Abstract
The 32 telomeres in the budding yeast genome cluster in three to seven perinuclear foci. Although individual telomeres and telomeric foci are in constant motion, preferential juxtaposition of some telomeres has been scored. To examine the principles that guide such long-range interactions, we differentially tagged pairs of chromosome ends and developed an automated three-dimensional measuring tool that determines distances between two telomeres. In yeast, all chromosomal ends terminate in TG1–3 and middle repetitive elements, yet subgroups of telomeres also share extensive homology in subtelomeric coding domains. We find that up to 21 kb of >90% sequence identity does not promote telomere pairing in interphase cells. To test whether unique sequence elements, arm length, or chromosome territories influence juxtaposition, we reciprocally swapped terminal domains or entire chromosomal arms from one chromosome to another. We find that the distal 10 kb of Tel6R promotes interaction with Tel6L, yet only when the two telomeres are present on the same chromosome. By manipulating the length and sequence composition of the right arm of chr 5, we confirm that contact between telomeres on opposite chromatid arms of equal length is favored. These results can be explained by the polarized Rabl arrangement of yeast centromeres and telomeres, which promote to telomere pairing by allowing contact between chromosome arms of equal length in anaphase.
Long-range interactions between chromosomal loci and their regulatory elements guide genomic function. It is well established that in higher eukaryotes contact between enhancers and promoters occurs over distances of 100 kb to regulate higher eukaryotic gene expression. Boundary elements, which restrict enhancer directionality, interact over similar distances (for review, see Burgess-Beusse et al. 2002), as do insulator elements such as the Drosophila Fab-7 or gypsy elements, which protect genes from encroaching heterochromatin (for review, see Celniker and Drewell 2007). A further example of preferred interaction in trans is that of coordinately expressed tissue-specific genes that can coordinately occupy the same transcription factory in differentiating hematopoietic cells (Osborne et al. 2004). Finally, mouse T- and B-cell-specific genes were found juxtaposed to centromeric heterochromatin in appropriate cell types (for review, see Fisher and Merkenschlager 2002). In mammals, the differentiation-specific repression that is mediated by such juxtaposition requires that centromeres cluster in so-called “chromo-centers,” which form a sink for heterochromatin factors. Functionally analogous to this is the clustering of silent telomeres in budding yeast (for reviews, see Scherf et al. 2001; Gasser et al. 2004).
In budding yeast, telomeric repeats and factors that bind them nucleate SIR-mediated silencing, a chromatin-based repression mechanism that propagates inward from chromosomal ends for 3 to 5 kb. Like centromeric heterochromatin, the transcriptionally silent budding yeast telomeres cluster in three to seven distinct foci (Palladino et al. 1993; Gotta et al. 1996). These foci associate with the nuclear envelope (NE) and sequester the silent information regulatory proteins, Sir2, Sir3, and Sir4, from potential binding sites in non-subtelomeric regions (Maillet et al. 1996; Hediger et al. 2002). Such clusters promote the repression of silencer-flanked genes brought into their vicinity by membrane-spanning anchors (Andrulis et al. 1998).
While telomeric foci have been studied for years, it remained unclear how reproducible their composition might be. The question is of interest, because such clustering events have functional repercussions not only for the expression of subtelomeric genes. Telomere tethering and clustering have been proposed to influence the rate of recombinational repair (Louis et al. 1994; Fabre et al. 2005) and to coordinate transcriptional programs that ensure evolutionary advantage (Turakainen et al. 1993; Halme et al. 2004; Fabre et al. 2005). Moreover, in Plasmodium and Trypanosoma, subtelomeric repeat-mediated clustering of telomeres may regulate the pattern of expression of the repetitive VSG genes. This allows the parasite to escape the host immune response and thus provides a major evolutionary advantage (for review, see Scherf et al. 2001). Given the importance of such long-range interactions, we have exploited the suitability of yeast for live imaging and its powerful reverse genetics to examine the principles that regulate the clustering of its telomeres. We have manipulated yeast chromosome architecture in order to identify elements that drive long-range interactions in interphase cells.
Both the anchorage of telomeres at the nuclear periphery and their interaction with other telomeres in trans contribute to focus formation. Two partially redundant pathways function in budding yeast to anchor telomeres at the NE (Hediger et al. 2002; Taddei et al. 2004b). One is dependent on Sir4 and its ligand Esc1, a peripheral inner membrane protein, and the second requires the end-binding factor yKu. The deletion of SIR4 did not significantly affect telomere interaction in trans, whereas deletion of YKU80 compromised both interaction and anchorage (Laroche et al. 1998; Gehlen et al. 2006). Mutations in a subset of nuclear pore proteins (Therizols et al. 2006) and the cohesion loading factors Ctf18 and Ctf8 (Hiraga et al. 2006) have also been shown to affect telomere anchoring. In the latter mutants, telomere clustering was also impaired, although it was unclear whether the effects were direct or indirect.
