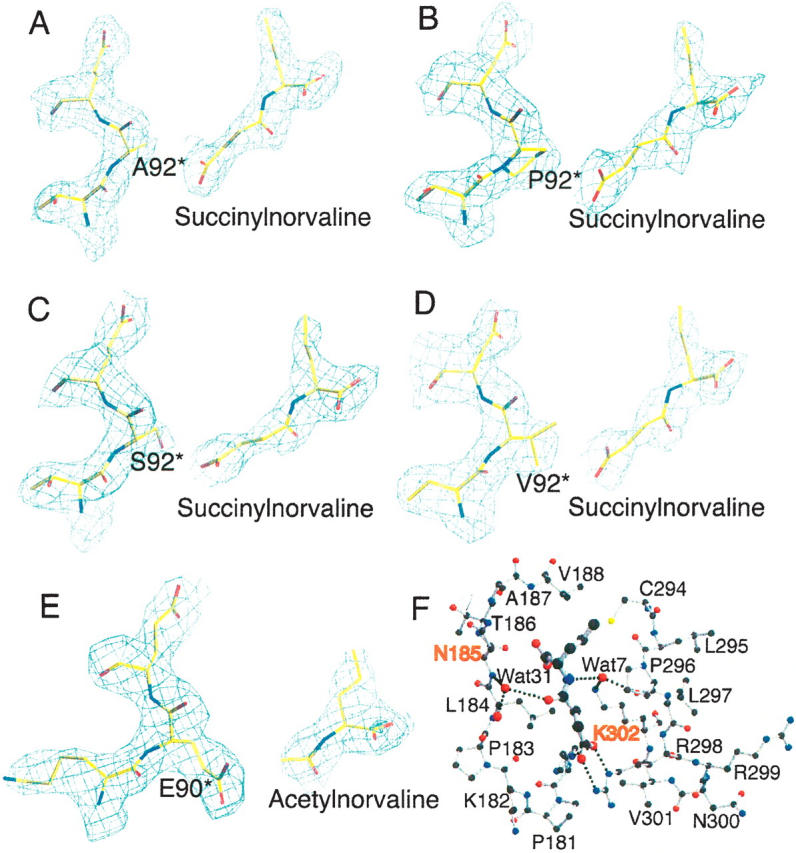
N-acetylnorvaline or N-succinylnorvaline binding of SOTCase or AOTCase mutants. (A–D) Contours of the electron density map (2Fo − Fc) (1.0 σ shown in blue cage) around N-succinylnorvaline and E92 region of X. campestris AOTCase mutants. (E) Contours of the electron density map (2Fo − Fc) (1.0 σ shown in blue cage) around N-acetylnorvaline and P90 region of B. fragilis SOTCase mutant. The final refined positions of the ligands and residues are represented as colored sticks. The residue indicated by * is from the adjacent subunit. The electron density clearly indicates the binding of substrate analogs and the expected amino acid substitutions. (F) N-succinylnorvaline binding site of X. campestris E92A mutant. SNOR is shown as thick ball-and-stick models, while the surrounding residues are shown as thin ball-and-stick models. Two water molecules, Wat7 and Wat31, form hydrogen-bonding bridges to link the ligand to protein residues. These water-mediated hydrogen-bonding networks can only be found in X. campestris AOTCase and its mutants, but not in B. fragilis SOTCase and mutants. The residues, N185 and K302 (X. campestris AOTCase numbering) shown in red, are proposed as the residues that distinguish SOTCase from AOTCase in addition to the key residue of E92.
