Abstract
ParD is the antidote of the plasmid-encoded toxin–antitoxin (TA) system ParD–ParE. These modules rely on differential stabilities of a highly expressed but labile antidote and a stable toxin expressed from one operon. Consequently, loss of the coding plasmid results in loss of the protective antidote and poisoning of the cell. The antidote protein usually also exhibits an autoregulatory function of the operon. In this paper, we present the solution structure of ParD. The repressor activity of ParD is mediated by the N-terminal half of the protein, which adopts a ribbon-helix-helix (RHH) fold. The C-terminal half of the protein is unstructured in the absence of its cognate binding partner ParE. Based on homology with other RHH proteins, we present a model of the ParD–DNA interaction, with the antiparallel β-strand being inserted into the major groove of DNA. The fusion of the N-terminal DNA-binding RHH motif to the toxin-binding unstructured C-terminal domain is discussed in its evolutionary context.
Keywords: NMR spectroscopy, ribbon-helix-helix, intermonomer NOEs, homodimeric protein, toxin–antitoxin systems, bacterial programmed cell death
Toxin–antitoxin (TA) modules, as encoded on the parDE operon, are highly abundant in plasmids and bacterial chromosomes (for recent reviews, see Engelberg-Kulka and Glaser 1999; Anantharaman and Aravind 2003; Hayes 2003; Buts et al. 2005; Gerdes et al. 2005; Pandey and Gerdes 2005). Typically, two open reading frames are encoded within one operon in these TA modules, where the first gene codes for the antidote and the second gene for the corresponding toxin. Many TA systems exhibit similar general biological functions, in terms of killing progenies that failed to inherit the TA module, but they are very different with respect to amino acid sequence, three-dimensional (3D) structures, and their mode of action. The different toxins were shown to interfere with DNA replication (e.g., by inhibiting DNA gyrase as shown for ParE [Jiang et al. 2002]) or protein synthesis (e.g., RelE cleaves mRNA bound to the ribosomal A site [Pedersen et al. 2003]), leading to growth arrest or cell killing. The antidote proteins form complexes with their respective toxins, thus inactivating their deleterious effects upon the host organism. The effect of TA modules is based on differential stability of the two components in these systems: The antidote is degraded by a cellular protease more easily than its toxic counterpart. If the pool of antidote proteins is not replenished continuously, as is the case after plasmid loss, the long-lived toxin exerts its function on the host organism. Generally, these modules are autoregulated at the level of transcription by binding of the antidote or the toxin–antidote complexes within their own promoter regions. The biological relevance of TA modules is associated with the stable inheritance of genetic elements (plasmid addiction), stress response, and more generic regulatory systems (Engelberg-Kulka and Glaser 1999; Gerdes 2000; de la Cueva-Mendez 2003; Hayes 2003).
While the different biological roles of addiction modules are still under investigation, the general properties of toxin–antitoxin elements provide promising potentials for biotechnological applications, e.g., in positive selection systems (Bernard 1996; Gabant et al. 2000), plasmid stabilization systems (Pecota et al. 1997), and antibacterial therapy, which has become especially important in view of the widespread multidrug resistance among clinically important bacterial species (Engelberg-Kulka et al. 2004).
The ParD–ParE system represents a TA module encoded on the broad host range, low copy number plasmid pRK2/RP4. ParE (103 amino acids) is the positively charged toxin, whereas ParD (83 amino acids) constitutes the negatively charged antidote able to neutralize ParE by forming a tight complex that is also effective in autorepression of the parDE operon. In contrast to other TA systems that require the complex for full negative regulation of the operon, ParD alone is sufficient for autorepression (Davis et al. 1992; Eberl et al. 1992). Our previous studies on ParD showed that the protein exists as a dimer in solution, and that ParD exhibits high thermal stability and excellent refolding properties after heat-induced denaturation (Oberer et al. 1999, 2002). Circular dichroism spectroscopy and preliminary characterization from NMR data indicated that the protein was composed of α-helical and β-strand regions. Chemical shift analysis as well as relaxation data revealed that ParD consists of two structurally distinct moieties, namely a well-ordered N-terminal half and an unstructured C-terminal half (Oberer et al. 2002). Among other TA systems, mutational and structural studies confirm the suggested separation into an N-terminal region, which is mainly responsible for autoregulation, and a C-terminal region, which functions in neutralization of the toxin (e.g., ParD/Pem [Ruiz-Echevarria et al. 1991a,b; Santos-Sierra et al. 2002], CcdA [Bernard and Couturier 1991; Salmon et al. 1994; Madl et al. 2006], Phd [Lehnherr et al. 1993; Smith and Magnuson 2004; McKinley and Magnuson 2005], MazE2MazF4 [Kamada et al. 2003], RelE2-RelB2 [Takagi et al. 2005], and YefM2-YoeB [Kamada and Hanaoka 2005]).
Here, we describe the dimeric structure of the bacterial antidote ParD, which was solved using multidimensional heteronuclear NMR spectroscopy. The N-terminal half of the protein displays the ribbon-helix-helix (RHH) fold, a DNA-binding motif found in different prokaryotic repressors with known 3D structures from both X-ray crystallography and NMR spectroscopy (Phillips 1994; Raumann et al. 1994; del Solar et al. 2002). The solution structure of ParD is compared with the recently solved structures of unbound prokaryotic repressors as well as their respective DNA complexes. Based on these structures and combined with the information from biochemical data, we built a model of the ParD–DNA complex to illustrate the mode of specific DNA binding by the ParD dimer.
Structural information of bacterial antidotes originating from similar systems has become available only very recently. This can be attributed to the intrinsic instability of the antidote and the toxic properties of the toxin, which render high-level expression, as required for structural studies, very difficult. Structures of uncomplexed toxins were solved for CcdB (Loris et al. 1999), Kid (Hargreaves et al. 2002), and YoeB (Kamada and Hanaoka 2005), whereas crystal structures of TA complexes have been solved for MazE2MazF4 (Kamada et al. 2003), RelE2-RelB2 (Takagi et al. 2005), YefM2-YoeB (Kamada and Hanaoka 2005), ɛ2ζ2 (Meinhart et al. 2003), and FitAB (Mattison et al. 2006). Structural information of uncomplexed antidotes has been available initially only from biophysical studies (e.g., for Phd [Gazit and Sauer 1999], CcdA [Van Melderen et al. 1996], YefM [Cherny and Gazit 2004], and ParD [Oberer et al. 1999]). Crystallization of the free antitoxins was often impaired due to low stability and largely unstructured regions of these proteins. 3D structural information of antitoxins was obtained by X-ray crystallography from complexes, either with their cognate DNA (e.g., ω–DNA complex [Weihofen et al. 2006] of the ω/ɛ/ζ module) or with an antibody (Lah et al. 2003; Loris et al. 2003). The use of NMR spectroscopy allowed us to determine the structures of antitoxins in solution, as was recently demonstrated with the free and DNA-complexed structures of CcdA (Madl et al. 2006) and the solution structure of ParD, as presented in this study.
