Abstract
To study intracellular pathways by which the human papillomavirus 16 oncogene E7 participates in carcinogenesis, we expressed an inducible chimera of E7 by fusion to the hormone-binding domain of the estrogen receptor. The chimeric protein (E7ER) transformed rodent fibroblast cell lines and induced DNA synthesis on addition of estradiol. In coimmunoprecipitation experiments, E7ER preferentially bound p130 when compared to p107 and pRb. After estradiol addition, E7ER localization changed to a more intense intranuclear staining. Induction of E7 function was not correlated with binding to p130 or pRb but rather with intranuclear localization and modest induction of binding to p107.
Keywords: retinoblastoma gene product, p107
Human papillomavirus (HPV) infection of genital tract epithelium is associated with dysplastic changes in epithelial growth and is an important prerequisite for the development of cervical cancer. Two viral genes, E6 and E7, mediate transformation by HPV (1–4). The majority of cervical carcinomas contain truncated forms of HPV, which retains E6 and E7, integrated into the cellular genome (5–9). HPV16 E7 induces growth of cells in soft agar (1), stimulates DNA synthesis (10, 11), cooperates with ras to transform primary cells (12, 13), and overcomes G1 arrest induced by serum deprivation, actinomycin D, or overexpression of p21 (14–17). The protein encoded by E7 has a region of shared sequence homology with both adenovirus E1A and SV40 large T antigen, which mediates binding to the retinoblastoma gene product pRb (13, 18). By binding to pRb, E7 alters the interaction of pRb with the transcription factor E2F-1, resulting in activation of E2F-responsive genes (19). E7 protein from high-risk HPV types, HPV16 and HPV18, binds with a higher affinity to pRb than E7 protein from low-risk HPV types such as HPV6 and HPV11 (20). Therefore, the prevailing model is that E7 protein, by binding to pRb, promotes progression through the cell cycle. The role of E7 binding to other Rb-family members such as p107 and p130 has not been entirely elucidated. However, it is known that induction of B-myb transcription by E2F-1 is the result of E7 binding to p107 rather than to pRb (21).
To assess the relationship between transformation and E7 binding to Rb-family members, we have constructed an inducible chimeric molecule consisting of HPV16 E7 fused in-frame to the hormone-binding domain of the estrogen receptor (ER), an approach used to render other oncogenes steroid-dependent (22–27). By using this inducible chimera of E7, we demonstrate that HPV16 E7 activity is correlated with intranuclear localization of the chimera. In addition, E7 binds to greater proportions of intracellular p130 and p107 than pRb in quiescent cells.
MATERIALS AND METHODS
Construction of E7ER.
The HPV16 viral genome (from J. Palefsky, Univ. of California, San Francisco) was used as the template to make E7 that lacked a stop codon by using PCR. E7 was cloned in-frame with ER, the hormone-binding domain of the estrogen receptor [HE14, from P. Chambon, Institute of Genetics and Molecular and Cellular Biology, Strasborg, France (28)]. E7 mutants were obtained from K. Vousden (National Cancer Institute, Frederick, MD) (29, 30), and were similarly cloned in-frame with ER to create mutant E7ER chimeras. A replication-defective murine retrovirus vector containing a neomycin-resistance gene under the control of the SV40 promoter [pMXE7ER (31)] was used to express the chimera (Fig. 1A). Viral stocks were generated by transfection into the packaging cell line Ψ2 (32) and were used to infect cultures of 3T3/C7 cells (a derivative of NIH/3T3 cells). The ER fragment was also subcloned alone into a mammalian expression vector LNCX (33); LNCX-ER was transfected directly into 3T3/C7 cells by using calcium phosphate precipitation and a clonal cell line selected with a high level of ER expression (C7/ER-3).
Figure 1.
Structure and function of the E7ER chimera. (A) pMXE7ER contains the E7ER chimera in a retroviral vector with the neomycin-resistance gene. X, Xho; B, BamHI; E, EcoRI; C, ClaI. (B) Lysates of 3 × 105 cells transfected with the ER construct (C7/ER-3) and E7ER (C7/E7ER 3-3) were analyzed by using Western blot analysis with anti-ER rabbit antiserum (S. Robbins). Positions of 45- and 29-kDa molecular mass markers are shown to the right. (C) [3H]Thymidine incorporation was measured in starved E7ER-expressing cells (C7/E7ER 1-1) or a pool of cells transfected with vector alone (C7/neo) in response to increasing doses of E2. Each point is the average of duplicate wells; duplicate cpm values varied by <10%. (D) Induction of DNA synthesis was measured in response to 1 μM E2 in clonal cell populations expressing the ER domain (C7/ER3), wild-type E7 in the context of the ER chimera (E7ER 1-1 and 3-3), and mutant E7 alleles containing a point mutation in the Rb-binding pocket (E7*ER 4-1 and 4-3) or a deletion of the Rb-binding pocket (E7*ER 5-4). Bar graphs represent the mean from 4 (C7/ER3), 12 (E7ER 1-1), 19 (E7ER 3-3), 3 (E7*ER 4-1 and 4-3), or 2 (E7*ER 5-4) independent assays. Error bars represent the SEM. (E) Cells (104) were plated in soft agar, and the total number of colonies greater than 6–8 cells in size was counted after 2 (C7/E7, C7/E7ER 1-1 and 3-3) or 4 (C7/ER3) weeks. Results are expressed as a ratio of the total number of colonies in the presence as compared with the absence of E2.
