Abstract
We have determined, by high resolution x-ray analysis, 10 structures comprising the mRNA cap-specific methyltransferase VP39 or specific mutants thereof in the presence of methylated nucleobase analogs (N1-methyladenine, N3-methyladenine, N1-methylcytosine, N3-methylcytosine) and their unmethylated counterparts, or nucleoside N7-methylguanosine. Together with solution affinity studies and previous crystallographic data for N7-methylguanosine and its phosphorylated derivatives, these data demonstrate that only methylated, positively charged bases are bound, indicating that their enhanced stacking with two aromatic side chains of VP39 (Tyr 22 and Phe 180) plays a dominant role in cap recognition. Four key features characterize this stacking interaction: (i) near perfect parallel alignment between the sandwiched methylated bases and aromatic side chains, (ii) substantial areas of overlap in the two-stacked rings, (iii) a 3.4-Å interplanar spacing within the overlapping region, and (iv) positive charge in the heterocyclic nucleobase.
The ability of proteins to discriminate alkylated from nonalkylated nucleic acids is of central importance in numerous biological processes (1, 2). VP39, a vaccinia virus protein, is an excellent system with which to study protein interactions with methylated nucleobases and mRNA recognition. In addition to serving as a processivity factor for the vaccinia poly(A) polymerase (3), VP39 acts at the mRNA 5′ end as a cap 0 [N7-methylguanosine (m7G) (5′)pppN⋅⋅⋅)]-specific (nucleoside-2′-O-)-methyltransferase (4). To perform this latter function, VP39 must specifically recognize the N7-methylguanine (m7Gua) moiety of the cap nucleotide while also binding the mRNA transcript in a sequence-nonspecific manner.
High resolution structures for VP39 complexed with its S-adenosylmethionine coenzyme product S-adenosylhomocysteine plus capped nucleotides m7G(5′)pppG, m7G(5′)pp, m7G(5′)p, or single-stranded RNA hexamer [m7G(5′)pppG(A)5] revealed a paradigm for alkylated nucleobase recognition and for the sequence-independent binding of single stranded mRNA (5–7). From these studies, three factors emerged as contributors to VP39’s specific recognition of the cap’s m7Gua moiety: (i) van der Waals contacts with the 7-methyl group itself; (ii) electrostatic interactions with guanine-specific polar functionalities, via hydrogen bonds and salt links; and (iii) enhanced stacking interactions arising from partial π orbital or charge transfer interactions between the highest occupied molecular orbital of each of the two donor rings (i.e., the aromatic side chains of Tyr 22 and Phe 180) and the lowest unoccupied molecular orbital of the acceptor ring [the N(7)-methylated guanine moiety, which possesses a net positive charge at pH values <7.5 (8)]. Similar features have more recently been observed in the structure of eIF4E, where m7G(5′)pp is found sandwiched between two Trp residues (9). The extension of our studies to complexes of wild-type and active mutant VP39 with other methylated, positively charged bases, as described herein, provides evidence that the major determinant of mRNA cap recognition is the enhanced stacking interaction. Additional features of methylated-base binding will also be described.
MATERIALS AND METHODS
Materials.
Site-directed mutations in the VP39 gene were made as described (10). VP39-ΔC26 and mutants thereof (Table 1) were expressed and purified as described (5). Bases, nucleosides, and nucleotides (Tables 1 and 2) were purchased from Sigma-Aldrich.
Table 1.
Statistics of crystallographic data and structure refinement
| Protein | Ligand, 10 mM | Nucleobase density* | Resolution, Å | Intensity data
|
R-factor | R-free | |
|---|---|---|---|---|---|---|---|
| R-sym, % | Completeness, % | ||||||
| VP39 | m3Ade | Complete | 1.83 | 5.1 | 88.2 | 0.217 | 0.251 |
| VP39 | m1Ade | Complete | 2.2 | 5.5 | 77.7 | 0.206 | 0.256 |
| VP39 | Ade | None | 1.92 | 8.2 | 90.7 | 0.236 | 0.251 |
| VP39 | m3Cyt | Complete | 2.0 | 6.7 | 98.8 | 0.234 | 0.285 |
| VP39 | m1Cyt | Complete | 1.86 | 8.8 | 90.7 | 0.227 | 0.265 |
| VP39 | Cyt | None | 1.86 | 6.7 | 90.2 | 0.218 | 0.261 |
| Y22A | m7G | None | 2.2 | 9.6 | 100 | 0.243 | 0.295 |
| D182A | m7G | Complete | 1.83 | 5.1 | 91.8 | 0.225 | 0.258 |
| E233A | m7G | None | 2.0 | 6.2 | 79.3 | 0.222 | 0.268 |
| E233Q | m7G | Complete | 2.2 | 4.7 | 89.7 | 0.211 | 0.255 |
Table 2.