Active site
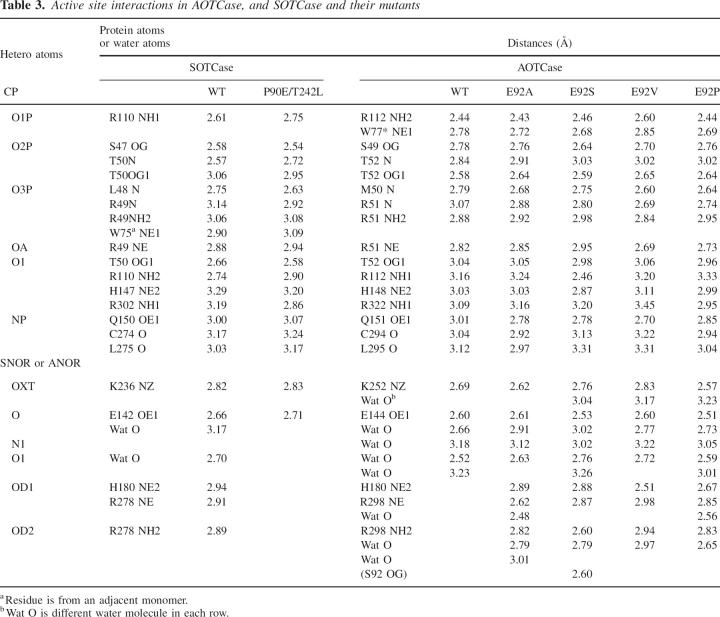
One CP and one SNOR were found in the active site of X. campestris AOTCase E92A, E92S, E92V, and E92P mutants, while one CP and one ANOR were identified in the B. fragilis SOTCase P90E mutant. Almost all equivalent active site residues that interact directly with the substrates in SOTCase are also involved in binding CP and SNOR to all AOTCase mutants (Table 3). As in B. fragilis WT SOTCase (Shi et al. 2006a), two residues, His180 and Arg298, which hydrogen bond to the Glu92 from the adjacent subunit in the X. campestris WT AOTCase structure, were found to interact via hydrogen bonds with the succinyl group of SNOR in all X. campestris AOTCase mutant structures. Likewise, the interactions of CP and ANOR with the active site residues in B. fragilis SOTCase P90E mutant are similar to those in X. campestris WT AOTCase (Shi et al. 2006b). The Glu90 (mutated from Pro90) interacts with His176 and Arg278 (B. fragilis numbering) in a manner similar to WT AOTCase, thus occupying the space that in WT SOTCase accommodates the succinyl moiety, and precluding SNOR binding to the P90E mutant of SOTCase.
Table 3.
Active site interactions in AOTCase, and SOTCase and their mutants
Even though the data quality, especially the presence of high resolution data, will influence the number of water molecules identified from the electron density map, four water molecules involved in binding ANOR or SNOR are conserved in X. campestris WT AOTCase and all its mutant structures (Table 3). In contrast, only two of them can be identified in the B. fragilis WT SOTCase structure and none were found in the P90E SOTCase mutant structure. Two residues in the active site, Asn185 and Lys302 (X. campestris numbering), appear to affect the role of water in substrate binding and catalytic activity. One molecule of water links the acyl O atom of ANOR or SNOR to the main-chain N atom of Asn185 in X. campestris WT or mutant structures (Fig. 1F). The equivalent residue in B. fragilis is Pro181, which is unable to contribute its main-chain N atom for hydrogen bonding. Consistent with this observation, all known sequences of SOTCases and AOTCases classified on the basis of the residue at the Glu92 position (X. campestris numbering) have an Asn in the equivalent position of AOTCase and a Pro in SOTCase (Fig. 2). Similarly, Lys302 may help a water molecule hydrogen bind to the imino N atom of AORN or SORN and the main-chain carbonyl O atom of Pro296 (Fig. 1F). The presence of Glu, Val, or Ile in the equivalent position of SOTCase appears to prevent water from participating in hydrogen bonding. The different ways in which water is involved in substrate binding in AOTCase and SOTCase appear to be one reason for the significantly lower catalytic efficiency of the mutants relative to the WT protein. Additional mutants such as N185P and K302I (X. campestris numbering) need to be studied to clarify this prediction.
Figure 2.