G418-resistant clonal cell lines of C7/E7ER were isolated by plating cells from transformed foci at low density and isolating individual colonies. Cells were grown in DMEM lacking phenol red with 10% stripped calf serum (complete medium). Serum was stripped of endogenous steroids by incubation with activated charcoal (1 g/50 ml of serum) for 20 minutes at 4°C.
Soft Agar Assays.
Cells were plated in 0.375% low melting agarose (FMC) in complete medium with 1 μM E2 or 0.1% ethanol and fed weekly with appropriate medium.
Stimulation of DNA Synthesis.
Cells were plated at a density of 105 cells per well into a six-well plate; after the cells reached confluence, the medium was changed to phenol red-free DMEM with 0.5% BSA and 5 μg/ml transferrin (Sigma), cultured for 36–48 hours, and treated with 1 μM E2, 0.1% ethanol, or 10% calf serum for 24 hours. [3H]Thymidine (1 μCi, Amersham Pharmacia; 1 Ci = 37 GBq) was added for 45 minutes, and incorporation into DNA was measured. For the dose–response curve, (Fig. 1C), all media contained a concentration of ethanol equivalent to that delivered with the highest dose of E2.
Immunoprecipitation and Western Blotting.
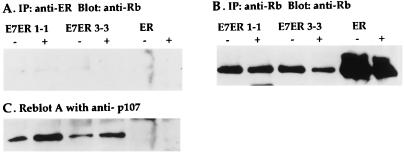
For the Western blot in Fig. 1B, cell pellets were lysed directly in sample buffer. For immunoprecipitation, cell pellets were lysed in 2 ml of lysis buffer (25 mM Tris⋅HCl, pH 7.4/150 mM NaCl/2% NP40/0.5% Na deoxycholate/0.2% Na dodecylsulfate) containing 1 mM Pefabloc, 0.1 mg/ml aprotinin, and 0.1 mg/ml leupeptin. Five microliters of rabbit polyclonal anti-ER antibody (from S. Robbins, University of Calgary) was added per cell pellet. Antigen–antibody complexes were isolated on Protein A-Sepharose beads (Sigma) and washed three times in wash buffer (25 mM Tris, pH 7.4/50 mM NaCl/0.5% Na deoxycholate/0.2% NP40). Proteins were resolved on 10% (Fig. 3) or 8% (Fig. 4) polyacrylamide gels, transferred to nitrocellulose (Schleicher & Schuell), and incubated with 5 μg/ml monoclonal anti-Rb antibody (clone G3–245, PharMingen) or 1 μg/ml rabbit anti-Rb IgG (Santa Cruz Biotechnology), anti-p107 IgG, or anti-p130 IgG (Santa Cruz Biotechnology), or 1:1,000-diluted anti-ER rabbit antiserum (from S. Robbins) in Tris-buffered saline. Detection was performed by enhanced chemiluminescence (Amersham Pharmacia) using horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit Ig (Amersham Pharmacia) or horseradish peroxidase conjugated-Protein A (Amersham Pharmacia) for detecting anti-ER antibodies. When sequential immunoprecipitations were performed, antibody (5 μg of mouse or rabbit IgG) was added (for 1–18 hours) to the supernatant collected from the first immunoprecipitation. For densitometry, films of Western blots were scanned with Adobe photoshop, and the intensities of bands representing pRb, p107, and p130 were quantitated by using imagequant (Molecular Dynamics).
Figure 3.
Minimal association of E7ER with pRb and induction of p107 association on E2 treatment. Confluent starved cultures were treated overnight with E2 (1 μM) (+) or ethanol (−) and immunoprecipitated with anti-ER antibodies (A and C) and then sequentially with anti-pRb antibodies (B). Western blots were probed with anti-Rb antibodies (A and B) or anti-p107 antibodies (C). Each lane consists of lysate from 2 × 107 cells expressing the E7ER construct (3-3 and 1-1) or the ER construct (ER-3).
Figure 4.