VP39-F180W binding affinities for m7G, m7G(5′)pp, and methylated nucleobases
| Protein mutant | Ligand | Kd, μM* |
|---|---|---|
| F180W | m7G | 7.6 |
| F180W | m7G(5′)pp | 14.6 |
| F180W | m7G(5′)pp† | 19.1 |
| F180W | m3Ade | 86.0 |
| F180W | m1Ade | 97.9 |
| F180W | m3Cyt | 85.5 |
| F180W | m1Cyt | 144.0 |
Determined by fluorescence titration at pH 5.8 (see Materials and Methods).
Reduced and reoxidized.
Structure Determination.
Co-crystals of VP39-ΔC26 and its mutants (Table 1) with bound S-adenosylhomocysteine were obtained as described (6). Crystals of mutant VP39 proteins were isomorphous with that of VP39-ΔC26 (5). For structure determination, one or two crystals were soaked in each nucleobase solution (Table 1). Soaking solutions were made by mixing 5 μl of 100 mM base (dissolved in 0.25 M HCl) with 1 μl of 100 mM S-adenosylhomocysteine in 5 mM HCl and 44 μl of 15% polyethylene glycol 8000 in 0.125 M ammonium sulfate and 0.1 M Na cacodylate (pH 6.5). The pHs of the resulting mixtures were typically between 5.6 and 5.8. Soaks were typically incubated for 2 days at room temperature. For x-ray data collection, crystals were flash-frozen in a cryoprotectant comprising the soaking solution (pH 5.8) and 25% glycerol at −160°C by using a cryogenic cooler. With the exception of the data set from the crystallized complex between VP39-E233Q and m7G, which was collected by using a Nonius (Bohemia, NY) DIP2030 two imaging plate detector system with double-mirror focusing optics, all data were collected by using a Siemens (Iseline, NJ) SMART 2K charge-coupled device detector with Göbel mirror (Table 1). Each detector system was mounted on a Rigaku (Tokyo) RU2000 rotating anode (Cu Kα) operated at 100 mA and 50 kV. Minimum resolution for the data collection was set at 9,999 Å on the charge-coupled device and 90 Å on the DIP2030. The software packages siemens saint and denzo-scalepack (11) were used to reduce and scale the charge-coupled device and DIP2030 data, respectively. Resolution and R-sym statistics for the various data sets are shown in Table 1.
The structures were phased directly by using the VP39-ΔC26 structure complexed with S-adenosylhomocysteine and with water molecules excluded (5). Data used in the refinement extended from 8 Å to the highest resolution, as indicated in Table 1. Water molecules were added to the models, and the structures were then refined through multiple rounds of positional and temperature-factor refinement by using the program xplor (12). Bound ligand was fitted to density in the final stages of the refinement. The final values of the R-factor and associated R-free for the refined structures are shown in Table 1. rms deviations for bond lengths vary from 0.010 to 0.015 Å, and bond angles vary from 1.550 to 1.804 degrees. The difference electron density maps shown in Figs. 1, 2, and 3 were calculated from the resulting refined structures after removing the coordinates for bound ligand. Figs. 1 a, b, and d, 2 a, b, and d, and 3 b, c, and d were generated by using midas (13) and Figs. 1c, 2c, and 3a were generated by using grasp (14).