Sequence alignments of the regions containing E92, N185, and K302 (shown in bold and indicated by a triangle) to define the substrate specificity of AOTCase and SOTCase. The sequences are from bacteria, Xanthomonas campestris, Xanthomonas oryzae, Xanthomonas axonopodis, Xylella fastidiosa, Parvularcula bermudensis, Maricaulis maris, Oceanicaulis alexandrii, Salinibacter ruber, Psychroflexus torquis, Tenacibaculum sp. Med152, Leeuwenhoekiella blandensis MED217, Polaribacter irgensii 23-P, Flavobacterium johnsoniae UW101, Prevotella ruminicola, Bacteroides thetaiotaomicron, Bacteroides fragilis, Flavobacteriales bacterium BBFL7, Robiginitalea biformata, Tanneralla forsythensis, Myxococcus xanthus, Cellulophaga sp. MED134, Cytophaga hutchinsonii, and Candidatus Sulcia Muelleri.
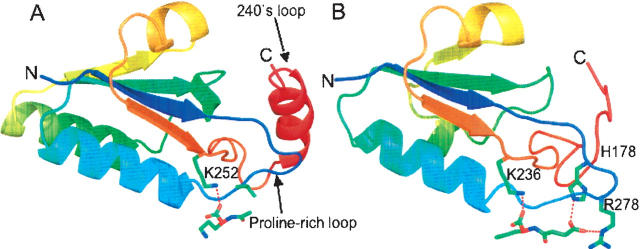
As pointed out recently by two independent groups there is a novel deep trefoil knot in AOTCase and SOTCase structures close to the active site that is not present in ATCase or OTCase structures (Fig. 3; Khatib et al. 2006; Virnau et al. 2006). It appears that the knot modifies the active sites to allow AOTCase and SOTCase to recognize acetylated or succinylated substrates and to use different amino acid motifs in substrate recognition. In ATCase and OTCase, the flexible 240's loop (termed SMG loop in OTCase) is the main recognition site for the second substrate, aspartate or ornithine (Krause et al. 1987; Gouaux and Lipscomb 1988; Ke et al. 1988; Ha et al. 1997; Shi et al. 1998, 2001), while in AOTCase and SOTCase, the unique proline-rich loop becomes part of the second substrate binding site.
Figure 3.
A ribbon diagram of the knotted region of X. campestris AOTCase (A) (residues 170–275) and B. fragilis SOTCase (B) (residues 166–255) with bound ligands shown in colored sticks. Colors change continuously from blue (first residue in the region) to red (last residue in the region). The proline-rich loop (from dark blue to light blue), in similar conformation for both AOTCase and SOTCase, appears to be critical for the knot formation. The 240's loop (from brown to red) was threaded through the proline-rich loop to form a trefoil knot.
Structural comparison of X. campestris AOTCase E92S, E92V, E92A, and E92P mutants
