Abstract
Improved ways to cleave peptide chains at engineered sites easily and specifically would form useful tools for biochemical research. Uses of such methods include the activation or inactivation of enzymes or the removal of tags for enhancement of recombinant protein expression or tags used for purification of recombinant proteins. In this work we show by gel electrophoresis and mass spectroscopy that salts of Co(II) and Cu(II) can be used to cleave fusion proteins specifically at sites where sequences of His residues have been introduced by protein engineering. The His residues could be either consecutive or spaced with other amino acids in between. The cleavage reaction required the presence of low concentrations of ascorbate and in the case of Cu(II) also hydrogen peroxide. The amount of metal ions required for cleavage was very low; in the case of Cu(II) only one to two molar equivalents of Cu(II) to protein was required. In the case of Co(II), 10 molar equivalents gave optimal cleavage. The reaction occurred within minutes, at a wide pH range, and efficiently at temperatures ranging from 0°C to 70°C. The work described here can also have implications for understanding protein stability in vitro and in vivo.
Keywords: protein cleavage, His tag, artificial protease, protein stability, Co(II), Cu(II)
Protein engineering allows changing the structure and thus the function of proteins in several ways. Proteins can be fused together to combine functionalities, for example, enzymatic activity and binding to specific surfaces or targets. One widely used application of protein engineering is to add tags to recombinant proteins that allow specific and easy purification. Examples of widely used tags are the His tag and glutathione S-transferase (Nilsson et al. 1997; Waugh 2005). A disadvantage that is often encountered is that it is often difficult to remove these tags after the purification step to ensure that they do not interfere with subsequent functionality and use. Most proteases lack the sequence specificity needed to cleave only at one desired position. This problem can be solved, for example, by using proteases that show high sequence specificity, such as blood clotting factors. However, large-scale use of these proteases is not feasible because of their high cost and often poor function (Arnau et al. 2006).
An easy way of cleaving proteins specifically at desired positions would be useful in many cases. It would be desirable that protein engineering could be used to insert the cleavage sequence, and that the cleavage would not be dependent on large molecules that could be sterically hindered to access the cleavage position. Such a cleavage system would allow the removal of purification or expression enhancement tags but could also be used as a tool for several other types of applications, for example, to inactivate or activate proteins or to study structure–function relations. Inactivation could be achieved by cleaving of a peptide sequence at a functional position, and activation by removing an added protein domain that blocked an enzyme from functioning.
Metal ion-dependent cleavage of proteins (and DNA) shows promise for the above-mentioned applications. The use of chelates of different metals has been widely studied for this purpose (Hegg and Burstyn 1998; Polzin and Burstyn 2001; Grant and Kassai 2006). Many complexes of metals have been studied that give significantly increased cleavage over the background reaction. The strategy typically relies on various chelates of metal ions of metals such as Ce, Co, Cu, Mo, Ni, Pd, Pt, Zn, and Zr, and can depend on either oxidative cleavage or hydrolysis. Several strategies depend on careful analysis of coordination chemistry and mimicking natural metalloproteases. Only a few such synthetic systems have, however, found practical applications in biochemical research. One successful example is the use of Fe2+ in combination with chelating molecules (Rana and Meares 1991b; Ermacora et al. 1992; Platis et al. 1993). The system is based on tethering the Fe2+ to a protein by a dual-function molecule that on the one hand binds Fe2+ by an EDTA-like chelate, and on the other hand has a reactive group for binding to proteins, for example, through coupling with free sulfhydryl groups. An oxidative cleavage reaction occurs when ascorbate and hydrogen peroxide is added. In the reaction the peptide backbone is cleaved, but only at positions very close to the tethered Fe2+. In this way, the reaction can be used to probe structure–function relations in proteins. The reaction is hydrolytic and proceeds through activated oxygen species coordinated by Fe2+ and requiring the presence of ascorbate and hydrogen peroxide (Rana and Meares 1991a).
In this work, we show that chemical cleavage of a peptide sequence can be obtained at a predetermined site. This cleavage does not depend on separate chelating molecules but relies on the use of free metal salts. For chelating the metal ions, His residues were engineered into the recombinant proteins, because the imidazole group of His is known to coordinate efficiently with soft (or borderline) metal ions (Ueda et al. 2003). Typically six or more His residues were used as a binding site for the metal ions. As with the Fe2+ chelates described above, the reaction was driven by the addition of ascorbate and optionally hydrogen peroxide. However, in this setup, addition of Fe2+ did not cause any reaction, but very clear reactions were obtained by the addition of Cu2+ or Co2+.
Results
An initial serendipitous finding that Cu(II) salts together with ascorbate can lead to the cleavage of engineered proteins containing multiple His residues encouraged a closer investigation of the phenomenon. In a systematic study, the effects of protein structure and other factors were studied. We used a simple setup in which two proteins were linked to each other by a linker sequence. Several constructs were made in which the number of His residues or the spacing of these was varied (Table 1).
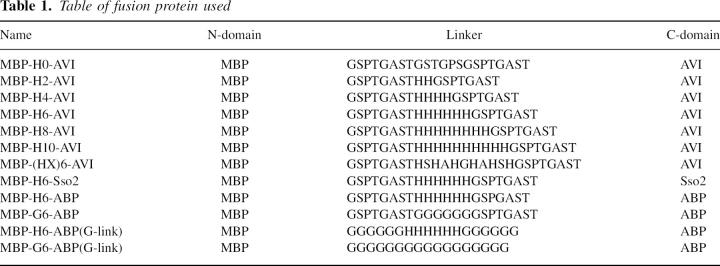
Table 1.
Table of fusion protein used
Cleavage conditions
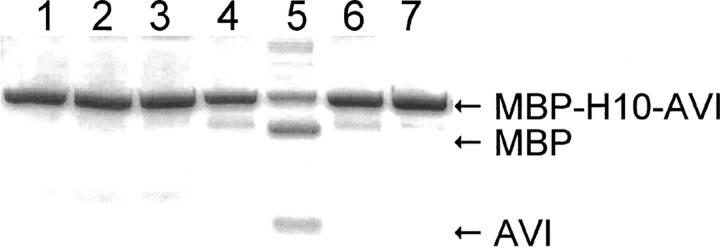
A typical cleavage experiment was performed by first mixing the protein, buffer, and metal salt. The ascorbate and hydrogen peroxide were mixed in a separate tube and then added to the protein/metal salt solution. To stop the reaction, a large excess of EDTA was added. The mixture was then typically analyzed by SDS-PAGE. For the SDS-PAGE, mercaptoethanol was added (in the case of the streptavidin fusions the samples were heated). In Figure 1, the result of a basic experimental setup is shown. The proteins used in this experiment were MBP-H10-AVI (10 His in the linker) and MBP-H0-AVI (no His in the linker) (see Table 1 and Materials and Methods). They were incubated with CoCl2, ascorbate, and hydrogen peroxide (3.3. μM protein, 33 μM CoCl2, 4.6 mM ascorbate, and 0.35 mM H2O2) at 21°C for 60 min. The results (Fig. 1) show that cleavage happens only when the protein with an H10 linker is mixed with CoCl2, ascorbate, and hydrogen peroxide. If the linker contains no His residues (H0) no cleavage occurs. Neither did we observe cleavage if CoCl2, or ascorbate, or hydrogen peroxide was left out (for experiments on the effect of concentrations, see below). However, it could often be observed (lane 4) that addition of ascorbate and hydrogen peroxide without metal ions did cause some minor cleavage of the fusion protein, which most probably was caused by contaminating metal ions in the reaction mixture, since such reactions were easily prevented by addition of EDTA. Thus, it was concluded that normal trace amounts of metal (probably copper) ions in distilled water and/or used reagents is enough to cause background cleavage. Addition of EDTA simultaneously as CoCl2 effectively hindered the cleavage reaction. This result also shows that no cleavage occurred during sample preparation for SDS-PAGE or during electrophoresis. Furthermore, results from size exclusion chromatography and mass spectroscopy (see below) also confirmed that the cleavage occurred before the SDS-PAGE sample preparation and verified that the preparation of samples for SDS-PAGE did not affect the cleavage.