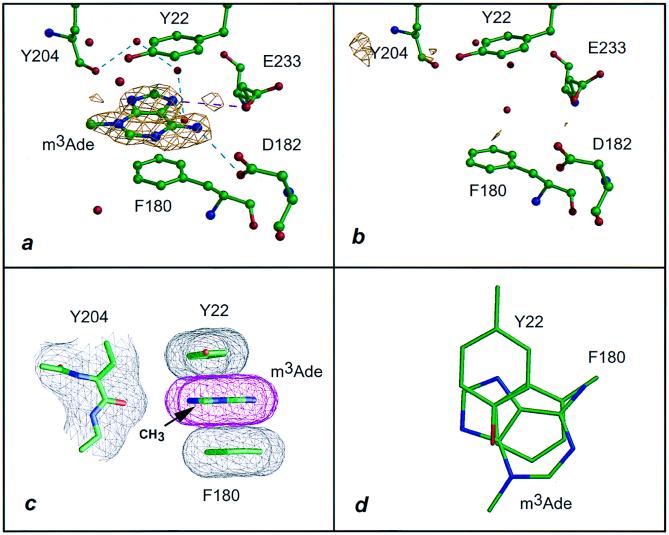
Figure 1.
Interaction of VP39 with m3Ade. (a) The 1.83-Å difference electron density of VP39 crystal soaked in 10 mM m3Ade contoured at the 2.5σ level (see Table 1 and Materials and Methods). Dashed lines represent hydrogen bonds (cyan) and salt links (magenta). (b) Equivalent to a but soaked in 10 mM adenine and with a 1.92-Å difference Fourier (Table 1). (c) Stacking of the adeninium ring of m3Ade with the aromatic side chains of Tyr 22 and Phe 180, projected parallel to the base ring. The atomic surface is shown in mesh representation. The (N)3-methyl group lies 6 Å from the peptide carbonyl oxygen of Tyr 204. (d) Same as c but projected perpendicular to the base ring with surface mesh and peptide backbone omitted for clarity.
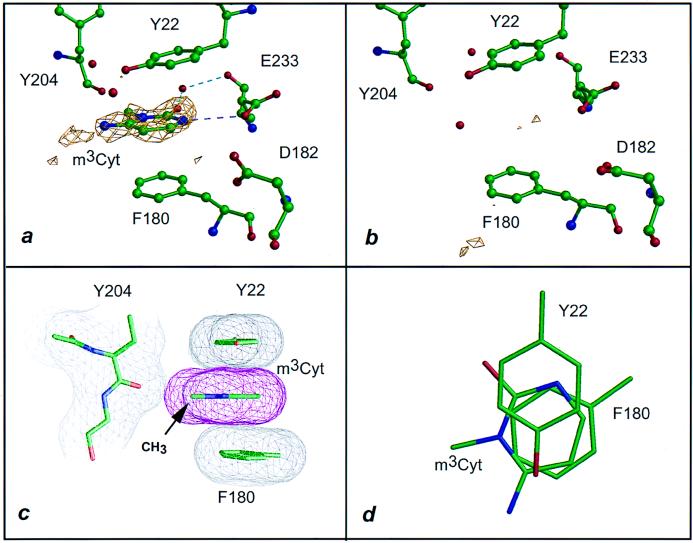
Figure 2.
Interaction of VP39 with m3Cyt. Dashed lines are colored as in Fig. 1. (a) The 1.86-Å difference electron density (2.5σ level) for VP39 crystal soaked in 10 mM m3Cyt (see Table 1 and Materials and Methods). (b) Identical to a but soaked in 10 mM Cyt (Table 1). (c) Stacking of cytosinium ring patterned after Fig. 1c. The (N)1-methyl group lies within 3.3 Å of the carbonyl oxygen of Tyr 204. (d) Same as c but fashioned after Fig. 1d.