Even though the specific SOTCase activities of these four mutants range from 66.9 μmol/min/mg for E92S to 0.22 μmol/min/mg for E92P, their structures are very similar with RMS <0.25 Å for all superimposed 332 Cα atoms. The relatively high activity of E92S may be a result of a favorable hydrogen-bonding interaction between the side chain of the mutated residue and the succinyl group of substrate which is not present in the other mutants. The reason for the extremely low activity of E92P mutant needs to be investigated further.
SOTCase and AOTCase in bacteria
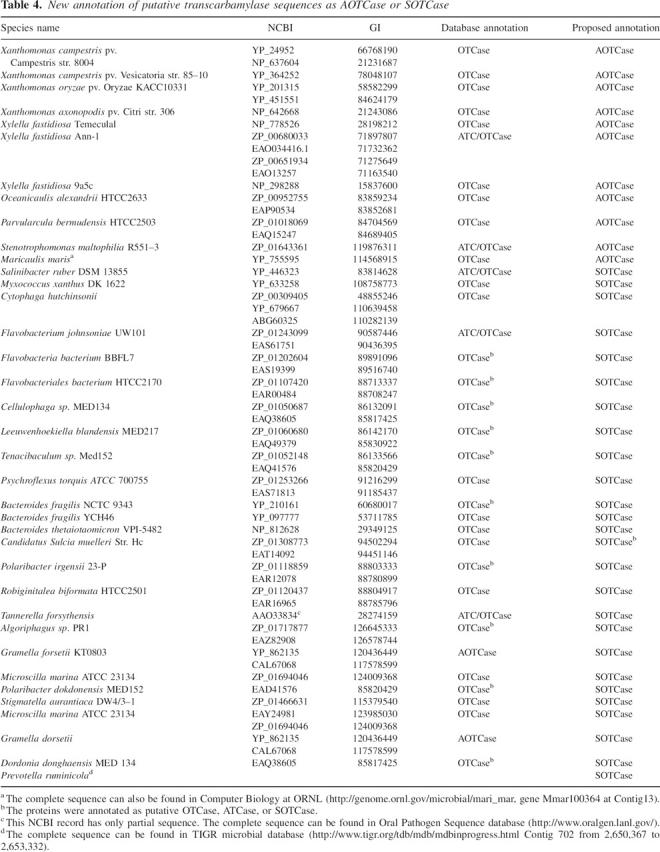
Although other subtle differences may affect the enzymatic activity of AOTCase and SOTCase, Glu92 (X. campestris numbering) plays a key role in distinguishing AOTCase from SOTCase. In addition to bacteria such as Bacteroides fragilis, Bacteroides thetaiotaomicron, Cytophaga fastidiosa, Tannerella forsythensis, Prevotella ruminicola, and Myxococcus xanthus in which SOTCase has been previously identified in the genomes (Shi et al. 2006a), other bacteria, including Robiginitalea biformata, Polaribacter irgensii, Flavobacterium sp., Flavobacteriales bacterium, Cellulophaga sp., and Salinibacter ruber, also have transcarbamylases that lack the signature Glu–Asn–Lys triad of AOTCase, suggesting that they are almost certainly SOTCases and that their arginine biosynthetic pathways likely use succinylated intermediates. Conversely, in addition to Xanthomonas campestris, Xanthomonas axonopodis, and Xylella fastidiosa, which we identified as AOTCases previously, Maricaulis maris, Oceanicaulis alexandrii, and Parvularcula bermudensis also have the AOTCase signature and are, therefore, likely to be AOTCases. These bacteria possess a modified pathway to synthesize arginine but still use acetyl intermediates as in the canonical pathway found in most microbes (Fig. 4). More interestingly, bacteria which use AOTCase to convert acetylornithine to acetylcitrulline in arginine biosynthesis have a unique bifunctional enzyme that catalyzes the first two steps of the pathway (Qu et al. 2007). The reannotation of these transcarbamylase sequences as AOTCase and SOTCase is provided in Table 4. The function of these transcarbamylases needs to be confirmed by enzymatic assay, particularly for the transcarbamylase (NBCI accession No. ZP_01308773) from Candidatus Sulcia muelleri, which has Gln in the position equivalent to Glu92 but lacks the Asn and Lys residues in the other two equivalent positions.
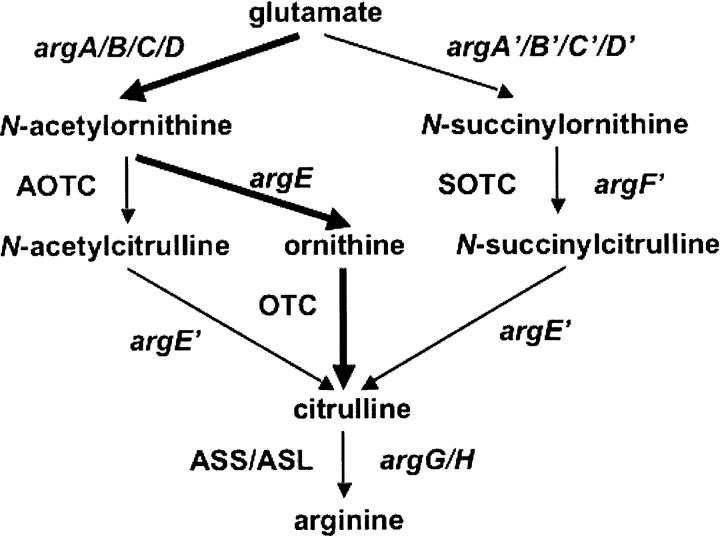
Figure 4.
Comparison of the canonical and two proposed linear arginine biosynthetic pathways. The canonical pathway is shown in thick arrows.
Table 4.