Figure 1.
Cleavage experiment of the fusion proteins MBP-H10-AVI and MBP-H0-AVI with CoCl2. Control protein MBP-H0-AVI in buffer (lane 1), with ascorbate and hydrogen peroxide (lane 2), and with ascorbate hydrogen peroxide and CoCl2 (lane 3). MBP-H10-AVI protein in only buffer with ascorbate and hydrogen peroxide (lane 4) and with CoCl2, ascorbate, and hydrogen peroxide (lane 5). In lane 6, the protein was treated as in lane 5 but with EDTA added prior to the CoCl2. In lane 7 the fusion protein was incubated with CoCl2 but without ascorbate or hydrogen peroxide.
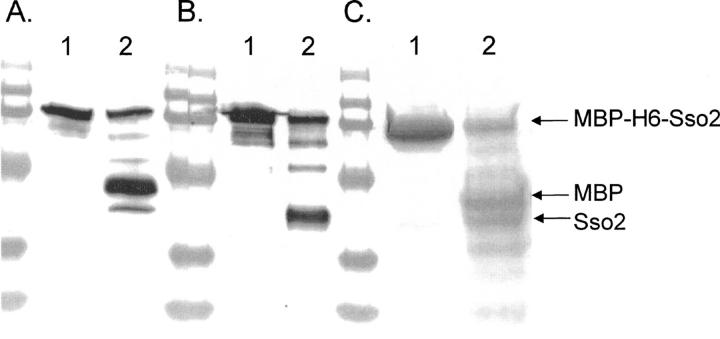
The cleavage reaction was confirmed using different types of fusion proteins with similar results (see Table 1). In Figure 2, we show the results for a protein containing the yeast cytosolic protein Sso2. The MBP-H6-Sso2 fusion protein (4.6 μM) was incubated with one molar equivalent amount of CuCl2, and the cleaved proteins were separated on a 12.5% SDS-polyacrylamide gel. For Western blot analysis the reaction mixture was diluted 50 times, and the fragments produced in the cleavage reaction were identified by specific antibodies. The antibody raised against maltose binding protein (MBP) (Fig. 2A), identified the separated MBP domain as well as the uncleaved fusion protein. Anti-Sso2 detected, in addition to the uncleaved fusion protein, the Sso2 domain of the cleaved fusion protein (Fig. 2B). In Figure 2C, the separated domains are shown in a Coomassie-stained gel. In lane 1, in which no ascorbate, hydrogen peroxide, or metal ions were used, the fusion protein remained intact, while the result of a cleavage reaction shown in lane 2 clearly visualizes the split of the fusion protein into its separate units.
Figure 2.
Coomassie and Western blot analysis of the cleavage of the fusion protein MBP-H6-Sso2. The gel has been blotted using anti-MBP (A) and anti-Sso2 (B), respectively, and a Coomassie-stained gel is shown in C. In all gels the samples in lane 1 are untreated controls and in lane 2 the same protein was incubated with one molar equivalent of CuCl2, 9 mM ascorbate, and 0.7 mM H2O2 at 21°C for 60 min. The result shows that the treatment splits the fusion protein into its two parts, MBP and Sso2.
Analysis of fragments using MALDI-TOF
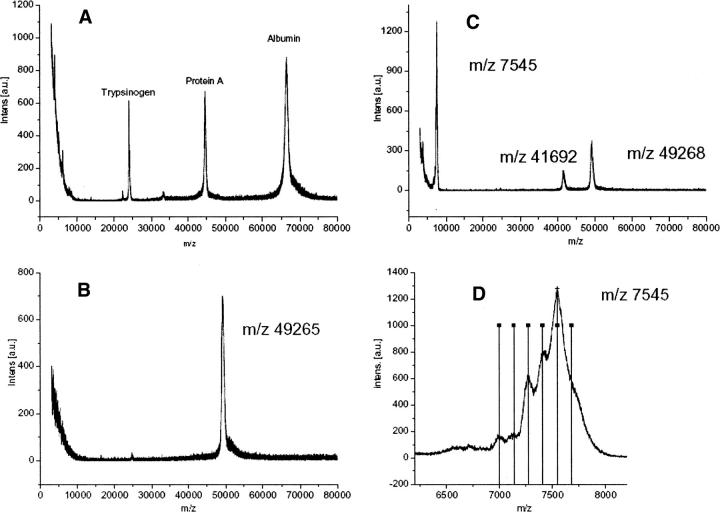
The fusion protein MBP-H6-ABP was subjected to cleavage reaction (10 molar equivalents of CoCl2, 60 min, 21°C, 4.6 mM ascorbate, 0.4 mM hydrogen peroxide) and the reaction mixture was analyzed by MALDI-TOF without any purification (Fig. 3). Mass standards are show in Figure 3A. The negative control without CoCl2 shows a single peak at 49,268 m/z (Fig. 3B). The sample subjected to cleavage showed two additional peaks at 41,692 m/z and 7545 m/z (Fig. 3C). The calculated mass for MBP-H6-ABP is 49,220 Da, while the mass for the MBP with half of the linker with three His residues is 41,785 Da and the mass for the ABP part with three His residues is 7453 Da. These measured mass peaks correspond with the expected fragments within typical error limits. The mass standards are shown here to demonstrate that peaks obtained for the standards and the samples are very similar and typical for the mass spectrometer used. There is no obvious peak broadening. The relative intensities of the peaks do not correspond to the abundance of each fragment but more probably reflect their tendency to ionize. Near the main ABP peak some additional peaks and shoulders can be noted (Fig. 3D). When lines are drawn with a spacing of 137.1 (the mass of a His residue), around the peak top, it can be noted that these lines coincide with the additional shoulders and peaks. Therefore, it seems that the cleavage can occur at the position of any His residue, although preferentially at a specific position.
Figure 3.
MALDI-TOF analysis of the cleavage of the MBP-H6-ABP fusion protein. The cleavage was performed using CoCl2, ascorbate and hydrogen peroxide, and the mass spectroscopy was performed directly on the sample without any intermediate purification. (A) Standard proteins (trypsinogen, 23,982 Da; protein A, 44,613 Da; albumin 66,431 Da). (B) Control sample without added CoCl2. (C) Reaction mixture where MBP, ABP, and the noncleaved protein peaks can be identified. (D) Close-up of the ABP peak shows additional peaks with a spacing corresponding to the mass of a His-residue (vertical lines drawn 137.1 Da apart).
In another experiment, the cleavage products from the MBP-H6-Sso2 fusion protein were analyzed by first separating the intact fusion protein and the released MBP and Sso2 by preparative size exclusion chromatography (data not shown). Cleavage was performed using one molar equivalent of CuSO4, 4.6 mM ascorbate, 0.25 mM hydrogen peroxide, and 90 mM Na actetate buffer at pH 5. Fractions were identified by SDS-PAGE and subjected to MALDI-TOF mass spectroscopy and Edman sequencing. The N-terminal sequencing of the MBP part of cleaved fusion proteins and of pure MBP-H6-Sso2 fusion protein showed the expected sequences. The N-terminal sequence was MKIEEGK in the MBP part of cleaved MBP-H6-AVI, MBP-H8-AVI, and MBP-H6-Sso2 fusion proteins, which is identical to the N-terminal sequence of the sequenced Eschericha coli transformants of the fusion proteins. No N-terminal sequence could be obtained from the Sso2 fragment, however, which is an indication that the N-terminal residue was modified. The molecular weights of the cleaved protein fragments as well as for the pure fusion protein were determined by MALDI-TOF mass spectrometry. The measured mass of the intact fusion protein was 73,164 m/z, 41,946 m/z, and 31,174 m/z for MBP and Sso2, respectively. The sum of the fragments differs only by 41 from the measured mass of the intact fusion protein, which is within the limits of the technique.