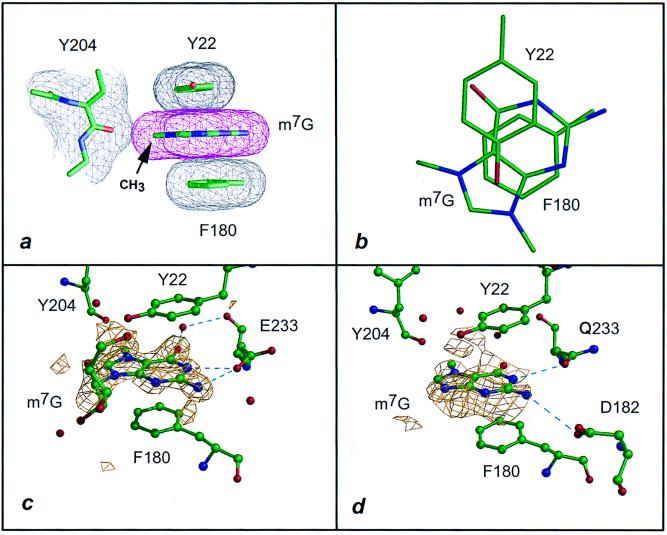
Figure 3.
Interaction of VP39 with the m7Gua moiety of m7G. Dashed lines are colored as in Fig. 1. (a) Stacking interaction of the methylated guaninium ring, fashioned after Fig. 1c (data from ref. 6, where this structure was mentioned but not shown). The methyl group lies within 3.1 Å of the carbonyl oxygen of Tyr 204. (b) Same as a but patterned after Fig. 1d. Note that, in a and b, the substituent on the N9 position of the base corresponds to the C1′ atom of the ribose. (c) The 1.83-Å difference electron density (2.5σ level) of m7G bound to VP39 mutant D182A (Table 1). As previously observed in the wild-type VP39-m7G structure (6), the ribose density is weak. (d) VP39 mutant E233Q soaked with m7G. The 2.2-Å difference electron density is shown for only the m7Gua moiety of m7G, contoured at the 2σ level (Table 1).
Seven sets of coordinates have been deposited in the Protein Data Bank, corresponding to the following complexes: VP39-ΔC26 with N3-methyladenine (m3Ade), m1Ade, N3-methylcytosine (m3Cyt), and m1Cyt (3mag, 1b42, 3mct, and 1bky, respectively); the D182A and E233Q mutants with m7G (4dcg and 1eqa, respectively); and the E233A mutant without bound ligand (1eam). Corresponding statistical details are given in Table 1.
Fluorescence Titration.
Fluorescence titration experiments used protocols very similar to those used with other proteins in our laboratory (e.g., refs. 15 and 16). By using an SLM–Aminco (Urbana, IL) model 4800 spectrofluorimeter with an excitation wavelength of 282 nm, titrations were conducted at 20°C by following the decrease in fluorescence at 330 nm after adding microliter aliquots of concentrated ligand solution to a 2 ml solution of 2.5 μM protein in 0.2 M NaCl and 0.1 M sodium cacodylate (pH 5.8). To correct for possible photooxidation, the fluorescence decrease at 330 nm was measured for an equivalent protein solution to which aliquots of ligand buffer alone had been added. For each addition of ligand or buffer, 10 individual measurements were recorded and averaged. Kd values were determined by Scatchard analysis. The fluorescence studies are described elsewhere (G.H., P.D.G., and F.A.Q., unpublished work).
Nucleobase pKa Determination.
Equivalent amounts of concentrated ligand solution were added to 1-ml aliquots of each of the following seven buffers (0.1 M): sodium cacodylate (pH 5.5, 6.0, 6.5), Hepes-NaOH (pH 7.0, 7.5), and Tris⋅HCl (pH 8.0, 8.5). The resulting solutions were scanned at wavelengths between 240 and 330 nm by using a UV-VIS Scanning Spectrophotometer (Shimadzu). pH changes correlated with significant changes in intensity and position of the major absorption peak. pKa values were equated to the midpoints in plots of pH vs. integrated peak area.
Reduction of m7G(5′)pp to 8-hydro-m7G(5′)pp and Subsequent Reoxidation.
m7G(5′)pp was reduced to 8-hydro-m7G(5′)pp by treatment with potassium borohydride as described (17). A sample of the reduced compound was reoxidized with H2O2, also as described (17).