New annotation of putative transcarbamylase sequences as AOTCase or SOTCase
Classification of transcarbamylases
The three-dimensional structures of four distinct members of the transcarbamylase superfamily, ATCase, OTCase, AOTCase, and SOTCase, are now available. The structures, especially for those bound with substrates or substrate analogs, provide important information about how enzymes recognize their substrates. The classification scheme is shown in Figure 5. The highest conservation across the transcarbamylase superfamily involves three regions: the SxRT motif, the HPxQ motif, and the HxLP motif (Allewell et al. 1999; Shi et al. 2001). These conserved motifs define the CP binding site characteristic of all known transcarbamylases including putrescine transcarbamylase (PTCase) (Naumoff et al. 2004) and other unannotated transcarbamylases in the genome databases. Therefore, these three motifs can be used to screen for all transcarbamylases in genomic databases.
Figure 5.
A schematic diagram representing the classification of four known transcarbamylases, ATCase, OTCase, AOTCase, and SOTCase, based on structure-derived amino acid signature motifs.
The sequence variations in four loop regions, 80's loop, 120's loop, proline-rich loop, and 240's loop, confer specificity for second substrate binding. The presence of the extended 80's and 120's loops and the proline-rich loop seem to be characteristic of AOTCase and SOTCase. We can use these features to distinguish AOTCase and SOTCase from other transcarbamylases while AOTCase can be distinguished from SOTCase using three residues, Glu92, Asn185, and Lys302 (X. campestris numbering). As noted previously, the 240's loop is the main binding site for the second substrate in ATCase and OTCase. The specific features of the 240's loop, the RxQxER motif in ATCase and the DxxxSMG motif in OTCase, distinguish ATCase and OTCase from other transcarbamylases, as well as from each other. No structural information for the remaining members of the transcarbamylases, such as PTCase, is available. Sequence alignment indicates the putrescine transcarbamylases have high sequence similarity to OTCase with sequence identity ∼35%–45% but do not have the DxxxSMG motif which is characteristic of OTCase.
Disordered regions in proteins evolve more rapidly than ordered regions (Brown et al. 2002), and loop regions seem to evolve faster than regions which have secondary structures. This observation is consistent with the notion that all transcarbamylases have evolved from a common ancestor, with branches defined by the sequence diversity of their loop regions.
Three possible arginine biosynthesis pathways in bacteria
Our results demonstrate that arginine biosynthesis occurs via three slightly different pathways in prokaryotes (Fig. 4). Both newly discovered pathway variants follow similar routes in which N-succinyl-L-ornithine or N-acetyl-L-ornithine is first carbamylated to form N-succinyl-L-citrulline or N-acetyl-L-citrulline, respectively, then desuccinylated or deacetylated to produce citrulline (Shi et al. 2005a, 2006a). In contrast, in the canonical arginine biosynthetic pathway, N-acetyl-L-ornithine is first deacetylated to ornithine, then carbamylated to form citrulline. The acetyl pathway is utilized by most microorganisms and plants, while the succinyl pathway seems to be used only by certain bacteria.
Variant pathways have been observed in meso-diaminopimelate (m-DAP)/lysine biosynthesis (Born and Blanchard 1999). In prokaryotes, three slightly different pathways, which diverge after the production of tetrahydrodipicolinate, lead to the formation of m-DAP and lysine. Two follow a similar route, using succinylated intermediates in most bacteria but acetylated intermediates in certain Bacillus species. The third route, found in a few members of the firmicutes, uses m-DAP dehydrogenase to convert tetrahydrodipicolinate to m-DAP directly, circumventing the use of acyl intermediates and shortening the pathway. In contrast to the arginine biosynthetic pathway in which the acetyl pathway dominates, the succinyl pathway for m-DAP/lysine biosynthesis is used by all Gram-negative and many Gram-positive bacteria while the acetyl pathway is limited to certain Bacillus species.