MBP-H6-AVI was subjected to a similar procedure, giving a mass for the MBP of 41,988 m/z. However, streptavidin could not be isolated and analyzed, probably because streptavidin forms tetramers where intact fusion protein is mixed with cleaved streptavidin incorporated into heterotetramers under nondenaturing conditions.
Comparison of different metal ions
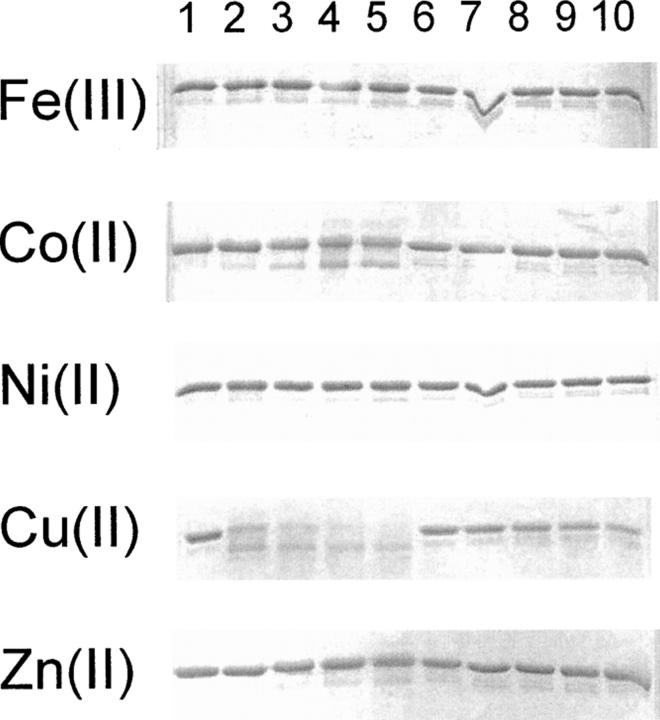
The effect of using different metal ions was compared by studying the cleavage of fusion proteins in the presence of FeCl3, CoCl2, NiCl2, CuCl2, and ZnCl2. These metals are the group 8–12 transition metals from period 4. In Figure 4, the cleavage of the fusion proteins MBP-H6-ABP or MBP-G6-ABP is shown. The reactions were performed in 50 mM ammonium carbonate buffer at pH 7.5 with 20 mM ascorbate and 1.5 mM hydrogen peroxide at 30°C for 15 min. The concentration of metal salts was either 1 μM, 3 μM, 6 μM, or 10 μM corresponding to 0.3×, 1×, 2×, and 3.3× molar ratios of metal ion to protein. With FeCl3, NiCl2, and ZnCl2 no effect of the treatment can be seen. Some effect can be seen with CoCl2, and clearly the largest effect is with CuCl2 (lanes 2–5). The control protein containing no His residues in the linker is not affected by the treatment (lanes 7–10), except for those with the highest CuCl2 concentration where the band seems to fade (lane 10). The excess of CuCl2 in these experiments seems to cause a nonspecific degradation. The effect of different metal ions was the same regardless of fusion protein (data not shown).
Figure 4.
Incubations of the fusion proteins MBP-H6-ABP and MBP-G6-ABP with different metal ions in the presence of ascorbate and hydrogen peroxide. Lanes 1–5 are with the His-containing linker (H6). In lane 1 no metal was added. In lanes 2–5 the metal concentration was 0.3, 1, 2, and 3.3 molar equivalents of metal to protein. Lanes 6–10 are with the Gly containing linker (G6) and the same concentrations of metal ions as for MBP-H6-ABP in lanes 1–5.
Effect of metal ion concentration
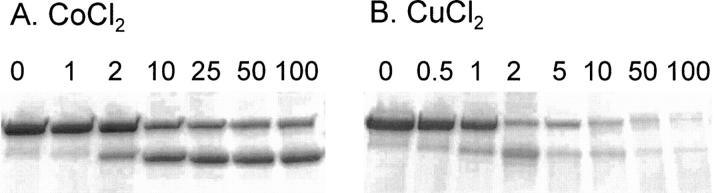
The effect of metal ion concentration on peptide cleavage was analyzed with CoCl2 and CuCl2. The CoCl2 ion concentration was increased from 1 to 100 molar equivalents of metal to protein (shown for MBP-H10-AVI in Fig. 5) (60 min, 21°C). At one molar equivalent concentration of CoCl2 only little cleavage took place, but already with double the concentration a clear cleavage occurred. The cleavage increased by increasing the CoCl2 concentration up to 10 times, but further increasing up to 100 times the molar ratio of CoCl2 to protein did not result in a more extensive reaction. The cleavage with CuCl2 differed from that of CoCl2. At a CuCl2 concentration of 0.5 molar equivalents some cleavage could be observed. When the CuCl2 concentration was increased, the amount of cleavage product increased but only up to 2 molar equivalents. At CuCl2 concentration above 2 molar equivalents, the band corresponding to fusion protein decreased in intensity but so did the bands corresponding to the cleavage products. Thus, CuCl2 seemed to cause nonspecific degradation at these concentrations.
Figure 5.
Effect of metal ion concentration on cleavage of MBP-H10-AVI. The fusion protein was incubated with increasing amounts of cobalt (A) or copper (B). The incubations were performed in the presence of ascorbate and H2O2. The molar equivalents of CoCl2 (A) and CuCl2 (B) added to protein are shown above each lane.
Reaction conditions
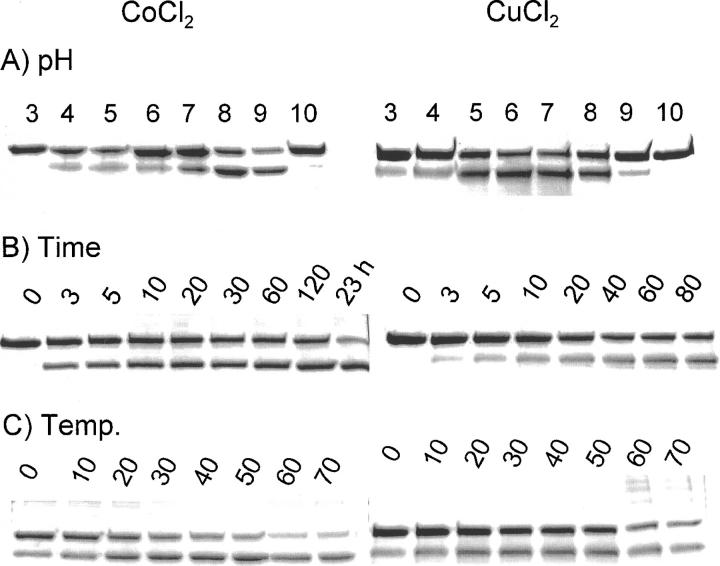
The effects of pH, buffer, reaction time, and temperature on the peptide bond cleavage with CoCl2 and CuCl2 were studied in several experimental series shown in Figure 6. The effects of pH and buffer on the cleavage of MBP-H10-AVI with CoCl2 or MBP-H6-AVI with CuCl2 are shown in Figure 6. The fusion protein was incubated with one molar equivalent of CuCl2 or 10 molar equivalents of CoCl2 in glycine-HCl buffer (pH 3), Na-acetate buffer (pH 4 and 5), Na-citrate buffer (pH 6), Hepes buffer (pH 7), Tris-HCl buffer (pH 8), and glycine-NaOH buffer (pH 9 and 10). The final concentration of the buffers was 86 mM. In the reactions in which CoCl2 was used, the most effective cleavage was at pH 8. For the reactions with CuCl2 ions, there was a broader optimum. The role of the nature of the buffer at pH 7 was studied by performing the cleavage reaction in ammonium carbonate, Tris, Hepes, and phosphate buffer. No significant differences could be seen between the different buffers (data not shown). Experiments with different His-containing linkers gave the same results (data not shown).