RESULTS AND DISCUSSION
VP39 can bind a variety of methylated nucleobases (Table 1) in the pocket previously identified as the binding site for the m7Gua portion of the cap dinucleotide m7G(5′)pppG and its nucleoside or nucleotide derivatives (6). This is clearly evident in Figs. 1a and 2a, which show the difference electron densities for bound m3Ade and m3Cyt, respectively. Similar well defined densities were observed for VP39-bound m1Ade and m1Cyt (Table 1; densities not shown). The x-ray structures were determined at pH 5.6–5.8, at which the heterocyclic ring of the bound ligand would be predominantly in the positively charged form. The structures (purine or pyrimidine), arrangements of polar groups, and locations of the methyl substituent differed for each methylated base. Nonetheless, the four methylated bases, each possessing a positive charge delocalized throughout the aromatic π ring as a common feature, interacted with the aromatic side chains of Tyr 22 and Phe 180 in equivalent two-stacked ring arrangements.
To assess VP39-binding by the various methylated bases in solution, we used a VP39 mutant in which Phe 180 [one of the two aromatic stacking residues (Fig. 1)] had been replaced with a Trp residue to provide a fluorescent reporter for ligand binding to the methylated nucleobase-binding pocket (see Materials and Methods). This mutation, as well as an equivalent substitution at Tyr 22, showed no apparent defects in methyltransferase catalytic rate or VP39-capped RNA interaction (A. Oguro and P.D.G., unpublished data). Solution binding data (Table 2) indicated Kd values for the complexes of VP39 with m7G and its phosphorylated derivatives or methylated nucleobases to be in the low-to-medium micromolar range. Electron density for Ade or Cyt (the unmethylated counterparts of the methylated bases used here) could not be detected in crystal-soaking experiments with VP39 (Table 1 and Figs. 1b and 2b), reflecting the absence of density observed previously for unmethylated cap dinucleotide [G(5′)pppG] or its mono and diphosphate derivatives [G(5′)p and G(5′)pp, respectively (6)]. Moreover, solution binding for the unmethylated bases was too weak to be accurately quantitated. Although the above experiments demonstrate an absolute requirement for the alkyl substituent, does the methyl group per se account for the discrimination between methylated and unmethylated bases? Our current studies clearly show that it does not. Thus, the N(7)-methyl group of m7G contacts only one protein atom: namely, the backbone carbonyl oxygen of Tyr 204, with which it makes a van der Waals contact at a distance of ≈3 Å (refs. 6 and 7; see also Fig. 3a). A similar interaction was observed for the methyl groups of m3Cyt (at a distance of 3.3 Å) (Fig. 2c) and m1Cyt (at a distance of 2.8 Å) (data not shown). However, no methyl–protein interaction whatsoever was observed for m3Ade (Fig. 1c), whose methyl group lay ≈6 Å from the Tyr 204 carbonyl oxygen.
Several lines of evidence argue against hydrogen-bonding interactions playing the dominant role in cap recognition. First, binding was not observed for unmethylated Ade, Cyt (Figs. 2b and 3b), or G(5′)pp or G(5′)pppG (6). Second, VP39 makes no direct hydrogen bonds with bound m1Cyt or m1Ade (Table 3). Although two hydrogen bonds are formed with m1Cyt and one with m1Ade via bound water molecules, the affinities for both nucleobases are similar to those for m3Cyt and m3Ade, each of which is engaged in a single direct hydrogen bond (Tables 2 and 3). Third, although the m7Gua portion of m7G forms fewer hydrogen bonds with the D182A mutant protein than with wild-type VP39 (Fig. 3c and Table 3), the mutant retains wild-type specificity for m7G-capped over uncapped RNA substrate (6, 18). With the exception of the mutated residue, the structures of the m7G-liganded wild-type and mutant proteins are essentially indistinguishable (comparing the data of Fig. 3c and ref. 6).
Table 3.