Results
Assignments and structure calculation
Based on the reported sequential backbone and side chain assignments for 1H, 13C, and 15N resonances of the antidote ParD (Oberer et al. 2002), the NOESY spectra were analyzed manually. The ParD protein has been established to be dimeric in the concentration range used for structure determination. The homodimeric nature of ParD is also indicated by the existence of a single set of resonances in the 1H-15N-HSQC spectra. Consequently, ambiguities in the assignment of NOEs are inherent, because inter- and intramolecular neighbors cannot be distinguished using standard NOE experiments. In order to discriminate inter- and intramolecular NOEs, either some prior knowledge of the three-dimensional structure (e.g., from a crystal structure or a homologous protein structure) is necessary, or a special experimental technique has to be used which provides this distinction de novo. For the case of ParD, we developed a new method for X-filtering (Zangger et al. 2003) and modeled the three-dimensional structure of ParD to assist in the correct assignment process of the observed NOEs.
A summary of the numbers of intraresidue, short- and long-range sequential, as well as intermolecular NOEs for each residue is shown in Figure 1. The lower number of NOEs found for the C-terminal portion of ParD is in agreement with the dynamic behavior already described for this protein, indicating an unstructured domain (Oberer et al. 2002).
Figure 1.
The experimentally derived restraints per residue reflect the two-domain architecture of ParD protein. The shading is hatched, light gray, dark gray, black, and dotted for intraresidue, sequential, short-range (dij, j < i + 5), long-range (dij, j > i + 4), and intermonomer NOEs, respectively.
Eight hundred eighty-seven intra- and 142 intermolecular distance restraints served as input for structure calculation, together with 59 dihedral restraints and 17 distance restraints representing the inferred H-bonds (Table 1). The NOEs used for the calculation were doubled and renumbered to take account of the dimeric structure of ParD (chain A Met1 to Ala83, chain B Met101 to Ala183). Overlapping of the proton resonances made it impossible to assign NOEs in the 2D-NOESY spectra other than the well resolved residues in the β-strand region and also made it difficult to assign long-range NOE cross peaks in the 3D-13C and 15N edited spectra. Based on the presence of a strong Phe36Hα–Pro37Hδ NOE, the Phe36–Pro37 bond was assumed to be in the trans configuration. Mainly intraresidual NOEs could be identified for the flexible C-terminal part of ParD.
Table 1.
Restraints and structural statistics of the resulting final structures of ParD

A total of 150 structures was calculated, and 24 structures out of the accepted ones were chosen on the basis of the lowest energy values (CNS potential energy [Etotal] <400 kcal/mol). In agreement with the previously identified secondary structure elements (Oberer et al. 2002), each monomer consists of a β-strand followed by α-helices A, B, and C, which are connected by turns and linker regions (Fig. 2). The root mean square deviations of these structures were 0.70 ± 0.14 Å for the backbone heavy atoms including residues Arg3 to Phe36 (including the β-sheet and α-helices A and B) of both chains. The conserved turn between α-helices A and B (Gly24 to Thr26) is well-defined, whereas the regions connecting helices B and C (Pro37-Asp41) are not ordered. When helix C of one monomer is calculated separately, the root mean square deviation is 1.13 ± 0.39 Å for the backbone heavy atoms Ala42 to Leu49. In agreement with our previous results, the structure contains 37% α-helix and 8% β-sheet calculated against the full-length protein (Oberer et al. 1999, 2002).
Figure 2.
Structure of ParD determined by NMR spectroscopy. (A) Sequence and secondary structure of ParD. (B) Ribbon presentation of a representative low-energy solution structure of the full-length ParD protein. Ribbon diagram of the closest-to-mean structure of ParD color-coded with the transverse 15N-relaxation times. Longer T2 times correspond to increased flexibility. The orientation of helix C with respect to the well-defined N-terminal core cannot be determined. (C) Stereoview of the backbone bundle of the 24 final structures of ParD (residues Met1 to Ala40). The RMSD of residues 3–36 and 103–136 of the dimeric protein are 0.70 and 1.44 Å for backbone and heavy atoms, respectively.
The dimeric ParD structure and its domain architecture
The ensemble of the 24 accepted structures shows that the bifunctional protein is a homodimer containing the RHH motif in the N-terminal domain. The RHH motif derives from a DNA-binding domain that was initially found in the Arc repressor (Breg et al. 1990). It includes two intertwined monomers with the N-terminal β-strands forming an antiparallel β-ribbon across the local twofold axis and two subsequent helices (Fig. 2). The dimer interface is characterized by numerous hydrophobic interactions, and the buried area (calculated for five representative low-energy structures of ParD using residues Ser2 to Ala40 of both chains) in the subunit–subunit interface was calculated to be 1263 ± 85.5 Å2. The hydrophobic core involves the residues Leu4, Ile6, and Met8 from the β-strand; residues His13, Leu16, Leu19, and Ala20 of helices A and A′; and residues Ile27, Tyr30, and Ala31 of helices B and B′. The residues from the β-strands and the helices B and B′ are contributing to the dimer interface, whereas hydrophobic residues from the helices A and A′ form mainly intramolecular contacts. A major exception is a patch of hydrophobic residues, Ala18, Ala21, and Leu22, located at the outside and close to the C-terminal end of helix A. Following helix B, the polypeptide chain extends into a flexible linker followed by helix C. The orientation of this helix with respect to the dimeric equivalent and to the N-terminal domain is most likely not strictly defined and could not be determined. In good agreement with our previous studies on the dynamic properties of ParD (Oberer et al. 2002), the remaining part of the protein (Thr51 to Ala83) is unstructured as seen by the almost complete absence of interresidual NOEs.
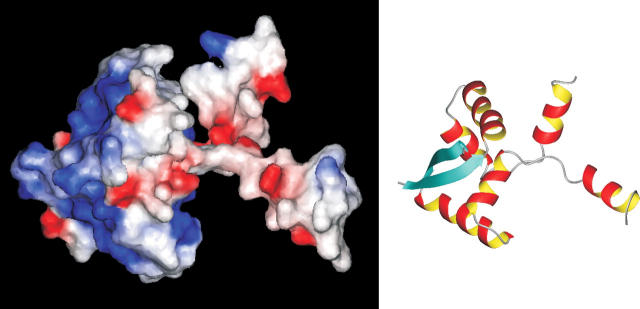
Inspection of the electrostatic surface (Fig. 3) of one representative of the calculated structures discloses a positively charged area (established by the residues Arg3, His13, Lys17, Lys25, and Lys28 of each monomer) at the N terminus of ParD, which is postulated to be responsible for protein–DNA interaction. The other regions of the core domain reveal a largely neutral surface area, whereas the C-terminal tail regions carry a surplus of negative charges (10 Glu and Asp residues compared with five Arg and Lys residues). Negative charges are accumulated in the linker between helix B and helix C (D39-ADAD-Q44) and in the very C-terminal amino acid sequence (E72-ILDEELSG-D81). This overall charge distribution is consistent with the DNA-binding function residing in the ordered N-terminal domain of ParD and the binding of the basic toxic protein ParE with the unstructured C-terminal region of ParD. We could also identify a region of (mainly negatively) charged and polar residues in the region connecting the DNA-binding domain of ParD with the toxin-interacting region including helix C (D39-ADADQAWQEL-K50). Based on this similarity to the interaction site4 of the MazE/F complex (Kamada et al. 2003) or to the charge distribution observed in the RelB–RelE complex (Takagi et al. 2005), it can be speculated that this region also plays an important role in binding of the toxin ParE.