E7ER preferentially binds to p130. Experiments were performed as in Fig. 3. In A, − or + refers to the absence or presence of E2. In C and D, untreated cell lysates were immunoprecipitated first with anti-ER antibodies and then sequentially with anti-p130 antibodies; immunoprecipitates were run in neighboring lanes to allow comparison of the proportion of protein by densitometric scanning of the autoradiograms (E). E7 mutER is a clonal population expressing a point mutation in the Rb-binding pocket (Gly-24). A second clonal cell line gave identical results. Data in E represents comparisons by densitometry scanning of the amounts of pRb, p107, and p130 immunoprecipitated with anti-ER antibodies compared with the sum of the amounts immunoprecipated with anti-ER and subsequently with the relevant Rb-family member antibody. Data from two (p107) or three (pRb and p130) independent experiments were averaged; error bars represent the SEM.
E7–Rb Interactions in a Mammalian Two-Hybrid System.
A mammalian two-hybrid system was constructed (based on ref. 34). cDNA encoding the retinoblastoma gene product (amino acids 379–792) was amplifed from a wild-type Rb allele (from W.-H. Lee, University of Texas Health Science Center, San Antonio) by using PCR and cloned in-frame into pGal0 (34) to create g4Rbp. A mutant Rb allele of the pRb pocket domain (Cys-to-Phe change at amino acid 706, from J. Horowitz, Duke University Medical Center Durham, NC) was similarly cloned in-frame into pGal1/0 to create g4Rbpm. Gal4–p107 was obtained from C. Dang, John Hopkins University, Baltimore. A Gal4/VP16-dependent luciferase reporter vector (g5E1Blux) was constructed by cloning five concatemerized Gal4-binding domains into a luciferase-reporter plasmid PXP1 (35). CV1 cells or PVU1 cells (CV1 cells containing a large tumor antigen allele with a deletion of the pRb-binding pocket) were transfected by using the calcium phosphate method with 2 μg of the luciferase reporter and 1 μg of a vector expressing chloramphenicol acetyltransferase (CAT) under control of the SV40 promoter (pSV40CAT) to quantitate transfection efficiency. In transfections designed to test for interactions between two proteins, the reporter and pSV40CAT were cotransfected with 10 μg each of the plasmids expressing the two fusion proteins (e.g., g4Rbp and E7ER). E2, tamoxifen, or ethanol were added 1 day after transfection, and the cells were harvested 48–72 hours later. Luciferase assays were performed by using Promega reagents. Liquid CAT assays were performed to quantitate transfection efficiency as described (36). To obtain relative transfection efficiencies, results of CAT assays from each plate were divided by results from a plate transfected with only the SV40CAT and the Gal4 luciferase reporter plasmids. Luciferase values from each plate were normalized for transfection efficiency by dividing by the corresponding relative transfection efficiency ratio. Transfections were carried out in duplicate or triplicate, and normalized luciferase values were averaged. Normalized luciferase values generally varied by <5% between plates transfected with the same set of plasmids.
Immunofluorescence.
Cells were plated on two-chamber culture slides (Lab-Tek). At confluence, the medium was changed to 0.5% BSA and 5 μg/ml transferrin in DMEM for 24 hours, at which time 1 μM E2 or 0.1% ethanol was added for 18 hours. Cells were rinsed, fixed in 3.7% formaldehyde in PBS, incubated in blocking solution (10% calf serum and 0.1% Triton X-100 in PBS) for 20–30 minutes, rinsed with PBS, and incubated for 1–2 hours with polyclonal rabbit anti-ER antibody (Santa Cruz Biotechnology; 1 μg/ml in blocking solution). As a negative control, parallel sets of slides were incubated with rabbit IgG at 1 μg/ml. The slides were then washed 3 times with PBS, incubated with Cy3-conjugated goat anti-rabbit IgG (The Jackson Laboratory) at 1:500 dilution for 40 minutes, rinsed, mounted, and viewed by using fluorescence optics with a Texas red filter. 4′,6-Diamidino-2-phenylindole (DAPI) (2 μg/ml) was added to the last wash for detection of nuclei. For in situ lysis, cells were incubated in hypotonic lysis buffer (10 mM Hepes, pH 7/10 mM KCl/0.1% Triton X-100/0.5 mM DTT/1.5 mM MgCl2) for 10 min at 4°C before fixation in 3.7% formaldehyde/PBS (37, 44).
RESULTS
Inducible Transformation in Cells Expressing E7ER.
Expression of E7ER in G418-selected populations was verified with Western blotting (Fig. 1B). Cells containing ER or E7ER constructs expressed proteins detected by anti-ER antibodies corresponding to the predicted sizes (38 kDa and 52 kDa, respectively). The E7ER chimera was also recognized by an anti-E7 mAb (data not shown), confirming that the chimeric molecule expressed the E7 epitope.