Figure 6.
Effect of pH, time, and temperature on cleavage of fusion protein with CoCl2 and CuCl2. MBP-H10-AVI was incubated with 10 molar equivalents of CoCl2 and MBP-H6-AVI with an equimolar amount of CuCl2 at standard reaction conditions. (A) Effect of pH on cleavage. The fusion proteins were incubated at pH values varying between 3 and 10 for 60 min at room temperature. The pH values of the cleavage reactions are shown above the lanes. The concentration of ascorbate and H2O2 were 4.6 mM and 0.35 mM, respectively. The incubations with copper were performed in 50 mM Na-acetate at pH 5 in the presence of 171 mM NaCl, and the incubations with cobalt were performed in water. (B) Effect of time on cleavage of fusion proteins under standard conditions for different times. The incubation times in minutes are shown above the lanes. (C) Effect of temperature on cleavage. The fusion proteins were incubated at different temperatures for 20 min (CuCl2) or 60 min (CoCl2), with 4.6 mM ascorbate and 0.35 mM hydrogen peroxide. The incubation temperatures in °C are shown above the lanes.
We also studied the effect of the reaction time on the peptide bond cleavage by incubating the fusion proteins with metal ions, ascorbate, and hydrogen peroxide for various reaction times. The rate of cleavage with CuCl2 or CoCl2 ions in the presence of ascorbate and hydrogen peroxide is fast. Cleavage products can be seen already within a few minutes of incubation (Fig. 6B), and the reaction proceeded only slowly after that.
The effect of the reaction temperature on peptide cleavage was investigated by incubating the fusion proteins in the presence of CoCl2 or CuCl2, ascorbate, and hydrogen peroxide. The effect of temperature was very small although the reaction was slightly slower at low temperatures than in higher reaction temperatures. Interestingly, high temperatures and CuCl2 resulted in some additional high-molecular-weight bands, which could be a result of a polymerization reaction with the protein.
Effect of ascorbate and hydrogen peroxide concentrations
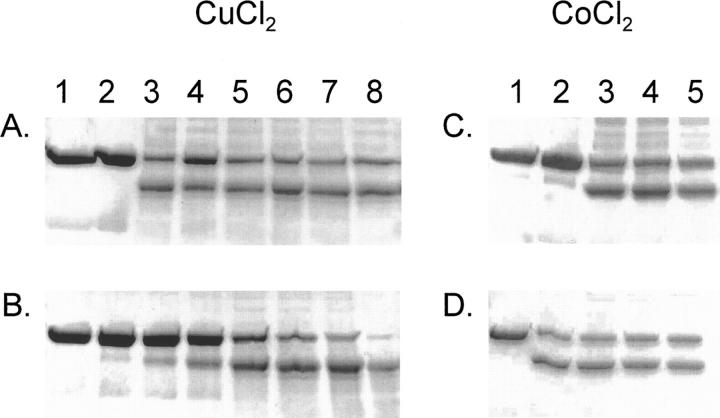
The effect of ascorbate and hydrogen peroxide on peptide cleavage was studied by varying their concentrations. In Figure 7, the results for cleaving MBP-H8-AVI in the presence of one molar equivalent of CuCl2 (60 min, 21°C) are shown. If neither ascorbate nor hydrogen peroxide was present, no reaction occurred. If only hydrogen peroxide but not ascorbate was present, no reaction occurred (Fig. 7A), and if ascorbate is present but not hydrogen peroxide, only a very slight reaction occurred (Fig. 7B). Already at concentrations as low as 1 mM ascorbate in the presence of 0.7 mM hydrogen peroxide, there was a clear reaction (Fig. 7A). The reaction was not significantly affected by increasing the concentration (up to 41 mM is shown in Fig. 7A). In Figure 7B, the effect of increasing the hydrogen peroxide concentration is shown. With ascorbate present at 9 mM, there was a clear increase in reaction efficiency with higher concentrations of hydrogen peroxide, although a minor reaction was observable even without hydrogen peroxide.
Figure 7.
Effect of concentration of ascorbate and H2O2 in the linker cleavage. (A, B) The cleavage of MBP-H8-AVI with a molar equivalent of CuCl2 and various amounts of ascorbate and hydrogen peroxide is shown. (A) Lane 1, only protein; lane 2, hydrogen peroxide at 0.7 mM. In lanes 3–8 the hydrogen peroxide concentration was kept constant at 0.7 mM and ascorbate concentration was increased as follows: 1.14 mM (lane 3), 2.3 mM (lane 4), 4.6 mM (lane 5), 9.1 mM (lane 6), 18.3 mM (lane 7), and 41.4 mM (lane 8). (B) Lane 1, only protein; lane 2, ascorbate at 9 mM. In lanes 3–8 the ascorbate concentration was kept at 9 mM and the hydrogen peroxide concentration was increased as follows: 0.07 mM (lane 3), 0.17 mM (lane 4), 0.7 mM (lane 5), 1.4 mM (lane 6), 2.7 mM (lane 7), and 5.5 mM (lane 8). (C, D) The cleavage of MBP-H10-AVI with 10 molar equivalents of CoCl2 is shown. (C) Lane 1, only protein; lane 2, hydrogen peroxide at 0.4 mM. In lanes 3–5, the hydrogen peroxide concentration was kept constant at 0.4 mM and ascorbate concentration was increased as follows: 35 μM (lane 3), 70 μM (lane 4), and 0.14 mM (lane 5). (D) Lane 1, only protein; lane 2, ascorbate at 4.6 mM. In lanes 3–8 the ascorbate concentration was kept at 4.6 mM and the hydrogen peroxide concentration was increased as follows: 0.09 mM (lane 3), 0.17 mM (lane 4), and 0.35 mM (lane 5).
In Figure 7, C and D, results are shown for cleaving MBP-H10-AVI using 10 molar equivalents of CoCl2 (60 min, 21°C) in the presence of varying concentrations of hydrogen peroxide and ascorbate. If neither hydrogen peroxide nor ascorbate was present, no reaction could be seen. However, in contrast to CuCl2, ascorbate without hydrogen peroxide caused a reaction although hydrogen peroxide alone did not result in any reaction. The effect of hydrogen peroxide was altogether much smaller for the CoCl2-mediated reaction than for the CuCl2-mediated one. With CoCl2 there was even a clear reaction with ascorbate present (no hydrogen peroxide) at 35 μM, which equals the molar amount of CoCl2 (data not shown).
In typical reactions, some intact fusion protein always remained. A complete cleavage of the fusion protein was not possible even by increasing the amount of metal salt or ascorbate and hydrogen peroxide. To get a more complete reaction we tested the reaction in cycles with fresh reagent and repeating the incubation several times. In one example, six Eppendorf tubes with 1.6 μM MBP-H10-AVI were incubated with a 10 molar equivalent of CoCl2 and 2.3 mM ascorbate at room temperature. After a 30-min incubation time, the reaction in one of the tubes was stopped with an excess of EDTA and a new dose of cobalt and ascorbate was added to the rest of the tubes. This procedure was repeated another five times so that the final cobalt concentration in the last tube was 60 times the concentration of the fusion protein and the ascorbate concentration 13.7 mM. The samples were then analyzed by SDS-PAGE (data not shown). In this experiment, the band corresponding to the uncleaved fusion protein and the product bands increased for each cycle. However, even in the last tube a clear band corresponding to the uncleaved protein could be seen. Thus, the cleavage efficiency can be improved up to a certain degree by repeated additions of metal ions and ascorbate.