Materials and Methods
Site-directed mutagenesis, protein expression, and purification
Site-directed mutagenesis was carried out using the “Quik-Change” mutagenesis kit (Stratagene) according to the manufacturer's protocol. The primers used to create E92P, E92V, E92A, and E92S mutants of X. campestris AOTCase and the P90E mutant of B. fragilis SOTCase are listed in Table 5. Preparation of the parents plasmids of X. campestris AOTCase and B. fragilis SOTCase was described previously (Shi et al. 2002, 2005a, 2006b). The correct mutants were confirmed by DNA sequencing. Recombinant proteins were expressed and purified as described previously (Shi et al. 2005a, 2006b). The protein purity was verified by SDS-PAGE (12% polyacrylamide gel) followed by Coomassie staining; a single band of the expected molecular mass was observed for each purified protein. Protein concentration was determined by the Bradford method using the BioRad protein assay dye reagent with bovine serum albumin as a standard (Bradford 1976).
Table 5.
Primers used for mutagenesis
Enzymatic assays
The modified colorimetric assay, which detects ureido groups, was used to test for enzymatic activity (Pastra-Landis et al. 1981). The method has been successfully used for the enzymatic assay of ATCase (Pastra-Landis et al. 1981), OTCase (Morizono et al. 1997), AOTCase (Morizono et al. 2006), and SOTCase (Shi et al. 2006a). With the exception of the E92P activity assay, the reaction mixtures contained in a final volume of 1 mL were 50 mM Tris-HCl buffer pH 8.3, 1 mM carbamyl phosphate (dilithium salt), 10 mM succinylornithine or acetylornithine or ornithine, and 0.2 μg of enzyme. Activity assay for E92P were performed by the same method with the exception that 0.12 mg of enzyme was used since its activity is much lower than that of the other proteins. All enzymes were diluted prior to the reaction with filtered storage buffer containing 20 mL Tris-HCl, 250 mM NaCl, and 1 mM EDTA pH 7.4. Reactions were initiated by adding the second substrate, run for 5 min at 25°C, and the reactions were quenched by adding 1 mL of quench reagent containing antipyrine in 50% H2SO4 and butadione monoxime in 2:1 ratio. Standard curves were prepared by adding varying amounts of succinylcitrulline or acetylcitrulline or citrulline in 50 mM Tris-HCl pH 8.3 to a total volume of 1 mL followed by the addition of 1 mL quench reagent. All reaction tubes were covered with foil and shaken in a dark chamber overnight before developing color at 42°C hot bath for 25 min. The absorbance was then measured at 466 nm.
Crystallization and data collection
The purified protein was concentrated to ∼10 mg/mL with an Amicon-Y30 membrane concentrator (Millipore). The E92P, E92A, E92V, and E92S mutants of X. campestris AOTCase complexed with CP and SNOR were crystallized in similar condition as wild-type AOTCase (Shi et al. 2005a), while the P90E mutant of B. fragilis SOTCase complexed with CP and ANOR was crystallized as described previously (Shi et al. 2006a).
During data collection, a crystal was dipped quickly into a mother solution containing 20% (v/v) ethylene glycerol for cryoprotection. The cryoprotected crystals were frozen by plunging crystals directly into liquid nitrogen. Data were collected on an R-axis IV image-plate diffractometer mounted on a Rigaku RU-200 rotating-anode generator with a Cu target (λ = 1.54178 Å). All data were processed using the HKL2000 package (Otwinowski and Minor 1997) and reduced using the program TRUNCATE in the CCP4 suite (Collaborative Computational Project, Number 4, 1994). The data collection statistics are summarized in Table 2.
Structure solution and refinement
The five structures reported herein were refined by slight variations of a basic protocol. The starting model for AOTCase mutants taken from the structure of wild-type AOTCase complexed with CP and ANOR (PDB code 2ZQ6) were stripped of the solvent atoms and hetero compounds. The starting model for the SOTCase P90E mutant was taken from the SOTCase structure complexed with CP and SNOR (PDB code 2FG7). The models were originally treated as single rigid bodies and refined with CNS 1.1 (Brünger et al. 1998) to 3 Å resolution. Further refinements involved simulated annealing, individual B-factor refinement manual with CNS 1.1, and model building with O (Jones et al. 1991). During the refinement process, 5% or 10% of the data, randomly selected, were used to monitor the free R-value (Brünger 1992). Water molecules were added using the WATERPICK protocol with CNS (Brünger et al. 1998). The final refinement statistics are listed in Table 2. The model quality was checked with the program PROCHECK (Laskowski et al. 1993), which indicated good stereochemistry for all structural models.