To evaluate whether E7 function is inducible in the context of E7ER, the amount of DNA synthesis was measured over a range of E2 concentrations and found to be maximal between 500 nM and 1 μM (Fig. 1C). These relatively high concentrations are consistent with the fact that the ER clone used in these experiments contains a mutation that results in a lower affinity for E2 than wild-type ER (38). Clonal populations of cells expressing E7ER reproducibly demonstrated an induction of incorporation of [3H]thymidine in response to 1 μM E2 ranging from 2- to 10-fold (Fig. 1D). Cells expressing chimeric molecules with a point mutation in the pRb-binding pocket (Gly-24), E7*ER 4-1 and 4-3, or a deletion of the pRb-binding pocket (amino acids 21–35), E7*ER 5-4, showed no induction of DNA synthesis in response to E2 (Fig. 1D), consistent with previously reported effects of these mutations.
Addition of E2 to soft agar assays of E7ER-expressing clonal cell lines resulted in a significant induction of growth of colonies in soft agar, as shown in Fig. 1E. E2 had little effect on cells carrying either the vector alone or wild-type E7; cloning efficiencies were either low (<1%) or high (12%), respectively. The estrogen receptor itself did not confer E2-dependent induction of DNA synthesis (Fig. 1D) or growth in soft agar (Fig. 1E), confirming that the observed effects are caused by the E7 moiety in the chimera.
Interaction of E7ER with pRb and p107 in a Mammalian Two-Hybrid System.
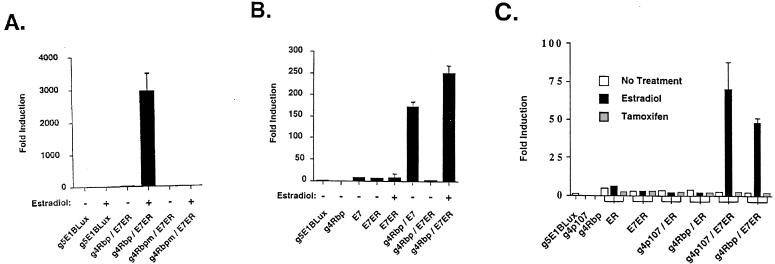
To document that chimeric E7ER molecules interact with pRb-family members in the intracellular environment, we constructed a modified mammalian two-hybrid system. The ER domain contains a hormone-dependent transactivation function that can be measured in the context of the Gal4 promoter (39); this property of the molecule was exploited to detect interactions between E7ER and pRb or p107 expressed as fusion proteins with a Gal4 DNA-binding domain. Thus, whether complexes form between E7ER and either Gal4pRb or Gal4p107, the ER transactivation domain will be brought into sufficient proximity of the transcriptional machinery by the Gal4 DNA-binding domain to activate transcription of a luciferase reporter gene under control of the Gal4 promoter (g5E1BLux). In cells transfected with the reporter construct alone, the addition of E2 did not increase the basal levels of luciferase activity (Fig. 2A). In cells cotransfected with the reporter construct, E7ER and the Gal4 fusion vector containing a truncated version of pRb retaining the Rb-pocket domain (amino acids 379–792) (g4Rbp), high levels of luciferase activity were induced by E2, whereas only background levels were found in its absence (Fig. 2A). A single point mutation in the Rb pocket known to result in loss of association of pRb with viral oncoproteins resulted in loss of the induction of luciferase activity (Fig. 2A, g4Rbpm). These results confirm that the interaction between E7ER and pRb is occurring through the expected domains in pRb. In addition, E7ER showed ligand-dependent transactivation of the Gal4 promoter in association with two other forms of Gal4–pRb fusion proteins, consisting of amino acids 2–928 and amino acids 379–928 of pRb (data not shown). The Gal4–p107 chimera (g4p107) similarly interacted with E7ER as measured by luciferase activity (Fig. 2C). However, the ER construct alone did not exhibit interaction with either pRb or p107 (Fig. 2C), and the E7ER chimera in the absence of E2 failed to induce luciferase activity (Fig. 2B). HPV16 E7 itself also contains a transactivation domain (13) which can function in the two-hybrid system; cotransfection of HPV16 E7 and Gal4–pRb results in induction of luciferase activity (Fig. 2B). In the presence of tamoxifen [which binds to the estrogen receptor but does not activate the transactivation domain (39)], no transactivation of the luciferase reporter was measured in cells cotransfected with E7ER and Gal4–pRb or Gal4–p107 (Fig. 2C). These results indicate that, in the context of the E7ER chimera, the endogenous transactivation domain of E7 is inert.
Figure 2.