Effect of the nature of the linker
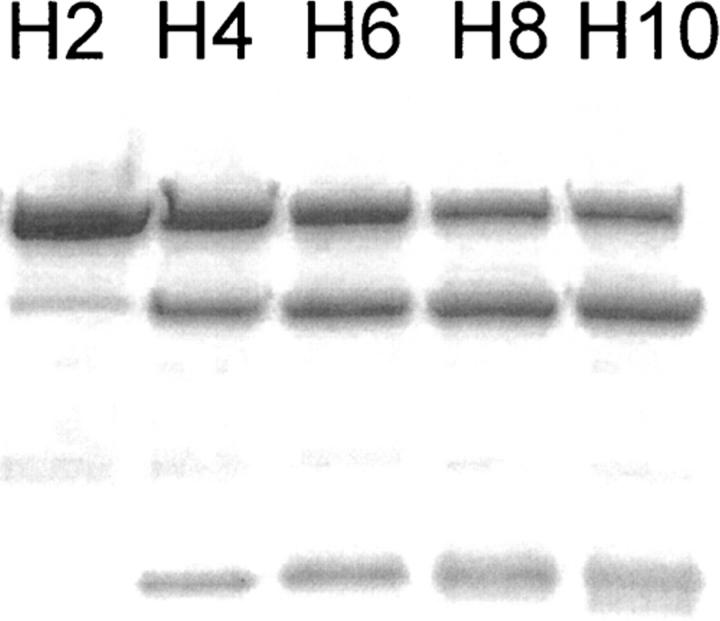
The role of the nature of the linker was studied by allowing proteins with different numbers of His residues to react under identical conditions (Fig. 8). Cleavage reactions were performed on MBP-Hn-AVI fusion proteins, where the subscript n indicates linkers containing 2, 4, 6, 8, or 10 His residues. The reactions were performed with 10 molar equivalents of CoCl2 to protein (60 min, 21°C). The results showed that the length of the linker clearly affects the cleavage result. Some minor cleavage products could already be observed if the linker of the fusion protein contained two His residues (MBP-H2-AVI). There was a slight but clear increase in reactivity with an increasing number of His residues. No cleavage occurred in the linker without His residues (MBP-H0-AVI) as shown in Figure 1, lane 3, or if the His in the linker were replaced by Gly (data not shown). Interestingly, inserting other amino acids in between His-residues (MBP-HX6-AVI) (see Table 1) resulted in a fusion protein that was cleaved as efficiently as the one with six consecutive His-residues.
Figure 8.
Effect of number of His residues in linker chain in MBP-Hn-AVI (n indicates the number of His residues). The reaction was with Co(II). Fusion protein (3.3 μM) was cleaved with 10 molar equivalents of CoCl2, 4.6 mM ascorbate, and 0.35 mM H2O2 in water for 60 min at room temperature. The number of His residues in the linker is shown above the lanes.
Functionality of domains after cleavage
The functionality of the cleaved domain was studied by analyzing how streptavidin bound to a biotin gel after the cleavage reaction. The fusion protein MBP-(HX)6-AVI (3.2 μM in 70 μL) was cleaved with one molar equivalent of CuCl2 under standard reaction conditions. The quenched reaction mixture was incubated with biotin gel (Biotin D-gel Immobilized, Pierce) prewashed with 0.2 M NaCl, 50 mM Tris-HCl at pH 7.5. The cleaved streptavidin was allowed to bind to the gel for 60 min at room temperature. After centrifugation the supernatant was analyzed by SDS-PAGE under reducing conditions. The results show that the uncleaved fusion protein as well as the cleaved streptavidin bound to the biotin gel (data not shown). As a negative control, glutathione beads were used (Glutathione Sepharose 4B, GE Healthcare), and they did not absorb any protein. Thus, the cleavage reaction did not affect the protein's ability to bind its ligand.
Discussion
In this work we have shown that protein engineering can be used to introduce sites into proteins where metal ions can bind and participate in a reaction that, under suitable conditions, results in the cleavage of the peptide backbone of the protein. In our experimental setup, the metal binding site was engineered into a peptide linker region that connected two otherwise independent proteins. After cleavage of the linker the two proteins were separated into individual chains, which were detected using electrophoresis (SDS-PAGE), chromatography, and mass spectroscopy.
The strategy for constructing metal binding sites was to add His residues into the linker region. We first demonstrated that cleavage of the connecting linker occurs, and that it is dependent on (1) the number of His residues, (2) the presence and nature of metal ions, and (3) the presence of redox agents, such as ascorbate and hydrogen peroxide. The effects of temperature, buffer composition, and pH on the kinetics of the reaction were then studied.
The typical cleavage reaction can be seen (Fig. 1) when a fusion protein with MBP linked to streptavidin by a linker that contains 10 consecutive His residues was mixed with a 10× molar excess of Co2+ and low concentrations of ascorbate and hydrogen peroxide for 60 min at room temperature. Adding EDTA was an efficient way to stop or inhibit the reaction. Treating a fusion protein without His residues in the linker or with Gly instead of His in the same way does not result in any detectable amount of cleavage. Sometimes a slight cleavage of His containing linkers could be seen even without adding metal. This was probably due to the presence of traces of metal ions from the purification steps or elsewhere. However, such cleavage could be inhibited by the addition of EDTA.
Having established the occurrence of the cleavage reaction, we proceeded to look more closely at parameters affecting reaction rates. The nature and amount of metal ion, the character of the linker, and reducing agents all affected the rate of reaction, as did temperature and pH but to a lesser extent.
There was a clear requirement for metal ions for the cleavage reactions to occur. We found that Cu2+ and Co2+ ions were the most reactive, with Cu2+ clearly causing a more extensive reaction. It is remarkable that only one molar equivalent of Cu2+ caused extensive cleavage of the fusion protein. For Co2+ a 10× excess was needed for an optimal yield (Fig. 5). A clear difference between the effect of Cu2+ and Co2+ ions was also that an excess of Cu2+ ions caused all the protein bands to decrease in intensity, indicating some kind of overall protein degradation. This overall degradation was not observed with Co2+ ions.
After a typical cleavage reaction some fusion protein typically remained uncleaved. Because each fusion protein potentially contains several potential binding sites within the His residues, it could be thought that an uneven distribution of metal ions between the protein molecules left some molecules without metal and hence remained intact. Surprisingly, however, increasing the amount of metal did not result in a complete cleavage, but some intact protein always remained. Repeated incubations with fresh reagents did cause the reaction to proceed, but a total cleavage of the initial material was difficult to achieve.
Salts of other metals tested, Zn, Ni, and Fe, did not cause any measurable cleavage of the proteins under the conditions tested. Fe is known to cause oxidative cleavage of proteins (see below) (Grant and Kassai 2006), but being a hard ion, it does not have a high affinity to His residues. This supports the idea that a close proximity of the metal and protein is needed for cleavage to occur under the conditions described here.