Figures were drawn using the programs MOLSCRIPT (Kraulis 1991), Raster3D (Merrit and Murphy 1994), PyMOL (DeLano 2002), and O (Jones et al. 1991). The coordinates have been deposited with the RCSB Protein Data Bank as entries 2G7M, 2G6C, 2G6A, 2G65, and 2G68.
Acknowledgments
This work was supported by Public Health Service grant DK-47870 (M.T.) and DK-067935 (D.S.) from the National Institute of Diabetes and Digestive and Kidney Diseases. We thank Dr. David Davies for facilitating our use of the diffraction equipment in the Molecular Structure Section of the National Institute of Health and Dr. Fred Dyda for help in data collection and processing.
Footnotes
Reprint requests to: Dashuang Shi, Children's Research Institute, Children's National Medical Center, 111 Michigan Avenue, NW, Washington, DC 20010-2970, USA; e-mail: dshi@cnmcresearch.org; fax: (202) 884-6014.
Abbreviations: ACDase, N-acetylcitrulline deacetylase; ANOR, N-acetylnorvaline; SNOR, N-succinylnorvaline; AODase, N-acetylornithine deacetylase; AOTCase, N-acetylornithine transcarbamylase; ATCase, aspartate transcarbamylase; CP, carbamyl phosphate; m-DAP, meso-diaminopime-late; OTCase, ornithine transcarbamylase; SOTCase, N-succinylornithine transcarbamylase; PTCase, putrescine transcarbamylase; WT, wild type.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072919907.
References
- Allewell N.M., Shi, D., Morizono, H., and Tuchman, M. 1999. Molecular recognition by ornithine and aspartate transcarbamoylases. Acc. Chem. Res. 32: 885–894. [Google Scholar]
- Born T.L. and Blanchard, J.S. 1999. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr. Opin. Chem. Biol. 3: 607–613. [DOI] [PubMed] [Google Scholar]
- Bradford M. 1976. Quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 1219–1223. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Takayama, S., Campen, A.M., Vise, P., Marshall, T.W., Oldfield, C.J., Williams, C.J., and Dunker, A.K. 2002. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 55: 104–110. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Crosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50: 760–763. [DOI] [PubMed] [Google Scholar]
- Cunin R., Glansdorff, N., Pierard, A., and Stalon, V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50: 314–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.H. 1986. Compartmental and regulatory mechanisms in arginine pathways of Neurospora crassa and Saccharomyces cerevisiae . Microbiol. Rev. 50: 280–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- Gouaux J.E. and Lipscomb, W.N. 1988. Three-dimensional structure of carbamoyl phosphate and succinate bound to aspartate carbamoyltransferase. Proc. Natl. Acad. Sci. 85: 4205–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y., McCann, M.T., Tuchman, M., and Allewell, N.M. 1997. Substrate-induced conformational change in a trimeric ornithine transcarbamylase. Proc. Natl. Acad. Sci. 94: 9550–9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and location of errors in these model. Acta Crystallogr. A47: 110–119. [DOI] [PubMed] [Google Scholar]
- Ke H.M., Lipscomb, W.N., Cho, Y.J., and Honzatko, R.B. 1988. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J. Mol. Biol. 204: 725–747. [DOI] [PubMed] [Google Scholar]
- Khatib F., Weirauch, M.T., and Rohl, C.A. 2006. Rapid knot detection and application to protein structure prediction. Bioinformatics 22: e252–e259, doi: 10.1093/bioinformatics/btl236. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- Krause K.L., Volz, K.W., and Lipscomb, W.N. 1987. 2.5 Å structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J. Mol. Biol. 193: 527–553. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Merritt E.A. and Murphy, M.E. 1994. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D50: 869–873. [DOI] [PubMed] [Google Scholar]