Confirming the idea that there may be reproducible patterns of telomere interactions in yeast, it was shown that Tel3R and Tel3L, and Tel6R and Tel6L tend to be juxtaposed in the W303 haploid background (Bystricky et al. 2005). The interaction between right and left telomeres of a single chromosome creates a chromosome loop, a structure that was initially proposed for chr 3 based on an intramolecular religation assay (Dekker et al. 2002). While the idea that chromosomes loop back upon themselves is attractive, it is clear that not all linked chromosome ends interact to a significant extent. In the same study, it was shown that the right and left ends of chr 5 are not juxtaposed, nor are those of chr 14 (Bystricky et al. 2005). Because all telomeres tested have conserved terminal TG1–3 sequences and subtelomeric X and/or Y′ elements (Chan and Tye 1983; Louis and Haber 1992), we can rule out that these semirepetitive elements are sufficient to promote selective interaction. On a sequence level, this leaves only the homology between subtelomeric genes, short unique sequences, or else peculiarities of chromosome architecture as elements that control telomere–telomere pairing.
To test systematically whether telomere-specific sequences influence juxtaposition in trans, we have developed a chromosome “swap” technique as well as novel automated distance-measuring software. The swap method allows us to exchange endogenous sequences between different chromosomes, altering the length of chromosome arms, without introducing foreign sequence elements. We scored the distance between a large number of tagged telomere pairs by measuring their separation in 3D space in living cells. We conclude from our results that the architecture of the chromosome itself, i.e., the position of the centromere and relative lengths of its chromosome arms, is a major determinant of long-range telomere–telomere interaction. This presumably reflects the relative positioning of telomeres in the Rabl orientation in anaphase and telophase cells.
Results
Dynamic organization of telomeric clusters
Yeast telomeres cluster in a variable number of foci that interact with the nuclear envelope. The clusters dissociate partially in metaphase (Laroche et al. 2000; Smith et al. 2003) and reform in early G1. Moreover, they are in constant random motion, within a confined and largely perinuclear domain (Heun et al. 2001; Hediger et al. 2002). Given this, it was unclear whether telomeric foci would reflect a reproducible subset of chromosomal ends or if focus composition would vary continuously. In an attempt to analyze the stability of telomeric foci, we monitored their dynamics using a fully functional fusion between GFP and the telomere binding factor Rap1 on a Nipkov spinning disc confocal microscope. Rap1-GFP is expressed at the endogenous gene locus and yields a pattern of labeling identical to the well-characterized Rap1 immunostaining (Gotta et al. 1996; Fig. 1A). This allowed us to visualize all telomeric foci in a given cell and track their movement over 90 min using the Imaris software (Bitplane; Fig. 1A; Supplemental Movie 1). Figure 1A shows a few frames from a typical deconvolved movie that tracks the telomere behavior for ∼6 min. We initially distinguish five separate foci, of which the two at the left (here labeled in green and yellow) fuse by 40 sec and then separate again by 1 min 40 sec. The same is true for the two foci at the right of the image, shown in red and blue (Fig. 1A). A large number of cells were examined, and in all cases the fusion and fission of telomeric foci occurred on a time scale of minutes. We conclude that this is a general feature of yeast nuclear organization (see Supplemental Movie 1).
Figure 1.
In vivo 4D fluorescence microscopy of telomeric foci. (A) Time-lapse 3D microscopy on haploid budding yeast in interphase carrying an integrated Rap1-GFP fusion under its own promoter. Microscopy was performed as described in the Methods, with stacks taken at 20-sec intervals over 90 min. Shown are 3D reconstructions from a typical 6-min series. Regions of maximal Rap1-GFP intensity were detected using the spot detection tool from the Imaris software from Bitplane, allowing each focus to be differentially labeled. When two foci fuse, they adopt one color, allowing us to score fusion and fission events. (B) Live time-lapse 3D microscopy and focus detection are as in A, but of a haploid yeast cell in interphase expressing Sir3-GFP under its own promoter. 3D stacks were taken at 30-sec intervals. (C) A kymograph of time-lapse imaging of Sir3-GFP as in B but spanning 30 min. The X-axis represents time. (Arrows) Branch or fusion points of Sir3-GFP foci. See Supplemental Movie 2 for rotation of the kymograph.
Fusion and fission is not restricted to nonsilenced populations of telomeres. Indeed, a similar behavior was observed in time-lapse movies of Sir3-GFP, which is a marker for telomere-associated silent chromatin (Fig. 1B). In a kymograph of 3D stacks taken at 30-sec intervals over 30 min, Sir3-GFP tagged telomeric foci repeatedly fuse or branch from each other (Fig. 1C). The full rotation of this kymograph (Supplemental Movie 2) rules out that the apparent fusion and fission of foci is an artefact of spatial projection. This behavior argues that telomeres may redistribute among foci; nonetheless, we can track distinct clusters for 5 to 10 min that have little or no visible telomere exchange.