The number of His residues in the linker region also clearly affected the cleavage reaction. Already two adjacent residues were sufficient for a noticeable reaction to occur, but increasing the number of His residues significantly increased the efficiency (Fig. 8). However, adding more than six residues only slightly improved the cleavage, as was demonstrated with fusion proteins containing eight or ten His residues. Linkers containing six His residues, but with one Gly or Ser in between each His residue, also behaved very similarly as the linker with six consecutive His residues, showing that the His residues do not have to be consecutive. The effect of linker length and composition was the same for cleavage with both CoCl2 and CuCl2. In this work we did not systematically study how side chains other than His would promote cleavage. However, it is likely that other types of side chains such as Cys or Trp also could have functioned similarly. The use of Cys was avoided because the reactivity of free sulfhydryl groups can cause experimental problems such as aggregation in protein production.
The cleavage reaction was very fast and occurred efficiently at a temperature of 4°C with only a few minutes of incubation. The pH dependence showed an optimum around 7–8. The presence of ascorbate was shown to be essential for cleavage to occur. Hydrogen peroxide increased the effect even at low concentration but was not essential. The CoCl2-mediated reaction proceeded even without hydrogen peroxide, having a strict requirement only for ascorbate.
There is a large interest in different metal-mediated cleavage reactions, and there are many reports on the subject (Grant and Kassai 2006). However, several findings suggest that the underlying mechanisms differ significantly. A work that is conceptually close to the current one was described by Humphreys et al. (1999, 2000). In that work it was found that proteins containing the sequence DKTH could be cleaved in the presence of Cu2+, and to a small extent by Ni2+. However, the mechanisms of cleavage seem to be very different. Cleavage occurred only at pH values over 7, and efficient cleavage only above pH 8. The temperature required was over 50°C, and 50% cleavage was obtained only after over 10 h. Most significantly, neither ascorbate nor hydrogen peroxide affected the cleavage rate. In view of the present work, this result is surprising. One possible reason for this difference is the affinity for the binding sequences for metals are different. In our work, longer sequences of His residues were used, giving a stronger affinity. Whereas only one molar equivalent of Cu was needed in the present work, several 100-fold was used by Humphreys et al. (1999, 2000). Results similar to those of Humphreys et al. (1999, 2000) have been obtained in another work (Allen and Campbell 1996). There it was noted that Ser-His and Thr-His are cleaved slowly by Cu2+ with a half-life of several hours at over 60°C and pH 8. Also in this work cleavage did not depend on added ascorbate.
Much work has been done on metal-mediated cleavage reactions that involve tethering the metal ion by synthetic chelating molecules (Rana and Meares 1991b; Ermacora et al. 1992). The metal tether was constructed so that it had one functionality binding a metal ion, typically a modified EDTA-based chelator used to bind Fe3+ ions. Another part of the tether was functionalized to bind to proteins through Cys residues. Using this type of molecule, it was possible to probe protein–protein interactions because the iron-mediated cleavage only occurred at very short range, and it could be concluded that the site of cleavage must be near the bound Fe ion. Also this iron-mediated cleavage required the presence of ascorbate and hydrogen peroxide. The reaction occurred quickly at pH 7. The Fe–chelate cleavage and the reaction described here are similar in that they both depend on ascorbate and hydrogen peroxide. In our work, the Fe ions did not bind to the proteins and therefore no effect of Fe ions was observed.
In another study by Kim et al. (1985) it was found that iron caused cleavage of some proteins without chelate-mediated localization. In that work it was found that a yeast–protein was degraded in the presence of Fe3+ and dithiothreitol (DTT), but the degradation was inhibited in the presence of EDTA. Since some reaction could also be detected in the presence of copper and ascorbate, and the reaction occurred at 30°C, pH 7, it seems that its underlying mechanism is similar to the one occurring in the current work. However, the reaction times required were much longer and the metal ion concentration higher, indicating that the engineered sites in the current work were able to bind the metal ions more efficiently.
Although cleavage reactions involving chelates of Fe3+/Fe2+, Ni2+, and Cu2+ have been described in the literature, there are not many reports on the use of cobalt ions. Chelates of cobalt ions have been used to achieve hydrolysis of some peptides and proteins (Kumar et al. 2000; Jeon et al. 2003; Jitsukawa et al. 2006). The reactions were very sensitive to the Co3+ chelate structure and did not depend on the use of ascorbate or hydrogen peroxide. The reaction rates were measured in tens of hours and required elevated temperatures and pH. The chelate concentrations used were typically 100-fold higher than the protein concentrations. In these reports, the cleavage sites in the peptide chains were not purposefully designed or engineered.
It is clear from the experimental data that the reaction depends on a close binding of the metal ion and the peptide chain. In principle, two types of reaction could be possible: either a radical reaction through reactive oxygen species or a hydrolysis type of reaction (Grant and Kassai 2006). The dependence of ascorbate and hydrogen peroxide indicates that a radical reaction is responsible for the observed cleavage. Bateman Jr. et al. (1985) suggested that hydroxyl radicals can cleave peptide bonds by a mechanism initiated by the abstraction of the hydrogen of the α-carbon in the peptide backbone.
Hydroxyl radicals can be produced by a Fenton-type reaction as described in Equations 1–3.
The reducing agent ascorbate leads to a cycle where Fe3+ is reduced back to Fe2+. This reaction is typically described for Fe, but other metals such as Cu and Co have also been reported to function in analogous ways but with different specificities and rates (Walling 1975; Stubbe and Kozarich 1987; Tung and Sawyer 1990; Sobkowiak et al. 1993; Sawyer et al. 1996). The Fe–chelate-mediated cleavage of peptide chains (Rana and Meares 1991b; Ermacora et al. 1992) resembles the one described here, but for that reaction it was shown that Fe ions coordinate the reactive oxygen species (Rana and Meares 1991a). We noted a clear difference between the Co2+- and Cu2+-mediated reactions, which could either be due to a different rate of activated oxygen radical production or more direct involvement of intermediates containing metal ions. However, we do not have data that would give a clear insight in the mechanism of the reaction. The mass spectroscopy (Fig. 3) shows that cleavage seems to happen preferentially at a specific His residue although it can occur at any of the His residues. The failure to achieve Edman sequencing from the C-terminal fragments indicates that the N-terminal residue is not in an intact amine.
The efficient and easy cleavage of proteins at specific sites can be of great potential in many applications and fundamental studies of proteins. Uses can be found, for example, in removal of purification tags, or for the specific activation or inactivation of proteins in controllable circumstances. The work described here has much in common with earlier work with metal ion-mediated cleavage of proteins but shows some clearly distinct new findings. The cleavage site can easily be engineered into a protein, the reaction needs only added metal salts, it occurs quickly at a broad pH range near neutral and at low temperature. Cu ions have been used in studies having some resemblance to the current one, but the finding that Co ions bound to proteins can cause a cleavage reaction is novel. The current work can also be of practical importance for protein purification using His tags, a method that is widely used. The His-tagged recombinant proteins (containing typically six His residues) are often purified with Cu ion-loaded chromatographic resins. Since only very low concentrations of Cu ions were needed for cleavage there is a risk that unexpected cleavage of the target protein could occur at any stage of the protein processing.
For practical use of the described reaction, it seems that Co-mediated cleavage has a larger potential since an excess of Cu ions could cause an overall degradation of the protein. We still have very little information on the exact mechanisms of the reaction. Future studies addressing the mechanism could help to develop the cleavage reaction into a practical tool in biochemistry and molecular biology.
Materials and Methods
Construction of vectors
Several different fusion proteins were used for analyzing the cleavage conditions. The proteins used are listed in Table 1. For construction of a vector expressing E. coli MBP (Pryor and Leiting 1997) as a fusion protein with a peptide linker and a fragment of the albumin binding protein (ABP) (MBP-linker-ABP) in E. coli, the DNA fragment encoding ABP was PCR amplified from vector pKN1 (Nord et al. 1995) with the 5′ oligo GCATTGGATTCGAATTCTTAGCTGAAGCTAAAGTCTTAGC (EcoRI sequence underlined) and 3′ oligo GCATTAAGCTTCTATTCGCTTTTTGCCGGAGTAG (HindIII sequence underlined). These oligos amplify fragment encoding amino acids 496–541 of the Staphylococcus carnosus cell surface protein (GeneBank U15515) albumin binding domain. The PCR fragment was ligated to the pCR2.1-TOPO vector (Invitrogen). This vector was cut with EcoRI and HindIII and the DNA fragment encoding ABP was ligated into EcoRI and HindIII cut pMal-c2X (New England Biolabs) vector to yield pLink1.