Figure 3.
(Left) Molecular surface of ParD colored according to its electrostatic potential. The surface potential of the ParD dimer (residues Met1–Met52) shows two charged regions: positive potential (blue) and negative potential (red). The surface at the side of the β-sheet is dominated by a positive electrostatic potential. The opposite side of the molecule displays an overall negative potential. (Right) Ribbon diagram depicts the protein in the same orientation.
Comparison of the ParD solution structure with other RHH proteins
The solution structure of ParD reaffirms that the protein is a member of the MetJ/Arc structural superfamily. The RHH superfamily includes several proteins with known structures, among them CopG (Gomis-Ruth et al. 1998), MetJ (Rafferty et al. 1989; Somers and Phillips 1992), Arc (Breg et al. 1990), Mnt (Burgering et al. 1994), protein ω (Murayama et al. 2001), NikR (Schreiter et al. 2003), ParG (Golovanov et al. 2003), HP0222 (Popescu et al. 2005), and the bacterial antidote CcdA (Madl et al. 2006). Despite their low sequence homologies, members of the RHH family often share their function as transcriptional repressors. However, they share a distinct conserved pattern of amino acids correlating with DNA interaction, structure stabilization, and function. These mainly hydrophobic residues form a rigid hydrophobic central core and are involved in the dimerization interface. The intertwined core structure of these proteins is highly conserved, yet they are quite divergent in terms of the absence (CopG, Arc) or presence of N- (MetJ, protein ω, ParG, HP0222) and C-terminal (ParD, MetJ, Mnt, CcdA) tails with completely different functions (see also Fig. 6 in Golovanov et al. [2003]).
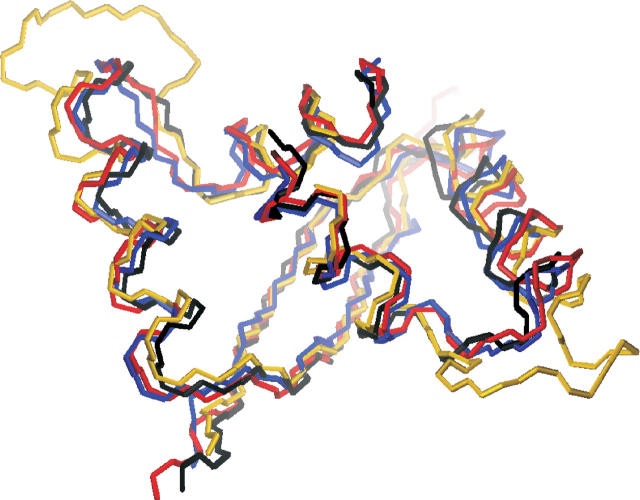
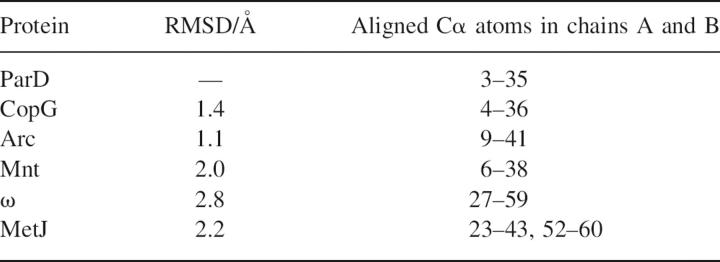
The superposition of ribbon-helix-helix (RHH) elements of ParD with RHH domains of other representative members of this family is shown in Figure 4. The root mean square deviations were calculated using the programs Profit and MOLMOL and are listed in Table 2.
Figure 4.
Structure comparison of the ParD with the RHH proteins Arc, CopG, and MetJ. The backbone of ParD was superimposed on the backbone atoms of other RHH domains using the program MOLMOL. The aligned residues can be seen in Table 2. A view from the top of the backbone traces of ParD (blue), CopG (black), Arc (red), and MetJ (yellow) is shown.
Table 2.
Superposition of ParD with other RHH proteins
The most striking difference in the general architecture of ParD to other members of the RHH family is the length of the inner helix B. These helices comprise residues Val33–Lys46 in Arc, Lys28–Lys41 in CopG, Asn31–Ser43 in Mnt, Asn53–Thr66 in MetJ, Val51–Tyr66 in protein ω, and Ile60–Glu74 in ParG where the “crossing-point” of the dimer-related helices always resides close to the center of the helical stretch. In ParD, however, the helix B appears to end right at the crossing-point, the last “α-helical” residue being Phe36, which is followed by the classical helix breakers Pro37, Gly38, and by a stretch of Asp–Ala repeats. This part of the structure was particularly difficult to assign due to resonance overlap and the lack of long-range NOEs. We assume that the third helix adopts a defined orientation only within the toxin–antitoxin complex.