E7 and E7ER interactions with pRb in a mammalian two-hybrid system. Cells were transfected with the luciferase reporter (g5E1Blux), the CAT reporter, and various constructs as indicated. Luciferase values were normalized to transfection efficiency as described in the text. Fold induction refers to the normalized luciferase value of the experimental cells (average of two plates) divided by the normalized luciferase value of cells transfected with the luciferase reporter and CAT reporter alone. E2 (1 μM), tamoxifen (1 μM), or ethanol was added as indicated. PVU1 cells were used in A, whereas CV1 cells were used in B and C; the enhanced induction of luciferase activity in A may be because of the fact that the large tumor antigen expressed by these cells replicates the Gal4-based plasmids that have an SV40 origin. Columns represent the average and error bars represent the SE of duplicate transfections. See Materials and Methods for descriptions of g5E1BLux, g4Rbp, and g4Rbpm.
Intracellular Binding of E7ER to pRb, p107, and p130.
By using polyclonal antisera to the ER domain, intracellular complexes containing E7ER were immunoprecipitated from cell lysates and analyzed with Western blotting. Comparison of Fig. 3A with Fig. 3B shows that pRb is measurable in the cells but that only a small proportion of the endogenous pRb is detectable in immunocomplexes with E7ER. In addition, equivalent proportions of pRb were complexed with E7ER in the presence or absence of 1 μM E2 (Fig. 3A and 4E), indicating that complex formation did not correlate with activation of E7 function. E7 is known to result in decreased levels of pRb protein because of proteolysis (40), and indeed the amount of pRb in E7ER-expressing cells was lower than the amount in ER-expressing cells (Fig. 3B, E7ER 1-1 and 3-3 vs. ER). However, the data shown in Fig. 4E reflect the amount of pRb immunoprecipitated with E7ER as a proportion of the total pRb in the cognate cell lysate. Therefore, the lack of induction of pRb binding on E7ER activation is independent of the effect of E7ER on pRb levels.
When the blot in Fig. 3A was reprobed with antibodies to p107, interaction of E7ER with p107 was readily detected (Fig. 3C). In addition, on treatment with E2, the amount of p107 in the immune complex with E7ER increased 2.6-fold (± 0.47, average of 3 independent experiments), whereas the amount of uncomplexed p107 was unchanged. The ER domain alone showed no detectable interaction with p107 in the presence or absence of E2 (Fig. 3C). Thus, complex formation between p107 and E7ER increased when E7 activity was induced compared with cells in which E7 was present but biologically inactive. This is in contrast to the binding of E7ER to pRb, which was difficult to detect and did not vary with the activation state of E7.
Significantly greater proportions of p130 than pRb or p107 were detected in complexes with E7ER. In Fig. 4A, the position of pRb migration is marked to the right of the figure, but no pRb complexed to E7ER was detectable in this experiment. However, on reprobing the blot in Fig. 4A with anti-p130 antibodies, p130 was easily detectable (Fig. 4B). Sequential immunoprecipitation of cell lysates, first with anti-ER antibodies and then with anti-p130 antibodies (Fig. 4C), revealed that significant proportions of the intracellular p130 (≈65%) were associated with E7ER (Fig. 4E).
In Fig. 4D, E7mutER, which contains a point mutation in the pRb-binding pocket (Gly-24), was also analyzed in coimmunoprecipitation experiments. This mutant allele was unable to bind to p130. A similar result was found by using a mutation of E7 lacking the pRb-binding pocket (amino acids 21–35) (data not shown). These results indicate that the domain of E7 involved in p130 binding overlaps with that used to bind pRb and p107.
Intracellular Localization of E7ER Changes On Activation of the Construct.
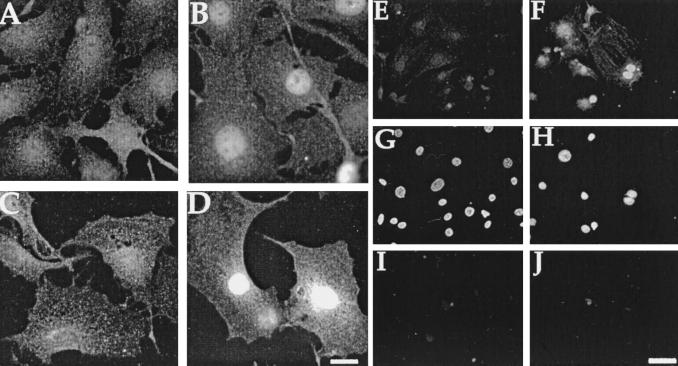
It was puzzling that induction of E7 effects were not correlated with induction of binding to pRb and p130. To test the possibility that the E7ER chimera might sequester Rb-family members within cellular compartment(s), where they are inactive, we compared the intracellular localization of E7ER before and after exposure to E2. The immunofluorescent staining pattern of E7ER changed to an intense nuclear signal after addition of E2 (Fig. 5B vs. Fig. 5A). This shift of E7ER localization mirrored the staining pattern of cells expressing the ER construct alone (Fig. 5 C and D), reflecting the known translocation of ER from the cytoplasm to the nucleus on ligand binding (27). Immunofluorescent staining with the anti-ER antibody of cells carrying the vector alone showed faint cytoplasmic staining that did not change in the presence of E2 (data not shown).