Hydrogen bonds and salt links between residues of VP39 and methylated bases or m7G
| Protein | Atom | Ligand | Atom | Distance, Å |
|---|---|---|---|---|
| Hydrogen bonds (≤3.5 Å) | ||||
| VP39 | Asp 182 OD1 | m3Ade | N(6)H2 | 3.4 |
| VP39 | m1Ade | None | ||
| VP39 | Glu 233 OE1 | m3Cyt | N(1)H | 3.3 |
| VP39 | m1Cyt | None | ||
| VP39* | Glu 233 OE1 | m7G | N(1)H | 3.0 |
| Glu 233 OE1 | N(2)H2 | 3.4 | ||
| Asp 182 OD1 | N(2)H2 | 3.2 | ||
| Asp 182 OD2 | N(2)H2 | 3.4 | ||
| D182A | Glu 233 OE1 | m7G | N(1)H | 2.8 |
| Glu 233 OE1 | N(2)H2 | 3.4 | ||
| E233Q | Gln 233 OE1 | m7G | N(1)H | 3.0 |
| Asp 182 OD1 | N(2)H2 | 3.5 | ||
| Salt links (≤4.0 Å) | ||||
| VP39 | Glu 233 OE1 | m1Ade | N(7) | 3.1 |
| VP39 | OE1 | m3Ade | N(7) | 4.0 |
| VP39 | OE1 | m1Cyt | N(3)H | 3.4 |
| VP39 | OE1 | m3Cyt | N(1)H | 3.3 |
| VP39* | OE1 | m7G | N(1)H | 3.0 |
| D182A | OE1 | m7G | N(1)H | 2.8 |
Data are from ref. 6.
pKa values below which the methylated base heterocycle is positively charged are known for m7G (8), m1Ade, and m3Cyt (19) and were determined here for m7G and each of the four nucleobases used in this study (Table 4). Each of the methylated nucleobases exhibits pKa values for protonation significantly above the pH (5.6–5.8) used for crystallographic soaking experiments. On binding of each of the methylated nucleobases, electrostatic interactions might therefore be possible. These might take the form of a salt link between the carboxylate side chain of Glu 233 and the positive charge delocalized in the nucleobase ring via the N(1)H of m7Guanosine, N(7) of m1Ade and m3Ade, N(3)H of m1Cyt, and N(1)H of m3Cyt (Table 3). However, the evidence indicates only a minor role for such salt links in cap recognition. First, comparison of Kd values for the complex between VP39-F180W and m7G in the absence of salt and at NaCl concentrations >0.1 M indicated that affinities were essentially unchanged or even slightly elevated at elevated ionic strength. This is contrary to the effect that might be expected if increased ionic strength were leading to the shielding of opposite formal charges. Second, the neutralization of Glu 233 by substitution with Gln did not eliminate m7G binding (Fig. 3d), the conformation of the cap-binding site in the m7G-liganded E233Q crystal structure being essentially identical to that of the liganded wild-type protein (6). Indeed, even the hydrogen-bonding of Glu 233’s OE1 with the N(1)H of m7G is recapitulated, being mimicked by the OE1 of the amide side chain of the Gln replacement (Table 3 and Fig. 3d). Given this degree of structural mimicry, it is, therefore, not surprising that the E233Q mutant retains cap-specific 2′-O-methyltransferase activity (data not shown).
Table 4.
pKa values for methylated nucleobases and m7G
Although the E233A substitution led to a loss of m7G binding (Table 1) and cap-specific RNA binding activity (6), the crystal structure of the E233A mutant indicates that this can be attributed to an altered conformation of Tyr 22, one of the two aromatics stacking with the m7Gua moiety. The altered conformation resulted from a disordering of the surrounding region (residues 25–30) [Table 1; data not shown, though atomic coordinates have been deposited in the Protein Data Bank (1eam)]. No such structural changes or disordering were observed with the E233Q mutant. Like VP39’s Glu 233 side chain, the Glu 103 side chain of translation initiation factor eIF4E also appears to be involved in hydrogen-bonding and charge-coupling interactions with a methylated base, as observed in a recent crystal structure of eIF4E with bound m7G(5′)pp (9). Although the phenotype of an E103A substitution mutant in eIF4E (20) resembles that of VP39’s E233A mutant with regard to its loss of ligand binding, it cannot be ascertained whether a similar structural anomaly underlies this similarity because no structural analysis for the eIF4E-E103A mutant has been reported.