ParD–DNA complex
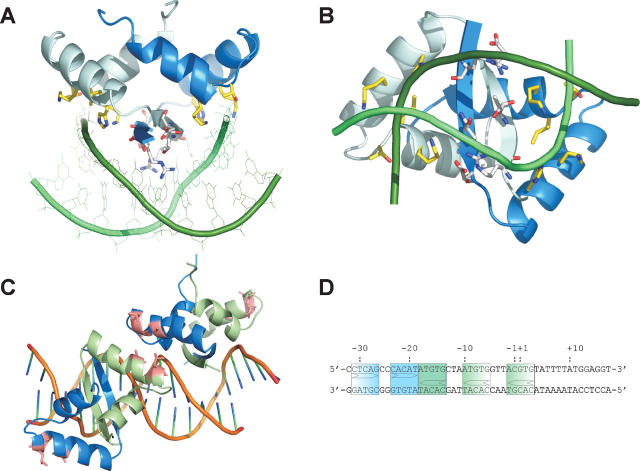
From comparison of the ParD structure with protein–DNA complexes of other members of the same fold family, it can be assumed that the binary complex consists of at least two dimers of the protein bound to adjacent major-groove binding sites of double-stranded DNA (dsDNA) (e.g., the Arc-repressor [Raumann et al. 1994], CopG [Gomis-Ruth et al. 1998], and MetJ [Somers and Phillips 1992]). The binding site of ParD could be narrowed down only to a stretch of 33 bp (corresponding to a molecular mass of 25 kDa) containing a direct and an inverted repeat sequence using gel-retardation experiments (see Supplemental Fig. S1). Preliminary biophysical experiments demonstrate that two to three ParD dimers bind to an oligonucleotide representing the parDE promoter (PparDE), resulting in a protein–DNA complex of 60–78 kDa in size that is not easily accessible by standard NMR experiments (data not shown). Therefore, we resorted to model building to gain first insights into the mode of this sequence-specific interaction. By homology with the RHH family of proteins, we assume that DNA binding is mediated by insertion of the β-sheet of ParD into the major groove of DNA. The model of a ParD dimer bound to a 10-bp inverted repeat was generated based on one half-site of the Arc-repressor–DNA complex (Raumann et al. 1994). First, the RHH motif of the ParD dimer (amino acids 3–35) was superimposed onto the Arc-repressor dimer. Then the bases of the Arc-binding site were mutated against the ParD site nucleotides, leaving the conformation of the Arc-site DNA intact. In order to remove steric clashes between the binding partners and to allow for a rearrangement of the ParD side chains necessary for a specific binding interaction, several cycles of energy minimization were performed. The conformational changes were checked after each cycle. The final energy minimization (Efin = −1218 kcal/mol) yielded a model without clashes and reasonable geometry (Laskowski et al. 1996). For the modeled oligonucleotide, a 10-bp palindromic DNA sequence (5′-C−5A−4C−3A−2T−1 • A1T2G3T4G5-3′) was used, corresponding to the inverted repeat sequence of the ParD operator. In the final model the ParD dimer is positioned directly over the major groove with the protein twofold axis coinciding approximately with the DNA symmetry axis (Fig. 5A,B). Helices A and A′ are positioned above the dsDNA approximately parallel to the phosphate backbone, and the β-ribbon is inserted in the major groove. Direct base contacts are observed for Arg3, which forms H-bonds to A1 and T2 bridging the two bases next to the palindrome center, and Thr5, which forms a H-bond to T−1. Corresponding ParD–DNA interactions occur in the symmetry-related monomer. In addition, Thr5 and Thr5′ side chains form an inter-dimer H-bond across the dimer axis, an interaction that can also be observed in the unliganded structure. Asp7′, which is also directed toward the major groove, does not form base-specific contacts, but stabilizes the Arg3 conformation instead, by forming a H-bond to Arg3_Nɛ. Helices B and B′ are positioned perpendicular to the phosphate backbone with their N-terminal ends pointing directly at phosphate groups. The main-chain amides of Ile27 and Lys28 of both protein chains form H-bonds to the phosphate groups of C−3. This helix-B–DNA interaction also occurs in a very similar manner in the Arc-repressor–DNA complex (Raumann et al. 1994) and appears to be a decisive feature of RHH–DNA complexes in general. In addition, the side chain of Thr26, which forms the cap of helix B in the unliganded structure, swings around and binds to the phosphate group of A−4. Additional putative interactions are feasible between the side chains of His13 and Lys17 and the A−2 phosphate group; however, side chain conformations would have to change upon DNA binding in order to accommodate strong H-bonds.
Figure 5.
Model of the ParD–DNA complex. The ParD–DNA complex is shown in a ribbon presentation: (blue and light blue) the two chains of the ParD dimer, and (green and pale green) the two strands of the 10-bp inverted repeat. Residues involved in protein–DNA interactions are shown in stick presentation: (gray) those pointing into the major groove of the DNA (Arg3, Thr5, and Asp7), (yellow) those interacting with the phosphate backbone. (A) View along the β-ribbon protruding the major groove. (B) View approximately along the twofold axis of the dimer. The line drawing of the nucleotides was omitted for clarity. (C) The superposition of two ParD dimers with the MetJ–DNA complex (PDB entry: 1mjo). The ParD dimers are shown as ribbon drawing: (blue) chain A, (pale green) chain B, (pink) the hydrophobic patches positioned at the dimer–dimer interface above the minor groove. The DNA represents the original 19-mer repressor site of MetJ (Garvie and Phillips 2000). (D) The ParD promoter sequence from −32 to +17, showing the inverted repeat (solid boxes) and the flanking half sites (half tone).
In order to visualize potential dimer–dimer interactions, the DNA-binding domain of ParD was superimposed onto the Arc–DNA complex (PDB code: 1par) as well as onto the MetJ–DNA complex (PDB code: 1mjo) consisting of two protein dimers bound to their operator DNA binding sites. Whereas the superposition of the ParD dimer onto the Arc repressor gave the best least-square fit (Table 2), the spacing of the binding sites in the Arc-operator is known to be 11 bp (Raumann et al. 1994). In contrast, the spacing of binding sites in the MetJ–DNA complex amounts to 8 bp, as observed for the inverted and direct repeat of the ParD operator sequence. The superposition of ParD dimers onto the MetJ–DNA complex positions the hydrophobic patches on the outside of helices A and A′ (Ala18, Ala 21, Leu 21) in close proximity (Fig. 5C). Interaction between adjacent ParD dimers bound to the operator sites is a likely mechanism for cooperative binding.
Discussion
The two different regions of ParD are devoted to two different functions
The solution structure of the homodimeric ParD protein presented here further confirms previous reports that ParD is organized in at least two structurally and functionally distinct regions. In this study, we show that the well-ordered N-terminal domain adopts the RHH fold whereas the C-terminal region is highly flexible in solution. These data are in excellent agreement with the two biological roles of ParD, namely its repressor and its antidote function. They also agree with biochemical data showing that the DNA-binding function resides in the N-terminal part of the protein whereas the toxin binding activity is suggested to reside in the C-terminal part. An N-terminal ParD mutant (ParD10) with a five-residue insertion after Asp10 was shown to have lost its DNA-binding activity but still functioned effectively in plasmid stabilization (Roberts et al. 1993). Similar to ParD, other proteins adopting the RHH fold also encode additional biological functions in the additional C-terminal parts, as in case of CcdA, MetJ, and Mnt, with the binding activity to the toxin, the corepressor SAM, and the tetramerization region, respectively. On the other hand Arc, CopG, and ω contain just the ribbon-helix-helix domain.
It is now well established that many proteins are intrinsically unstructured (for a recent review, see Dyson and Wright 2005). Many of these unfolded proteins or domains do not exist as statistical random coils but have propensities for defined conformational structures upon binding to the molecular target (specific nucleotide sequences, proteins, or small molecules). ParD seems to exhibit such features with its division into a highly ordered N-terminal domain and the less structured C-terminal domain with its propensities for the formation of α-helical regions. The internal mobility and regions of local structure of ParD were assessed previously (Oberer et al. 2002) by relaxation experiments with 15N nuclei and heteronuclear 1H-15N NOEs. Obviously, the backbone dynamics of amino acid residues located in the well-structured part of ParD and in the unstructured C terminus of the protein showed rather different behavior. An important biological implication of unstructured proteins is the respective half-life in vivo. The proteic and RNA-based killing modules are very much dependent on different stabilities of the toxin and the antidotes in the bacterial cells. The unstructured C-terminal half of ParD provides a vulnerable region for degradation of the antitoxin since unfolded proteins are far more accessible to proteolytic attack than densely packed regions of well-ordered domains. The crystal structure of MazE alone (Loris et al. 2003) also shows that the protein is partly unfolded, with only 44 out of the 88 residues observed in the crystal structure. Other antidote proteins reveal quite similar properties: CcdA is partly unfolded in solution (Madl et al. 2006), and the thermodynamic stability of CcdA is low enough to keep the protein close to the unfolded state under in vivo conditions, thereby facilitating its easy degradation by the cellular protease (Dao-Thi et al. 2000). Even more drastically, entire antitoxin proteins such as Phd (Gazit and Sauer 1999) and YefM (Cherny and Gazit 2004) are reported to be unstructured. The crystal structure of the MazEF complex (Kamada et al. 2003) features the N-terminal β-barrel core structure with an extended C terminus of MazE that wraps around a MazF homodimer. This observation is in agreement with our preliminary CD spectroscopic data, where no significant increase in secondary structure elements can be observed in the complex compared with the CD spectra of ParD and ParE alone (data not shown).