Figure 5.
Immunofluorescent localization of E7ER. (A–D) Altered intracellular localization on E2 treatment. Serum-starved confluent cultures of C7/E7ER 1-1 cells (A and B) or C7/ER-3 (C and D) were treated overnight with 1 μM E2 (B and D) or ethanol (A and C) and prepared for immunofluorescence as described. (Bar = 20 μm.) (E–J) In situ lysis reveals differential nuclear localization of E7ER on E2 treatment. Serum-starved cultures of C7/E7ER 1-1 cells were treated overnight with 1 μM E2 (F, H, and J) or ethanol alone (E, G, and I). Cells were lysed in hypotonic buffer before fixation and then prepared for immunofluorescence with anti-ER antibodies (E and F), DAPI (G and H) or rabbit IgG (I and J). (Bar = 50 μm.)
Discrepancies remain in the literature regarding the intracellular localization of E7: it appears to be cytoplasmic when cells are fractionated but nuclear when studied by using immunofluorescence (41–43), a paradox that has been explained by the hypothesis that cell fractionation disrupts the association of E7 with the nucleus (41). To study E7ER localization directly, we lysed cells in situ before fixation and examined nuclei on the substrate by using immunofluorescence (37, 44). We found a dramatic increase in the amount of E7ER associated with the nucleus in the presence as compared with the absence of E2 (Fig. 5 E and F).
DISCUSSION
HPV16 E7 Preferentially Binds p130.
Previous reports suggest that the capability of E7 to transform cells arises, at least in part, from its ability to complex with the nuclear phosphoprotein pRb (20, 45). E7 also interacts with the pRb-like protein p107 (21) and with a 130-kDa protein that resembles the E1A-associated protein p130 (46). In addition, E7 peptides compete with E1A for binding to p130 (47). Our results show that E7ER, like E7, complexes with the pRb-family members in both biochemical and in vivo assays. Surprisingly, the relative proportion of intracellular p130 bound to E7ER is far greater than the proportions of pRb or p107. Indeed, ≈65% of the p130 in the cell was present in complexes with E7ER, compared with 5% of pRb and 25% of p107 (Fig. 4E).
Recent reports have indicated that the Rb-family members have different functions in regulating cell growth and differentiation and that p130 is the predominant member present in quiescent cells (recently reviewed in ref. 48). In addition, p130 has been implicated in the G1–G0 transition (49, 50). The predominant intracellular E2F activity, E2F-4, is bound primarily to p130 in quiescent cells (51, 52) and switches to association with p107 and pRb as cells pass through the G1–S transition (52).
In skin, the Rb-family members are expressed in different histological compartments, with pRb and p107 localized predominantly in the basal and parabasal levels and p130 in the upper, differentiated cell layers (50). When HPV infects epithelium, productive viral replication occurs in the same differentiated layers where p130 is expressed. The histology of cervical dysplasia demonstrates aberrant differentiation and an increased number of mitotically active cells, consistent with an inability of cervical keratinocytes to undergo terminal differentiation. Our work suggests that the effect of HPV16 E7 in human disease may derive in part from interactions with p130, which may modify keratinocyte differentiation and thereby facilitate viral replication.
Nuclear Localization Is Important for E7-Induced Transformation.
The data presented here indicate that properties of E7, such as induction of DNA synthesis and growth of cells in soft agar, are rendered E2-dependent in the context of the E7ER allele. In a mammalian two-hybrid assay (Fig. 2), the biochemical readout for interactions between E7ER and pRB or p107 were dependent on the presence of E2. However, results from the coimmunoprecipitation experiments suggested that the amounts of endogenous p130 and pRb associated with E7ER were unchanged on activation of E7, and the amount of p107 was modestly induced. It is possible the results of the immunoprecipitation experiments represent indiscriminate accessibility of Rb family members to E7ER secondary to lysis of cells, but this seems unlikely given the pronounced differences in the proportions of Rb-family members bound to E7ER within the same cell lysate and the observation that binding to p107 was consistently induced on activation of E7ER.
Our results suggest two explanations for E2 dependence of E7ER function. One possibility is that the transformation elicited by E7 in these assays depended on induction of association with p107 and was independent of association with pRb and p130. This interpretation is supported by data showing that the induction of B-myb expression by E7 depended on p107 but not pRb binding (21); it is weakened by our observation that interaction with p107 was only modestly induced (2.6-fold) on E7ER activation. A second explanation is that binding of E7 to Rb-family members per se is not as important for E7 activity as is the intracellular localization of these complexes. After the addition of E2, E7ER localized to the nucleus (Fig. 5). Our data further support the hypothesis that the nucleus is the physiologically important site of E7 action. The E7 sequence has no obvious nuclear localization signal, and the mechanism by which wild-type E7 is targeted to the nucleus is unknown. Mutations in E7 that bind pRB but are nontransforming have been described (53–55). The phenotype of these mutants may be caused by their inability to localize to the correct intracellular compartment. Because E7ER is localized to the nucleus at least in part because of the ER domain, it will be interesting to test these mutants in the context of the E7ER chimera to determine whether restoration of nuclear localization can restore transforming capability.