We are left considering the stacking interaction, possibly reinforced by some degree of interplanar π–π charge transfer from the electron-rich aromatic side chains to the electron-deficient methylated bases as the dominant factor in cap recognition and binding. It should be emphasized that quaternization of the base by methylation lowers its lowest unoccupied molecular orbital energy closer to the highest occupied molecular orbital energy of the aromatic side chains and, consequently, would be expected to promote and strengthen the stacking interaction. The requirement of a positively charged nucleobase is buttressed by the finding that reduction of m7G(5′)pp to 8-hydro-m7G(5′)pp (see Materials and Methods) abolished VP39-F180W binding, as indicated by fluorescence quenching (data not shown). Affinity could be restored (within the limits of experimental variation) by reoxidation of the 8-hydro-m7G(5′)pp. Thus, a Kd of 19.1 μM was determined for the reoxidized m7G(5′)pp, comparable to the Kd of 14.6 μM determined for the untreated compound (Table 2).
In each of the three VP 39 structures with bound methylated nucleobase and nucleoside shown (Figs. 1 c and d, 2 c and d, and 3 a and b), each side of the base ring stacks with an aromatic side chain, the A-face side with Tyr 22 and the B-face side with Phe 180. This arrangement of two stacks (named stacks A and B) exhibits three notable common features in each of the three complexes: They have nearly perfect parallel alignment (Figs. 1c, 2c, and 3a), the areas of overlap (especially for stack B) are substantial (Figs. 1d, 2d, and 3b), and the interplanar spacing within the overlapping region is 3.4 Å. These features, which were also observed in the m1Ade- and m1Cyt-liganded structures (data not shown), have previously been posited as essential for enhanced stacking interaction in experimental studies and theoretical analyses of stacked model aromatic compounds and positively charged methylated nucleobase derivatives, including nucleosides and nucleotides (21, 22). The stacking interaction may be viewed as a “cation–π interaction” (23), although the positive charge of the methylated bases would be more delocalized than those of the cationic metals and ammonium groups often observed interacting with the π faces of benzene and aromatic side chains.
A major role for stacking interactions in cap recognition is consistent with the finding that substitution of either of the two stacking side chains (Tyr 22 and Phe 180) with Ala eliminates specificity for m7G-capped over uncapped RNA in a BIAcore-based assay (6). Consistent with this, no detectable m7G binding to the Y22A mutant was observed crystallographically (Table 1). However, the Y22A and F180A mutants each retain detectable methyltransferase activity (6, 18), which is specific for m7G-capped over uncapped RNA substrate (18). This would suggest that the two mutants each retain tiny amounts of m7G-binding activity, which, although below the detection limits of the crystallographic and BIAcore assays, is sufficient for RNA methylation to be catalyzed at a low rate—provided the RNA substrate is m7G-capped. This would therefore suggest that the Tyr 22 and Phe 180 side chains can individually confer some degree of specificity for the m7Gua moiety over its Gua counterpart, by stacking with the positively charged base, but that the two side chains combine to confer high affinity.
The observation that the overlap of the heterocylic ring of the nucleobases and of m7G with Phe 180 (in stack B) is larger than that with Tyr 22 (in stack A) (Figs. 1d, 2d, and 3b) is consistent with the finding that the F180A mutation led to a significantly greater reduction in VP39 methyltransferase activity than the Y22A mutation (6, 18). That Phe 180 exerts a greater influence in the stacking interaction is further indicated by the greater overlap it makes with the pyrimidine portions of the adeninium and guaninium rings than with the imidazole portions (Figs. 1d and 3b). Studies of model compound systems also indicate stronger stacking interaction with the pyrimidine portion of the adeninium and guaninium rings than with the imidazole portion (22, 24).
In conclusion, we have provided evidence for the major role of enhanced stacking interactions with two aromatic residues in protein recognition of methylated nucleobases. Hydrogen bonds, salt links, and steric factors further contribute to molecular recognition and rotational orientation of the bases about an axis perpendicular to that of the plane of the ring. These results may have significant ramifications in mRNA cap recognition and DNA repair enzyme function.
Acknowledgments
We thank W. E. Meador and S. W. Lockless for technical assistance. G.H. is a Predoctoral Fellow of the Welch Foundation through a grant to F.A.Q. This work was supported by grants from the National Science Foundation to P.D.G. (Grant MCB-9604188) and to the Biomedical Computation and Visualization Laboratory, Baylor College of Medicine (Grant STI-9512521). F.A.Q. is an Investigator in the Howard Hughes Medical Institute. This paper is dedicated to the memory of Prof. David C. Phillips.