The ParD–DNA model provides insights into cooperativity of binding and the mode of regulation of the PparDE promoter
The repression of the parD and parE gene products is regulated by the ParD protein level. In contrast to other toxin–antitoxin pairs, where the toxin plays a vital role as corepressor, it has been shown that ParD is sufficient for regulation of expression. Furthermore, it has been shown that ParD binds to its promoter in a concentration-dependent manner, occupying the central promoter region at submicromolar concentrations and covering the flanking regions at increasing concentrations, as shown by DNase I footprint analysis (Roberts et al. 1993). The promoter region contains an inverted repeat and one direct repeat. In addition there are two less conserved repeats flanking the inverted repeat region (Fig. 5D). We generated a ParD–DNA model using the central inverted repeat, showing that the ParD dimer can accommodate the putative binding site in a manner very similar to other RHH proteins with known structures (Fig. 5). Side chains (Arg3 and Thr5) of the N-terminal β-ribbon are responsible for base-specific interactions like in the Arc-repressor–DNA structure. In contrast to the Arc structure (Raumann et al. 1994), Arg3 and Thr5 contact the central A–T base pairs of the palindromic sequence (A−2T−1 • A1T2). Additional nonspecific interactions with the phosphoribose backbone are formed by side chains of Helix A, Thr26, and the main-chain amides of V33 and N34. This capping of Helix B by a phosphate group (C−3 in the ParD–DNA complex) appears to be one of the main features of the RHH–DNA interactions and has been observed in all RHH–DNA structures so far. In the case of the ω2–DNA complex, the investigators argued that the relative position of the β-ribbon to the ends of helices B determine the extent of bending of the DNA: A less protruding β-ribbon like that in the ω2–DNA complex would result in straight DNA, whereas a protruding β-ribbon enforces a bent DNA structure as observed in the Arc-, MetJ-, and CopG-complex structures (Weihofen et al. 2006). ParD exhibits a structural similarity to Arc, and it can be assumed that a similar DNA bending will be promoted upon binding.
Interestingly, a hydrophobic patch is observed at the outside of helices A and A′. The corresponding residues are obviously not involved in the formation of the hydrophobic core of the ParD dimer, and this patch is positioned above the minor groove of the DNA in the model of the ParD–DNA complex. In other RHH proteins like the ω- or the MetJ-repressor, this hydrophobic patch has also been observed and promotes a strong dimer–dimer interaction in the respective complex structures (Somers and Phillips 1992; Weihofen et al. 2006). Using the Arc–DNA complex as a template, the hydrophobic patches of two adjacent ParD dimers do not come in contact, because the spacing of the original Arc binding sites is 11 bp (Raumann et al. 1994). In the ParD promoter the distance of the half-sites to the center of the inverted repeat is 8 bp (Fig. 5D), comparable to the distance of binding sites of the MetJ repressor (8 bp) and the ω-repressor (7 bp). Therefore, we used the MetJ–DNA complex as a template for the superposition of the ParD structure to visualize that adjacent ParD dimers are appropriately placed along the dsDNA to allow this interaction (Fig. 5C). According to the concentration-dependent mode of DNA binding, we propose that the sites at the inverted and direct repeat are occupied by two or three ParD dimers in a cooperative manner (see also Supplemental Fig. S1), and that additional dimers are recruited to the flanking regions at higher concentrations of ParD only (as observed in Roberts et al. [1993]). This mode of covering the whole promoter region including the transcription start may be necessary for fine-tuning the regulation. The hydrophobic patch at the end of helix A is obviously not involved in the hydrophobic core and is the potential point of attachment for dimer–dimer interactions of DNA-bound molecules, similar to Val22 in CcdA (Madl et al. 2006).
The fusion of different DNA- and toxin-binding domains as observed in various TA modules provides arguments against a common ancestor theory
The discovery of TA systems gave rise to many questions concerning the evolution of these systems. Initially, a common ancestor to all TA systems was postulated, despite the diversity in sequence and different biological targets of the various toxins. A more recent systematic study of gene neighborhood and sequence profile searches of TA systems suggests that most TA systems have resulted from mixing and matching of a relatively small pool of antidotes and toxins (Anantharaman and Aravind 2003; Arcus et al. 2005): In situ displacement of the genes encoding either the toxin or the antidote ensured proper function of the TA module as long as the ability of the repressor and/or the cellular toxicity was ensured. Anantharaman and Aravind (2003) were also able to unify the functionally quite different toxin families defined by ParE (acting on replication) and RelE (acting on translation). While the investigators provided new evolutionary insights for different repressor–toxin combinations in general, the importance of the intact interaction between the antidote and the toxin remained elusive in their study. Most likely this gap will be filled in the future upon the availability of more biochemical and structural data targeting toxin–antitoxin interactions. However, it is tempting to speculate that the in situ displacement targeted not only the different genes, but also the C-terminal regions of the respective antidotes as separate moieties. The low sequence homology between full-length antidotes of the various addiction systems may reflect the necessity of developing high specificity for DNA and target binding, respectively, in order to be useful to the host organism. These (mostly unstructured) toxin-binding tails are clearly more restrained to the neighboring toxin than to the repressor. Sequence comparison of the unstructured moiety of ParD with the toxin-interacting region of RelB reveals similar charge distribution, suggesting a related mode of interaction between ParD and ParE as observed in the RelB–RelE complex (Takagi et al. 2005) and MazE–MazF complex (Kamada et al. 2003). In a very recent study, a C-terminal 24-residue acidic peptide of MazE was shown to be sufficient to bind strongly to the toxin MazF (Li et al. 2006). In both complex structures, the C-terminal part of the antitoxin wraps around the toxin and establishes an array of polar, van der Waals, and salt-bridge interactions. As such, the negative charges (Asp39, Asp41, Asp43) in the unstructured linker region and the following helix of ParD are reminiscent of the negatively charged, surface-exposed linker region (Glu31, Asp33, Asp35, and Glu40) of RelB, which interacts with a positive patch on the surface of RelE. The important relationship of the toxin-interaction moieties of the antidotes and the fact that repressor proteins also have other functions fused to their DNA-binding domain (e.g., MetJ, Mnt) could be indicative of an in situ displacement of the C-terminal region of the antitoxin concomitantly with its respective toxin. Such a “mix-and-match” scheme could explain the existence of TA systems with related toxins yet completely unrelated repressors within one operon, as identified for ParE and RelE (Anantharaman and Aravind 2003). Consequently, this relationship would completely uncouple the repressor from the toxin. Moreover, this evolutionary scheme would easily accommodate fully functional TA systems consisting of three separate proteins: the repressor, the antidote, and the toxin. For full functioning of the TA system, the expression levels of all three proteins have to be regulated in a coordinated manner. The ω/ɛ/ζ module (Zielenkiewicz and Ceglowski 2005) could be seen in light of this evolutionary branch. This addiction system comprises three proteins with experimentally determined 3D structures: the repressor molecule ω, adopting the RHH fold; the antidote ɛ; and the toxic protein ζ (Meinhart et al. 2003; Weihofen et al. 2006). Clearly, a more definite answer on the evolution of TA systems can be provided only when more TA systems are characterized comprehensively.