To add the sequence encoding the linker GSPTGASTHHHHHHGSPTGAST in between MBP and ABP in pLink1, synthetic oligos were kinased with T4 polynucleotide kinase, annealed, and ligated directly into SacI and EcoRI cut pLink1. This generated pLink2. The sequences of all linker oligos are shown in the Supplemental data. Identically, a plasmid expressing MBP-GSPTGASTGGGGGGGSPTGAST-ABP was generated by a pair of oligos encoding GSPTGASTGGGGGGGSPTGAST. This generated pLink3. For expression of the cytosolic fragment of Sso2 (Sso2-ma, amino acids 1–270) (Aalto et al. 1993) as a MBP fusion separated by a linker, Sso2-ma fragment was amplified by PCR with the oligos GCATTGAATTCATGAGCAACGCTAATCCTTATGA and GCATTAAGCTTTTATCATCTTATTTTGTTTTTTCTTGC using a genomic fragment of Sso2 as the template followed by ligation into pCR2.1-TOPO. SSO2-ma was then cloned as an EcoRI/HindIII fragment into EcoRI/HindIII cut pLink2 to replace streptavidin with Sso2-ma and generate pLink14 for expression of MBP-GSPTGASTHHHHHHGSPTGAST-Sso2.
For construction of a vector for expression of MBP-GSPTGASTHHHHHHGSPTGAST-AVI in E. coli, a DNA fragment of the Streptomyces avidinii gene for amino acids 25–183 of streptavidin (GeneBank X03591) was amplified by PCR by using the vector pK501–1 (Oker-Blom et al. 1996) as a template. The streptavidin (AVI) specific 5′ oligo was GCATTGAATTCGACCCCTCCAAGGACTCGAAGG and the 3′ oligo was GCATTAAGCTTCTACTGCTGAACGGCGTCGAGC. The PCR fragment was TA-ligated to the pCR2.1-TOPO vector (Invitrogen) and subsequently after cutting with EcoRI and HindIII enzymes ligated into EcoRI and HindIII cut pLink2 vector to yield pLink6. To generate pLink7 vector expressing MBP-GSPTGASTGSTGPSGSPTGAST-AVI, the corresponding oligos were kinased, annealed, and ligated into SacI and EcoRI cut pLink6. Identically, pLink8 expressing MBP-GSPTGASTHHHHGSPTGAST-AVI was generated by linker oligo ligation into SacI and EcoRI cut pLink6. The pLink10 expressing MBP-GSPTGASTHHGSPTGAST-AVI, pLink12 expressing MBP-GSPTGASTHHHHHHHHGSPTGAST-AVI, and pLink13 expressing MBP-GSPTGASTHSHAHGHAHSHGSPTGAST-AVI were generated identically to pLink8.
Production and purification of fusion proteins
The purification of MBP fusion proteins was carried out using the pMAL Protein Fusion and Purification System (New England Biolabs Inc.); this was used in the production and purification of the fusion proteins in Table 1. XL1 Blue E. coli cells transformed with the vectors encoding the various fusion proteins were grown in a rotary shaker (200 rpm) in LB medium with 0.1 mg/mL ampicillin at 37°C. IPTG was added to a final concentration of 0.3 mM when the absorbance at 600 nm was 0.5, and the incubation was continued for 3 h. After centrifugation, 5000 rpm, 15 min at 4°C, the cells were resuspended in 31.5 mL of MAL buffer (50 mM Tric-Cl at pH 7.5, 0.2 M NaCl, and 1mM EDTA at pH 8) and broken by sonication (Soniprep 150 sonicator, MSE Scientific Instruments) 3 × 15 sec on ice. The cell debris was removed by centrifugation at 9000g for 20 min at 4°C. The fusion proteins were purified by affinity chromatography on a 5-mL amylose column (New England Biolabs Inc.). The column was equilibrated with MAL buffer, and the protein was eluted with 10 mM maltose in MAL buffer. The fusion protein containing fractions was pooled, and the buffer was changed by using Econo-Pac 10 DG columns (Bio-Rad Laboratories) to 50 mM ammonium acetate at pH 7.5. The buffer exchange was repeated in Vivaspin concentrators (10,000 MWCO, PES, Vivascience Ltd.). The purity of the fusion proteins was verified by SDS-PAGE and Coomassie staining.
Cleavage of fusion proteins by using metal ions
Unless otherwise stated, a general cleavage reaction was performed by mixing 3.3 μM fusion protein with 3.3–33 μM metal salt (corresponding to 1–10 molar equivalents), typically in 10 mM ammonium acetate at pH 7.5 or water. The metal salts used were FeCl3, CoCl2, MnCl2, ZnCl2, CuCl2, or CuSO4. The protein solution was mixed and incubated for 2 min at room temperature. All metal ions used were from 10× stocks dissolved in MilliQ-purified water (18.2 MΩcm resistivity). In a separate tube, freshly prepared ascorbate adjusted to neutral pH with Na2CO3 was mixed with hydrogen peroxide just before use. The cleavage reaction was initiated by adding the mixed ascorbate and hydrogen peroxide to give final concentrations of 4.6 mM and 0.35 mM, respectively (or as stated). The reaction volume was typically 35 μL in a 1.5-mL Eppendorf tube, and the cleavage reaction was typically carried out for 60 min at room temperature in darkness. The reaction was quenched by adding a 10,000-fold molar excess of EDTA at pH 7.5. For SDS-PAGE, the quenched reaction solutions were heated for 5 min at 95°C together with SDS-PAGE loading buffer containing β-mercaptoethanol: Electrophoresis was typically performed using Tris-HCl 10%–20% SDS-polyacrylamide gels (Bio-Rad). The protein bands were visualized by staining the gels with Coomassie brilliant blue (Amersham Pharmacia Biotech) for 60 min, after which the gels were destained before they were scanned and dried between cellulose membranes for storage.
Western blot analysis
For Western blot analysis, proteins were separated by SDS-PAGE and transferred to a nitrocellulose filter by electroblotting. The filter was blocked with 2% skim milk powder in buffer [10 mM Tris-HCl at pH 8, 150 mM NaCl, and 0.05% (v/v) Tween-20]. The primary antibodies used were anti-MBP antiserum (New England Biolabs) or anti-Sso2 antibody (Jäntti et al. 1994), both diluted 1:10000 in TBST buffer, and as a secondary antibody, goat anti-rabbit IgG (H + L) coupled to alkaline phosphatase (Bio-Rad) diluted 1:1000 was used.
Size exclusion chromatography
To separate the protein fragments produced in the cleavage reaction with fusion protein, size exclusion chromatography on a Superdex 200 HR 10/30 column (Amersham Pharmacia Biotech) was performed. The buffers used were 0.2 M NaCl in 50 mM acetate buffer at pH 5.0. Proteins from a Gel Filtration Calibration Kit (Amersham Pharmacia Biotech) were used as standards.
Mass analysis and N-terminal sequencing
The mass analyses were performed on MALDI-TOF (Matrix-Assisted Laser Desoption-Ionization Time-of-Flight) (Autoflex II, Bruker Daltonics). A small aliquot of protein sample was put on the sample plate and sinapic acid were added as the matrix. Ionization and data collection were performed by standard procedures. N-terminal sequencing was performed by Edman degradation on a Procise 494A sequencer (Applied Biosystems).