- Morizono H., Tuchman, M., Rajagopal, B.S., McCann, M.T., Listrom, C.D., Yuan, X., Venugopal, D., Barany, G., and Allewell, N.M. 1997. Expression, purification and kinetic characterization of wild-type human ornithine transcarbamylase and a recurrent mutant that produces “late onset” hyperammonaemia. Biochem. J. 322: 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono H., Cabrera-Luque, J., Shi, D., Gallegos, R., Yamaguchi, S., Yu, X., Allewell, N.M., Malamy, M.H., and Tuchman, M. 2006. Acetylornithine transcarbamylase: A novel enzyme in arginine biosynthesis. J. Bacteriol. 188: 2974–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumoff D.G., Xu, Y., Glansdorff, N., and Labedan, B. 2004. Retrieving sequences of enzymes experimentally characterized but erroneously annotated: The case of the putrescine carbamoyltransferase. BMC Genomics 5: 52. doi: 10.1186/1471-2164-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pastra-Landis S.C., Foote, J., and Kantrowitz, E.R. 1981. An improved colorimetric assay for aspartate and ornithine transcarbamylases. Anal. Biochem. 118: 358–363. [DOI] [PubMed] [Google Scholar]
- Qu Q., Morizono, H., Shi, D., Tuchman, M., and Caldovic, L. 2007. A novel bifunctional N-acetylglutamate synthase-kinase from Xanthomonas campestris that is closely related to mammalian N-acetylglutamate synthase. BMC Biochem. 8: 4. doi: 10.1186/1471-2091-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Morizono, H., Ha, Y., Aoyagi, M., Tuchman, M., and Allewell, N.M. 1998. 1.85 Å resolution crystal structure of human ornithine transcarbamylase complexed with N-phosphonacetyl-L-ornithine. J. Biol. Chem. 273: 34247–34254. [DOI] [PubMed] [Google Scholar]
- Shi D., Morizono, H., Yu, X., Tong, L., Allewell, N.M., and Tuchman, M. 2001. Human ornithine transcarbamylase: Crystallographic insights into substrate recognition and conformational changes. Biochem. J. 354: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Gallegos, R., DePonte III, J., Morizono, H., Yu, X., Allewell, N.M., Malamy, M., and Tuchman, M. 2002. Crystal structure of a transcarbamoylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 Å resolution. J. Mol. Biol. 320: 899–908. [DOI] [PubMed] [Google Scholar]
- Shi D., Morizono, H., Yu, X., Roth, L., Caldovic, L., Allewell, N.M., Malamy, M.H., and Tuchman, M. 2005a. Crystal structure of N-acetylornithine transcarbamoylase from Xanthomonas campestris: A novel enzyme in a new arginine biosynthetic pathway found in several eubacteria. J. Biol. Chem. 280: 14366–14369. [DOI] [PubMed] [Google Scholar]
- Shi D., Yu, X., Roth, L., Morizono, H., Hathout, Y., Allewell, N.M., and Tuchman, M. 2005b. Expression, purification, crystallization and preliminary X-ray crystallographic studies of a novel acetylcitrulline deacetylase from Xanthomonas campestris . Acta Crystallogr. F61: 676–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Morizono, H., Cabrera-Luque, J., Yu, X., Roth, L., Malamy, M.H., Allewell, N.M., and Tuchman, M. 2006a. Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis . J. Biol. Chem. 281: 20623–20631. [DOI] [PubMed] [Google Scholar]
- Shi D., Yu, X., Roth, L., Morizono, H., Tuchman, M., and Allewell, N.M. 2006b. Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogues imply mechanisms for substrate binding and catalysis. Proteins 64: 532–542. [DOI] [PubMed] [Google Scholar]
- Shi D., Yu, X., Roth, L., Tuchman, M., and Allewell, N.M. 2007. Structure of a novel N-acetyl-L-citrulline deacetylase from Xanthomonas campestris . Biophys. Chem. 126: 86–93. [DOI] [PubMed] [Google Scholar]
- Slocum R.D. 2005. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 43: 729–745. [DOI] [PubMed] [Google Scholar]
- Virnau P., Mirny, L.A., and Kardar, M. 2006. Intricate knots in proteins: Function and evolution. PLoS Comput. Biol. 2: e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]