Materials and Methods
Protein expression and purification
Unlabeled and uniformly 15N- and 13C-labeled ParD protein was expressed and purified in Escherichia coli as previously described (Oberer et al. 2002). Three types of ParD samples were generated for the structural studies. One sample was uniformly 15N-labeled, one was uniformly 15N/13C-labeled, and another was a 1:1 mixture of uniformly 15N/13C-labeled and unlabeled protein. The mixing of labeled and unlabeled protein was carried out by thermally denaturing and refolding, employing the refolding properties of ParD described earlier (Oberer et al. 1999, 2002). A 1:2:1 equilibrium for unlabeled, asymmetrically labeled, and fully labeled protein was assumed after slow cooling to room temperature. The protein samples were dialyzed against 20 mM potassium phosphate buffer, pH 6.0, containing 50 mM KCl and 0.1% NaN3. The dialyzed samples were concentrated to ∼0.6 mM, and the NMR spectroscopic studies were performed in this buffer after the addition of 10% D2O.
NMR spectroscopy
All NMR data were acquired on a Varian Unity INOVA 600 MHz spectrometer equipped with a triple-resonance z-gradient probe at 30°C. On the basis of the two- and three-dimensional experiments used for the complete sequential assignment of the protein (Oberer et al. 2002), the NOE connectivities were assigned with the help of 2D NOESY, 3D 15N-edited NOESY, 3D 13C-edited NOESY, 3D 13C-edited NOESY optimized for the aromatic region, and a 3D 15N,13C-edited, 15N,13C-filtered NOESY-HSQC for the separation of intermolecular from intramolecular NOEs. The latter experiment relies on an efficient X-filter to suppress 12C- and 14N-bound protons. 13C-bound 1H magnetization is observed after a NOESY period in a 3D-13C-filtered, 13C-edited NOESY-HSQC experiment. In the resulting spectrum of a molecular complex consisting of a 13C,15N double-labeled and an unlabeled protein, only intermolcular NOEs are detected (Zangger et al. 2003). The spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed using the program NMRView (Johnson 2004).
Structure calculation
Resolved cross-peaks in all NOESY experiments were picked and assigned manually. Subsequently, they were integrated using NMRView and converted into distance restraints employing the built-in feature of the display program. The restraints were classified as strong, medium, weak, and very weak, and the corresponding upper bounds were set to 2.8, 3.4, 5.0, and 6.0 Å, respectively. The lower distance bounds were all effectively 1.8 Å.
To assist in the resonance assignment procedure for structure calculation, we also built a model of ParD using the comparative modeling program MODELER4 (Marti-Renom et al. 2000). The aligned and modeled sequences comprised residues Met1 to Trp46 of ParD. Hydrogen atoms were added, and the energies were minimized using SYBYL, version 7.1 (Tripos Inc.).
The assignment procedure of the NOE cross-peaks and structure calculation was performed in two stages: First, intramolecular NOEs, which unambiguously did not arise from an intermolecular contact, were taken as input in the standard protocol for simulated annealing of CNS (version 1.1). The calculation was started from an extended monomeric structure (Brunger et al. 1998). These peaks were identified and assigned based on the proximity in the amino acid sequence (intraresidual peaks, short-range NOEs typical for the corresponding structure elements as determined via secondary structure predictions, NOE patterns, chemical shift index (CSI) [Wishart and Sykes 1994], and TALOS [Cornilescu et al. 1999]). Long-range intramolecular NOEs were identified based on their absence in the 3D 15N,13C-edited, 15N,13C-filtered NOESY-HSQC (X-filtered) experiment (Zangger et al. 2003) and their agreement with the modeled structure. This monomeric structure was not completely refined but calculated merely to confirm the secondary structure elements of each chain.
In the second stage the biologically relevant dimeric structure of ParD was calculated: Residues with intermolecular long-range NOEs were identified and assigned using the newly developed efficient X-filtering on 2D and 3D spectra with the heterolabeled dimeric ParD protein. Inspection of the dimeric model of ParD independently obtained via comparative modeling corroborated their assignments.
Cross-peaks corresponding to permanently violated NOEs were re-examined and re-introduced correctly into the calculation. Some cross-peaks finally had to be excluded on the grounds of either spectral overlap that prevented proper assignment or of being too close to the noise level of the spectra. Additional distance restraints representing H-bonds inferred from slowly exchanging amide protons and observed secondary structural elements of ParD (Oberer et al. 2002) were obtained. ϕ torsion angles were derived from an HNHA experiment and were combined with ψ and ϕ torsion angles obtained from TALOS (Cornilescu et al. 1999) predictions.
These restraints were used as input files for the calculation of the dimeric structures with the simulated annealing protocol of the software package Crystallography & NMR System (CNS) version 1.1 (Brunger et al. 1998). Here, the calculation was started from two noninteracting extended structures of both chains. The resulting structures and Ramachandran plots were analyzed with CNS, PROCHECK-NMR (version 3.5.4), and AQUA (version 3.2) (Laskowski et al. 1996). Secondary structure elements and root mean square deviations were calculated using MOLMOL (version 2K.1) (Koradi et al. 1996). Hydrogen bonds, van der Waals interactions, and salt bridges were analyzed using HBPlus and CONTACSYM (Connolly 1983). Structure and folding similarity searches in the PDB (Berman et al. 2000) were performed using the DALI server (Holm and Sander 1993; http://www.ebi.ac.uk/dali/) and yielded the expected hits with the ribbon-helix-helix proteins CopG (pdb accession code 2cpg), Arc (1baz), NikR (1q5v), ω (1irq), and MetJ (1cmb) (with Z-scores between 3.4 and 2.1). The coordinates of 24 3D structures with lowest energies were deposited in the Protein Data Bank (http://www.rcsb.org/pdb/) under the accession code 2AN7.