ABBREVIATIONS
- m7G
N7-methylguanosine
- m7Gua
N7-methylguanine
- m3Ade
N3-methyladenine
- m3Cyt
N3-methylcytosine
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3mag, 1b42, 3mct, 1bky, 4dcg, 1eqa, and 1eam).
References
- 1.Yamagata Y, Kato M, Odawara K, Tokuno Y, Nakashima Y, Matsushima N, Yasumura K, Tomita K, Ihara K, Fujii Y, et al. Cell. 1996;86:311–319. doi: 10.1016/s0092-8674(00)80102-6. [DOI] [PubMed] [Google Scholar]
- 2.Labahn J, Scharer O D, Long A, Ezaz-Nikpay K, Verdine G L, Ellenberger T E. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 3.Gershon P D, Moss B. J Biol Chem. 1993;268:2203–2210. [PubMed] [Google Scholar]
- 4.Barbosa E, Moss B. J Biol Chem. 1978;253:7698–7702. [PubMed] [Google Scholar]
- 5.Hodel A E, Gershon P D, Shi X, Quiocho F A. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 6.Hodel A E, Gershon P D, Shi X, Wang S-M, Quiocho F A. Nat Struct Biol. 1997;4:350–354. doi: 10.1038/nsb0597-350. [DOI] [PubMed] [Google Scholar]
- 7.Hodel A E, Gershon P D, Quiocho F A. Mol Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 8.Hendler S, Furer E, Srinivasan P R. Biochemistry. 1970;9:4141–4153. doi: 10.1021/bi00823a017. [DOI] [PubMed] [Google Scholar]
- 9.Marcotrigiano J, Gingras A-C, Sonnenberg N, Burley S K. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 10.Shi X, Bernhardt T G, Wang S-M, Gershon P D. J Biol Chem. 1997;272:23292–23302. doi: 10.1074/jbc.272.37.23292. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Brünger A T. xplorVersion 3.1. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 13.Huang C C, Pettersen E F, Klein T E, Ferrin T E, Langridge R. J Mol Graphics. 1991;9:230–235. doi: 10.1016/0263-7855(91)80016-s. [DOI] [PubMed] [Google Scholar]
- 14.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 15.Miller D M, Olson J S, Pflugrath J W, Quiocho F A. J Biol Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- 16.Vermersch P S, Tesmer J J, Lemon D D, Quiocho F A. J Biol Chem. 1990;265:16592–16603. [PubMed] [Google Scholar]
- 17.Adams B L, Morgan M, Muthukrishnan S, Hecht S M, Shatkin A J. J Biol Chem. 1978;253:2589–2595. [PubMed] [Google Scholar]
- 18.Lockless S W, Cheng H-T, Hodel A E, Quiocho F A, Gershon P D. Biochemistry. 1998;37:8564–8574. doi: 10.1021/bi980178m. [DOI] [PubMed] [Google Scholar]
- 19.Fasman G D. Handbook of Biochemistry and Molecular Biology: Nucleic Acids. Vol. 1. Cleveland: CRC; 1975. [Google Scholar]
- 20.Morino S, Hazama H, Ozaki M, Teraoka Y, Shibata S, Doi M, Ueda H, Ishida T, Uesugi S. Eur J Biochem. 1996;239:597–601. doi: 10.1111/j.1432-1033.1996.0597u.x. [DOI] [PubMed] [Google Scholar]
- 21.Ishida T, Doi M, Ueda H, Inoue M, Scheldrick G M. J Am Chem Soc. 1988;110:2286–2296. [Google Scholar]
- 22.Ishida T, Shibata M, Fujii K, Inoue M. Biochemistry. 1983;22:3571–3581. doi: 10.1021/bi00284a006. [DOI] [PubMed] [Google Scholar]
- 23.Dougherty D A. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 24.Pullman B, Pullman A. Proc Natl Acad Sci USA. 1958;44:1197–1202. doi: 10.1073/pnas.44.12.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]