Electronic supplementary material
A table containing the DNA oligomers used for constructing the recombinant proteins is listed in the Supplemental material.
Acknowledgments
We thank Riita Suihkonen and Outi Könönen for technical assistance. This work was supported by Tekes (The Finnish Funding Agency for Technology and Innovation).
Footnotes
Supplemental material: www.proteinscience.org
Reprint requests: to Markus B. Linder, VTT, Technical Research Centre of Finland, Tietotie 2, Espoo FIN-02044 VTT, Finland; e-mail: markus.linder@vtt.fi; fax: 358-20-7227071.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072846407.
References
- Aalto M.K., Ronne, H., and Keränen, S. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12: 4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G. and Campbell, R. 1996. Specific cleavage of histidine-containing peptides by copper(II). Int. J. Pept. Protein Res. 48: 265–273. [DOI] [PubMed] [Google Scholar]
- Arnau J., Lauritzen, C., Petersen, G.E., and Pedersen, J. 2006. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr. Purif. 48: 1–13. [DOI] [PubMed] [Google Scholar]
- Bateman R., Youngblood, W., Busby Jr, W., and Kizer, J. 1985. Nonenzymatic peptide α-amidation. Implications for a novel enzyme mechanism. J. Biol. Chem. 260: 9088–9091. [PubMed] [Google Scholar]
- Ermacora M., Delfino, J., Cuenoud, B., Schepartz, A., and Fox, R. 1992. Conformation-dependent cleavage of staphylococcal nuclease with a disulfide-linked iron chelate. Proc. Natl. Acad. Sci. 89: 6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant K.B. and Kassai, M. 2006. Major advances in the hydrolysis of peptides and proteins by metal ions and complexes. Curr. Org. Chem. 10: 1035–1049. [Google Scholar]
- Hegg E.L. and Burstyn, J.N. 1998. Toward the development of metal based synthetic nucleases and peptidases: A rationale and progress report in applying the principles of coordination chemistry. Coord. Chem. Rev. 173: 133–165. [Google Scholar]
- Humphreys D., Smith, B., King, L., West, S., Reeks, D., and Stephens, P. 1999. Efficient site specific removal of a C-terminal FLAG fusion from a Fab′ using copper(II) ion catalysed protein cleavage. Protein Eng. 12: 179–184. [DOI] [PubMed] [Google Scholar]
- Humphreys D., King, L., West, S., Chapman, A., Sehdev, M., Redden, M., Glover, D., and Smith, B. 2000. Improved efficiency of site-specific copper(II) ion-catalysed protein cleavage effected by mutagenesis of cleavage site. Protein Eng. 13: 201–206. [DOI] [PubMed] [Google Scholar]
- Jäntti J., Keranen, S., Toikkanen, J., Kuismanen, E., Ehnholm, C., Soderlund, H., and Olkkonen, V. 1994. Membrane insertion and intracellular transport of yeast syntaxin Sso2p in mammalian cells. J. Cell Sci. 107: 3623–3633. [DOI] [PubMed] [Google Scholar]
- Jeon J.W., Son, S.J., Yoo, C.E., Hong, I.S., and Suh, J. 2003. Toward protein-cleaving catalytic drugs: Artificial protease selective for myoglobin. Bioorg. Med. Chem. 11: 2901–2910. [DOI] [PubMed] [Google Scholar]
- Jitsukawa K., Mabuchi, T., Einaga, H., and Masuda, H. 2006. Site-specific recognition of dipeptides through non-covalent inter-ligand interactions for the hydrolysis of dipeptide to amino acid ligands mediated by ternary cobalt(III) complexes. Eur. J. Inorg. Chem. 2006: 4254–4263. [Google Scholar]
- Kim K., Rhee, S., and Stadtman, E. 1985. Nonenzymatic cleavage of proteins by reactive oxygen species generated by dithiothreitol and iron. J. Biol. Chem. 260: 15394–15397. [PubMed] [Google Scholar]
- Kumar C.V., Buranaprapuk, A., Cho, A., and Chaudhari, A. 2000. Artificial metallopeptidases: Regioselective cleavage of lysozyme. Chem. Comm. doi: 10.1039/a907477e. [DOI]
- Nilsson J., Stahl, S., Lundeberg, J., Uhlen, M., and Nygren, P.-A. 1997. Affinity fusion strategies for detection, purification, and immobilization of recombinant proteins. Protein Expr. Purif. 11: 1–16. [DOI] [PubMed] [Google Scholar]
- Nord K., Nilsson, J., Nilsson, B., Uhlen, M., and Nygren, P.-A. 1995. A combinatorial library of an α-helical bacterial receptor domain. Protein Eng. 8: 601–608. [DOI] [PubMed] [Google Scholar]
- Oker-Blom C., Orellana, A., and Keinanen, K. 1996. Highly efficient production of GFP and its derivatives in insect cells for visual in vitro applications. FEBS Lett. 389: 238–243. [DOI] [PubMed] [Google Scholar]
- Platis I.E., Ermacora, M.R., and Fox, R.O. 1993. Oxidative polypeptide cleavage mediated by EDTA-iron covalently linked to cysteine residues. Biochemistry 32: 12761–12767. [DOI] [PubMed] [Google Scholar]
- Polzin G.M. and Burstyn, J.N. 2001. Synthetic Cu(II) and Ni(II) peptidases. Met. Ions Biol. Syst. 38: 103–143. [PubMed] [Google Scholar]
- Pryor K.D. and Leiting, B. 1997. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 10: 309–319. [DOI] [PubMed] [Google Scholar]
- Rana T. and Meares, C. 1991a. Transfer of oxygen from an artificial protease to peptide carbon during proteolysis. Proc. Natl. Acad. Sci. 88: 10578–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T.M. and Meares, C.F. 1991b. Iron chelate mediated proteolysis: Protein structure dependence. J. Am. Chem. Soc. 113: 1859–1861. [Google Scholar]
- Sawyer D.T., Sobkowiak, A., and Matsushita, T. 1996. Metal [MLx; M = Fe, Cu, Co, Mn]/hydroperoxide-induced activation of dioxygen for the oxygenation of hydrocarbons: Oxygenated Fenton chemistry. Acc. Chem. Res. 29: 409–416. [Google Scholar]
- Sobkowiak A., Qui, A., Liu, X., Llobet, A., and Sawyer, D.T. 1993. Copper(I)/(tert-BuOOH)-induced activation of dioxygen for the ketonization of methylenic carbons. J. Am. Chem. Soc. 115: 609–614. [Google Scholar]
- Stubbe J. and Kozarich, J.W. 1987. Mechanisms of bleomycin-induced DNA degradation. Chem. Rev. 87: 1107–1136. [Google Scholar]
- Tung H.C. and Sawyer, D.T. 1990. Cobalt-induced activation of hydrogen peroxide for the direct ketonization of methylenic carbons [c-C6H12.fwdarw. c-C6H10(O)], the oxidation of alcohols and aldehydes, and the dioxygenation of aryl olefins and acetylenes. J. Am. Chem. Soc. 112: 8214–8215. [Google Scholar]
- Ueda E.K.M., Gout, P.W., and Morganti, L. 2003. Current and prospective applications of metal ion-protein binding. J. Chromatogr. A. 988: 1–23. [DOI] [PubMed] [Google Scholar]
- Walling C. 1975. Fenton's reagent revisited. Acc. Chem. Res. 8: 125–131. [Google Scholar]
- Waugh D.S. 2005. Making the most of affinity tags. Trends Biotechnol. 23: 316–320. [DOI] [PubMed] [Google Scholar]