Our results indicate that the formation of E7ER complexes with the Rb-family members is not sufficient for transformation and suggest that intranuclear concentration of the complexes is essential for cell cycle reentry. This interpretation runs counter to the currently held view that E7 acts by leading to release of transcriptionally active E2F on binding to the Rb-family members. Recent data have indicated that the activity of endogenous E2F4 (the predominant E2F partner of p130) is regulated by changes in its intracellular localization, from nuclear in G1/G0 cells to cytoplasmic in cycling cells (56). In addition, pRb-E2F, p107-E2F, and p130-E2F species have distinct patterns of intracellular localization that vary independently during the cell cycle (56). E2F response elements in B-myb, cyclin A, and cdc2 are occupied in G0 (57), suggesting that repression by E2F needs to be actively overcome to initiate cell cycle reentry. Our data suggest a model in which E7-Rb family member complexes play an active, and perhaps direct, role in E2F transciptional regulation. The experimental system described in this report will allow us to study the effects of E7 activation on intracellular localization of the Rb and E2F family members and the transcriptional repression or activation of E2F sites in response to these changes.
Acknowledgments
We thank P. Chambon for providing the HE14 ER clone, C. Dang for the Gal4–p107 construct, J. Horowitz for the mutant Rb allele, W.-H. Lee for the wild-type Rb clone, M. McMahon for the retroviral vector, J. Palefsky for the HPV16 clone, S. Robbins for antibody to ER, K. Vousden for E7 mutants, and A. Alberts, L. Deiss, M. McMahon, S. Robbins, E. Shtivelman, and S. Schirm for helpful suggestions. This research was supported by the G. W. Hooper Research Foundation, the Irwin Foundation, the University of California Cancer Research Coordinating Committee, and Public Health Service Grants HD31057 and CA61797. S.S. was supported by a grant from the Cowell Foundation. K.S.-M. acknowledges past support of the Reproductive Scientist Development Program of the National Institute of Child Health and Human Development and the American Gynecological and Obstetrical Society.
ABBREVIATIONS
- E2
estradiol
- HPV
human papillomavirus
- ER
estrogen receptor
- CAT
chloramphenicol acetyltransferase
References
- 1.Vousden K, Doniger J, DiPaolo J A, Lowy D R. Oncogene Res. 1988;3:167–175. [PubMed] [Google Scholar]
- 2.Yutsudo M, Okamoto Y, Hakura A. Virology. 1988;166:94–97. doi: 10.1016/0042-6822(88)90532-6. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka A, Noda T, Yajima H, Hatanaka M, Ito Y. J Virol. 1989;63:1465–1469. doi: 10.1128/jvi.63.3.1465-1469.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson J B, Bedell M A, McCance D J, Laimins L A. J Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durst M, Kleinheinz A, Hotz M, Gissman L. J Gen Virol. 1985;66:1515–1522. doi: 10.1099/0022-1317-66-7-1515. [DOI] [PubMed] [Google Scholar]
- 6.Pater M M, Pater A. Virology. 1985;145:313–318. doi: 10.1016/0042-6822(85)90164-3. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Nature (London) 1985;314:111–113. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 8.Yee C, Krishnan-Hewlett I, Baker C C, Schlegel R, Howley P M. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 9.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banks L, Barnett S C, Crook T. Oncogene. 1990;5:833–837. [PubMed] [Google Scholar]
- 11.Sato H, Furuno A, Yoshike K. Virology. 1989;168:195–199. doi: 10.1016/0042-6822(89)90423-6. [DOI] [PubMed] [Google Scholar]
- 12.Matlashewski G, Schneider J, Banks L, Jones N, Murray A, Crawford L. EMBO J. 1987;6:1741–1746. doi: 10.1002/j.1460-2075.1987.tb02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phelps W C, Yee C L, Munger K, Howley P M. Cell. 1988;53:39–47. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 14.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones D L, Alani R M, Munger K. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morozov A, Shiyanov P, Barr E, Leiden J M, Raychauduri P. J Virol. 1997;71:3451–3457. doi: 10.1128/jvi.71.5.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruesch M, Laimons L A. J Virol. 1997;71:5570–5578. doi: 10.1128/jvi.71.7.5570-5578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson N, Howley P M, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 19.Phelps W C, Bagchi S, Barnes J A, Raychaudhuri P, Kraus V, Münger K, Howley P M, Nevins J R. J Virol. 