ParD—DNA model
For the ParD–DNA model, a 10-bp palindromic DNA sequence 5′-CACATATGTG-3′ was used that is identical to the inverted repeat from the ParD operator (positions −23 to −14) (Davis et al. 1992). The initial model was built by least-square fitting ParD (residues Arg3 to Leu35 of both chains) onto one Arc-repressor dimer complexed with its cognate DNA using the program MOLMOL (Koradi et al. 1996). The dsDNA comprising the nonpalindromic Arc-binding site was extracted from the Arc–DNA complex structure (1par.pdb) and mutated into the palindromic ParD binding site using the SYBYL program suite. The DNA structure was regularized by energy minimization and combined with the ParD dimer, comprising the DNA-binding domain only (residues 1–40). Hydrogen atoms were added at calculated positions and charges were calculated in SYBYL using the Gasteiger–Hückel method. Several rounds of energy minimization were performed with intermittent visual checks and manual adjustment of side chains involved in clashes or exhibiting poor conformation. The quality of the final energy-minimized model was analyzed with PROCHECK (Laskowski et al. 1993).
For the superposition of the ParD dimers with the MetJ–DNA complex (Garvie and Phillips 2000), the ParD dimers (residues 3–36 of both chains) were least-square fitted to the corresponding residues of the MetJ dimers (chains A–B and chains C–D).
Acknowledgments
We thank Donald Helinski for expression strains and Stefan Prytulla for help at the initial stages of the project. This work was supported by the Austrian Science Fund (FWF) projects P15040, P19794 (W.K.) and the EU-FP5 project QLK2-2000-31624 (W.K.).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Walter Keller, Institut für Chemie, Arbeitsgruppe Strukturbiologie, Karl-Franzens-Universität Graz, Heinrichstrasse 28, A-8010 Graz, Austria; e-mail: walter.keller@uni-graz.at; fax: 43–316–380-9850.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062680707.
References
- Anantharaman V. and Aravind, L. 2003. New connections in the prokaryotic toxin–antitoxin network: Relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 4: R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus V.L., Rainey, P.B., and Turner, S.J. 2005. The PIN-domain toxin–antitoxin array in mycobacteria. Trends Microbiol. 13: 360–365. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P. 1996. Positive selection of recombinant DNA by CcdB. Biotechniques 21: 320–323. [DOI] [PubMed] [Google Scholar]
- Bernard P. and Couturier, M. 1991. The 41 carboxy-terminal residues of the miniF plasmid CcdA protein are sufficient to antagonize the killer activity of the CcdB protein. Mol. Gen. Genet. 226: 297–304. [DOI] [PubMed] [Google Scholar]
- Breg J.N., van Opheusden, J.H., Burgering, M.J., Boelens, R., and Kaptein, R. 1990. Structure of Arc repressor in solution: Evidence for a family of β-sheet DNA-binding proteins. Nature 346: 586–589. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Burgering M.J., Boelens, R., Gilbert, D.E., Breg, J.N., Knight, K.L., Sauer, R.T., and Kaptein, R. 1994. Solution structure of dimeric Mnt repressor (1–76). Biochemistry 33: 15036–15045. [DOI] [PubMed] [Google Scholar]
- Buts L., Lah, J., Dao-Thi, M.H., Wyns, L., and Loris, R. 2005. Toxin–antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30: 672–679. [DOI] [PubMed] [Google Scholar]
- Cherny I. and Gazit, E. 2004. The YefM antitoxin defines a family of natively unfolded proteins: Implications as a novel antibacterial target. J. Biol. Chem. 279: 8252–8261. [DOI] [PubMed] [Google Scholar]
- Connolly M.L. 1983. Solvent-accessible surfaces of proteins and nucleic acids. Science 221: 709–713. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Dao-Thi M.H., Messens, J., Wyns, L., and Backmann, J. 2000. The thermodynamic stability of the proteins of the ccd plasmid addiction system. J. Mol. Biol. 299: 1373–1386. [DOI] [PubMed] [Google Scholar]
- Davis T.L., Helinski, D.R., and Roberts, R.C. 1992. Transcription and autoregulation of the stabilizing functions of broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens and Pseudomonas aeruginosa . Mol. Microbiol. 6: 1981–1994. [DOI] [PubMed] [Google Scholar]
- de la Cueva-Mendez G. 2003. Distressing bacteria: Structure of a prokaryotic detox program. Mol. Cell 11: 848–850. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- del Solar G., Hernandez-Arriaga, A.M., Gomis-Ruth, F.X., Coll, M., and Espinosa, M. 2002. A genetically economical family of plasmid-encoded transcriptional repressors involved in control of plasmid copy number. J. Bacteriol. 184: 4943–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H.J. and Wright, P.E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Eberl L., Givskov, M., and Schwab, H. 1992. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol. Microbiol. 6: 1969–1979. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H. and Glaser, G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53: 43–70. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Sat, B., Reches, M., Amitai, S., and Hazan, R. 2004. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 12: 66–71. [DOI] [PubMed] [Google Scholar]
- Gabant P., Van Reeth, T., Dreze, P.L., Faelen, M., Szpirer, C., and Szpirer, J. 2000. New positive selection system based on the parD (kis/kid) system of the R1 plasmid. Biotechniques 28: 784–788. [PubMed] [Google Scholar]
- Garvie C.W. and Phillips, S.E. 2000. Direct and indirect readout in mutant Met repressor–operator complexes. Structure 8: 905–914. [DOI] [PubMed] [Google Scholar]
- Gazit E. and Sauer, R.T. 1999. Stability and DNA binding of the phd protein of the phage P1 plasmid addiction system. J. Biol. Chem. 274: 2652–2657. [DOI] [PubMed] [Google Scholar]
- Gerdes K. 2000. Toxin–antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Christensen, S.K., and Lobner-Olesen, A. 2005. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 3: 371–382. [DOI] [PubMed] [Google Scholar]
- Golovanov A.P., Barilla, D., Golovanova, M., Hayes, F., and Lian, L.Y. 2003. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50: 1141–1153. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F.X., Sola, M., Acebo, P., Parraga, A., Guasch, A., Eritja, R., Gonzalez, A., Espinosa, M., del Solar, G., and Coll, M. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 17: 7404–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D., Santos-Sierra, S., Giraldo, R., Sabariegos-Jareno, R., de la Cueva-Mendez, G., Boelens, R., Diaz-Orejas, R., and Rafferty, J.B. 2002. Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure 10: 1425–1433. [DOI] [PubMed] [Google Scholar]
- Hayes F. 2003. Toxins–antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301: 1496–1499. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Pogliano, J., Helinski, D.R., and Konieczny, I. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44: 971–979. [DOI] [PubMed] [Google Scholar]
- Johnson B.A. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278: 313–352. [DOI] [PubMed] [Google Scholar]
- Kamada K. and Hanaoka, F. 2005. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19: 497–509. [DOI] [PubMed] [Google Scholar]