1991;65:6922–6930. doi: 10.1128/jvi.65.12.6922-6930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heck D V, Yee C L, Howley P M, Munger K. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam E W, Morris J D, Davies R, Crook T, Watson R J, Vousden K H. EMBO J. 1994;13:871–878. doi: 10.1002/j.1460-2075.1994.tb06330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehmelt G, Walker A, Kabrun N, Mellitzer G, Beug H, Zenke M, Enrietto P J. EMBO J. 1992;11:4641–4652. doi: 10.1002/j.1460-2075.1992.tb05566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burk O, Klempnauer K-H. EMBO J. 1991;10:3713–3719. doi: 10.1002/j.1460-2075.1991.tb04939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eilers M, Picard D, Yamamoto K, Bishop J M. Nature (London) 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 25.Superti-Furga G, Bergers G, Picard D, Busslinger M. Proc Natl Acad Sci USA. 1991;88:5114–5118. doi: 10.1073/pnas.88.12.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuels M L, Weber M J, Bishop J M, McMahon M. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picard D, Salser S J, Yamamoto K R. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Green S, Staub A, Chambon P. EMBO J. 1986;5:2231–2236. doi: 10.1002/j.1460-2075.1986.tb04489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edmonds C, Vousden K. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris J D, Crook T, Bandara L R, Davies R, LaThangue N B, Vousden K H. Oncogene. 1993;8:893–898. [PubMed] [Google Scholar]
- 31.McMahon M, Schatzman R C, Bishop J M. Mol Cell Biol. 1991;11:4760–4770. doi: 10.1128/mcb.11.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 33.Miller D A, Rosman G J. BioTechniques. 1989;7:80–90. [PMC free article] [PubMed] [Google Scholar]
- 34.Fearon E R, Finkel T, Gillison M L, Kennedy S P, Casella J F, Tomaselli G F, Morrow J S, Van Dang C. Proc Natl Acad Sci USA. 1992;89:7958–7962. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordeen S. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 36.Gorman C M, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittnacht S, Weinberg R A. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 38.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. EMBO J. 1989;8:1981–1989. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster N J, Green S, Jin J R, Chambon P. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 40.Berezutskaya E, Yu B, Morozov A, Raychaudhuri P, Bagchi S. Cell Growth Diff. 1997;8:1277–1286. [PubMed] [Google Scholar]
- 41.Sato H, Watanabe S, Furuno A, Yoshike K. Virology. 1989;170:311–315. doi: 10.1016/0042-6822(89)90386-3. [DOI] [PubMed] [Google Scholar]
- 42.Smotkin D, Wettstein F O. J Virol. 1987;61:1686–1689. doi: 10.1128/jvi.61.5.1686-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oltersdorf T, Seedorf K, Rowekamp W, Gissman L. J Gen Virol. 1987;68:2933–2938. doi: 10.1099/0022-1317-68-11-2933. [DOI] [PubMed] [Google Scholar]
- 44.Alberts A, Thorburn A M, Shenolikar S, Mumby M C, Feramisco J R. Proc Natl Acad Sci USA. 1993;90:388–392. doi: 10.1073/pnas.90.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Münger K, Yee C L, Phelps W C, Pietenpol J A, Moses H L, Howley P M. J Virol. 1991;65:3943–3948. doi: 10.1128/jvi.65.7.3943-3948.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies R, Hicks R, Crook T, Morris J, Vousden K. J Virol. 1993;76:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dyson N, Guida P, Munger K, Harlow E. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulligan G, Jacks T. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 49.Kiess M, Gill R M, Hamel P A. Cell Growth Differ. 1995;6:1287–1298. [PubMed] [Google Scholar]
- 50.Parasio J M, Lain S, Segrelles C, Birgit Lane E, Jorcano J L. Oncogene. 1998;17:949–957. doi: 10.1038/sj.onc.1202031. [DOI] [PubMed] [Google Scholar]
- 51.Mayol X, Csarriga J, Grana X. Oncogene. 1996;13:237–246. [PubMed] [Google Scholar]
- 52.Moberg K, Starz M A, Lees J A. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banks L, Edmonds C, Vousden K H. Oncogene. 1990b;5:1383–1389. [PubMed] [Google Scholar]
- 54.Barbosa M S, Edmonds C, Fisher C, Schiller J T, Lowy D R, Vousden K. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Firzlaff J M, Luscher B, Eisenman R N. Proc Natl Acad Sci USA. 1991;88:5187–5191. doi: 10.1073/pnas.88.12.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. Mol Cell Biology. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwicker J, Ningshu L, Engeland K, Lucibello F C, Muller R. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]