- Kamada K., Hanaoka, F., and Burley, S.K. 2003. Crystal structure of the MazE/MazF complex: Molecular bases of antidote–toxin recognition. Mol. Cell 11: 875–884. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 29–32. [DOI] [PubMed] [Google Scholar]
- Lah J., Marianovsky, I., Glaser, G., Engelberg-Kulka, H., Kinne, J., Wyns, L., and Loris, R. 2003. Recognition of the intrinsically flexible addiction antidote MazE by a dromedary single domain antibody fragment. Structure, thermodynamics of binding, stability, and influence on interactions with DNA. J. Biol. Chem. 278: 14101–14111. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Laskowski R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8: 477–486. [DOI] [PubMed] [Google Scholar]
- Lehnherr H., Maguin, E., Jafri, S., and Yarmolinsky, M.B. 1993. Plasmid addiction genes of bacteriophage P1:doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233: 414–428. [DOI] [PubMed] [Google Scholar]
- Li G.Y., Zhang, Y., Chan, M.C., Mal, T.K., Hoeflich, K.P., Inouye, M., and Ikura, M. 2006. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J. Mol. Biol. 357: 139–150. [DOI] [PubMed] [Google Scholar]
- Loris R., Dao-Thi, M.H., Bahassi, E.M., Van Melderen, L., Poortmans, F., Liddington, R., Couturier, M., and Wyns, L. 1999. Crystal structure of CcdB, a topoisomerase poison from E. coli . J. Mol. Biol. 285: 1667–1677. [DOI] [PubMed] [Google Scholar]
- Loris R., Marianovsky, I., Lah, J., Laeremans, T., Engelberg-Kulka, H., Glaser, G., Muyldermans, S., and Wyns, L. 2003. Crystal structure of the intrinsically flexible addiction antidote MazE. J. Biol. Chem. 278: 28252–28257. [DOI] [PubMed] [Google Scholar]
- Madl T., Van Melderen, L., Mine, N., Respondek, M., Oberer, M., Keller, W., Khatai, L., and Zangger, K. 2006. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J. Mol. Biol. 364: 170–185. [DOI] [PubMed] [Google Scholar]
- Marti-Renom M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. 29: 290–325. [DOI] [PubMed] [Google Scholar]
- Mattison K., Wilbur, J.S., So, M., and Brennan, R.G. 2006. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin–antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 281: 37942–37951. [DOI] [PubMed] [Google Scholar]
- McKinley J.E. and Magnuson, R.D. 2005. Characterization of the Phd repressor–antitoxin boundary. J. Bacteriol. 187: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A., Alonso, J.C., Strater, N., and Saenger, W. 2003. Crystal structure of the plasmid maintenance system ɛ/ζ: Functional mechanism of toxin ζ and inactivation by ɛ 2 ζ 2 complex formation. Proc. Natl. Acad. Sci. 100: 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K., Orth, P., de la Hoz, A.B., Alonso, J.C., and Saenger, W. 2001. Crystal structure of ω transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J. Mol. Biol. 314: 789–796. [DOI] [PubMed] [Google Scholar]
- Oberer M., Lindner, H., Glatter, O., Kratky, C., and Keller, W. 1999. Thermodynamic properties and DNA binding of the ParD protein from the broad host-range plasmid RK2/RP4 killing system. Biol. Chem. 380: 1413–1420. [DOI] [PubMed] [Google Scholar]
- Oberer M., Zangger, K., Prytulla, S., and Keller, W. 2002. The anti-toxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA-binding proteins. Biochem. J. 361: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey D.P. and Gerdes, K. 2005. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecota D.C., Kim, C.S., Wu, K., Gerdes, K., and Wood, T.K. 1997. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 63: 1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Zavialov, A.V., Pavlov, M.Y., Elf, J., Gerdes, K., and Ehrenberg, M. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140. [DOI] [PubMed] [Google Scholar]
- Phillips S.E. 1994. The β-ribbon DNA recognition motif. Annu. Rev. Biophys. Biomol. Struct. 23: 671–701. [DOI] [PubMed] [Google Scholar]
- Popescu A., Karpay, A., Israel, D.A., Peek Jr, R.M., and Krezel, A.M. 2005. Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators. Proteins 59: 303–311. [DOI] [PubMed] [Google Scholar]
- Rafferty J.B., Somers, W.S., Saint-Girons, I., and Phillips, S.E. 1989. Three-dimensional crystal structures of Escherichia coli met repressor with and without corepressor. Nature 341: 705–710. [DOI] [PubMed] [Google Scholar]
- Raumann B.E., Rould, M.A., Pabo, C.O., and Sauer, R.T. 1994. DNA recognition by β-sheets in the Arc repressor–operator crystal structure. Nature 367: 754–757. [DOI] [PubMed] [Google Scholar]
- Roberts R.C., Spangler, C., and Helinski, D.R. 1993. Characteristics and significance of DNA binding activity of plasmid stabilization protein ParD from the broad host-range plasmid RK2. J. Biol. Chem. 268: 27109–27117. [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Berzal-Herranz, A., Gerdes, K., and Diaz-Orejas, R. 1991a. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol. Microbiol. 5: 2685–2693. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., de Torrontegui, G., Gimenez-Gallego, G., and Diaz-Orejas, R. 1991b. Structural and functional comparison between the stability systems ParD of plasmid R1 and Ccd of plasmid F. Mol. Gen. Genet. 225: 355–362. [DOI] [PubMed] [Google Scholar]
- Salmon M.A., Van Melderen, L., Bernard, P., and Couturier, M. 1994. The antidote and autoregulatory functions of the F plasmid CcdA protein: A genetic and biochemical survey. Mol. Gen. Genet. 244: 530–538. [DOI] [PubMed] [Google Scholar]
- Santos-Sierra S., Pardo-Abarrio, C., Giraldo, R., and Diaz-Orejas, R. 2002. Genetic identification of two functional regions in the antitoxin of the parD killer system of plasmid R1. FEMS Microbiol. Lett. 206: 115–119. [DOI] [PubMed] [Google Scholar]
- Schreiter E.R., Sintchak, M.D., Guo, Y., Chivers, P.T., Sauer, R.T., and Drennan, C.L. 2003. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10: 794–799. [DOI] [PubMed] [Google Scholar]
- Smith J.A. and Magnuson, R.D. 2004. Modular organization of the Phd repressor/antitoxin protein. J. Bacteriol. 186: 2692–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W.S. and Phillips, S.E. 1992. Crystal structure of the met repressor–operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature 359: 387–393. [DOI] [PubMed] [Google Scholar]
- Takagi H., Kakuta, Y., Okada, T., Yao, M., Tanaka, I., and Kimura, M. 2005. Crystal structure of archaeal toxin–antitoxin RelE–RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 12: 327–331. [DOI] [PubMed] [Google Scholar]
- Van Melderen L., Thi, M.H., Lecchi, P., Gottesman, S., Couturier, M., and Maurizi, M.R. 1996. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J. Biol. Chem. 271: 27730–27738. [DOI] [PubMed] [Google Scholar]
- Weihofen W.A., Cicek, A., Pratto, F., Alonso, J.C., and Saenger, W. 2006. Structures of ω repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 34: 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. and Sykes, B.D. 1994. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4: 171–180. [DOI] [PubMed] [Google Scholar]
- Zangger K., Oberer, M., Keller, W., and Sterk, H. 2003. X-filtering for a range of coupling constants: Application to the detection of intermolecular NOEs. J. Magn. Reson. 160: 97–106. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz U. and Ceglowski, P. 2005. The toxin–antitoxin system of the streptococcal plasmid pSM19035. J. Bacteriol. 187: 6094–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]