To better estimate the amount of time a single telomere remains associated with a given telomeric cluster, we combined a general label for telomeres (Rap1 fused to YFP) with the integration of lacO repeats at Tel14L, in a cell expressing LacI-CFP. The colocalization of Tel14L with a large telomeric cluster could be scored by time-lapse 3D microscopy, since Rap1-YFP is known to label all telomeres thanks to its affinity for the TG repeat (Fig. 2; Supplemental Movie 3). As shown in Figure 2B, the association of Tel14L with a Rap1-staining focus persisted for up to 5 min (from ∼3 to 8 min). Association was not continuous, however, and at times Tel14L could be seen to move into and out of foci (Supplemental Movie 4). A quantitative 3D analysis of 220 frames (five time-lapse series totaling 110 min from five different cells) showed that Tel14L is either within or adjacent to a telomeric focus 70% of the time. The average residence time is on the order of minutes, not seconds, which is sufficient to allow preferential interactions between telomeres to be scored. To analyze specific telomere–telomere interactions, we next differentially tagged specific pairs of telomeres with lacO and tetO repeats, in order to monitor the distribution of distances separating them over a population of cells. The choice of telomeres was made based on the sequence homology as described below.
Figure 2.
Dynamics of telomeric clusters relative to a single telomere. (A) 3D reconstruction of a yeast nucleus expressing Rap1-YFP, which labels all telomeres, and LacI-CFP, which recognizes a lacO array inserted on Tel14L (GA-2558). The larger view is rotated to allow visualization in the form of two other versions using Imaris software. The position of Tel14L (green) is scored relative to the Rap1 foci (red). (c) Tel14L colocalizing with a Rap1 focus, (ad) Tel14L adjacent to a Rap1 focus, (ap) Tel14L apart from Rap1 foci. Two-hundred-twenty nuclei reconstituted in 3D were analyzed, and in 70% of the nuclei Tel14L was either colocalizing or adjacent to the Rap1 focus. (B) Time-lapse imaging of the same yeast strain presented in A (GA-2558). 3D image stacks were taken at 30-sec intervals, deconvolved, and are shown in a 3D reconstitution. The top view is shown, and the juxtaposition of Tel14L to Rap1 was scored and indicated as in A. Continuous juxtaposition can be seen over 5 min.
Yeast telomeres fall into subgroups based on subtelomeric homology
The pairwise analysis of subtelomeric sequences from the Saccharyomyces cerevisiae S288c background shows regions of >90% identity scored over 1-kb intervals, extending up to 21 kb (Supplemental Fig. 3). If chromosomes are grouped on the basis of this subtelomeric homology, we find that all but three telomeres can be partitioned into seven groups of two or more telomeres, based on sequence alone. Only Tel5R, Tel6R, and Tel16R have no extensive homology with any other telomere, apart from the ubiquitous TG1–3 repeats and telomere-associated sequences (TAS), which include the Y′, STR, and X elements (Supplemental Fig. 3).
Various studies have shown that the conserved subtelomeric regions are not repetitive, but contain highly homologous gene families that are often implicated in alternative carbon source metabolism. Falling into this category are the PAU, COS, and HXT families, which encode seriPAUperin, integral membrane proteins, or hexose transporters, respectively (Viswanathan et al. 1994; Ozcan and Johnston 1999; Poirey et al. 2002). Because the number of sequence-derived telomere groups roughly matches the number of telomeric foci detected by microscopy, we tested whether the homology of subtelomeric gene families drives telomere juxtaposition. Appropriate pairs of yeast telomeres were differentially labeled with lacO or tetO arrays, which can be inserted without deleting endogenous sequences. These were visualized by coexpression of YFP- or CFP- tagged LacI and TetR proteins (Robinett et al. 1996; Michaelis et al. 1997; Bystricky et al. 2004).
Homology between subtelomeric sequences does not determine clustering
Using 3D imaging of live S288C yeast cells bearing differentially tagged telomere pairs, we acquired 21-plane through-focal stacks of haploid cells on media-containing agar (Fig. 3A). Cell cycle stages were classified based on cell morphology, and distances between the two telomere spots in G1- or S-phase cells were measured in 3D using an automated SpotDistance plug-in for ImageJ software (see Methods; Supplemental Figs. 1, 2). Between 150 and 300 distance measurements were analyzed for each telomere pair using R (www.r-project.org; see box plot, Fig. 3A). Because of the constant motion inherent to yeast DNA (Heun et al. 2001), we obtained a distance distribution for each pair, which is presented as a box plot. To determine whether two distributions were significantly different, we used a two-sample Kolmogorov–Smirnov (KS) test (Kolmogorov 1956).
Figure 3.
Extensive homology does not confer interaction in telomeric foci. (A) 3D image stacks were acquired at 0.2-μm spacing along the Z-axis of the indicated yeast strains having targeted integration of lacO and tetO arrays and expressing LacI-CFP and TetR-YFP. Image stacks were analyzed by the SpotDistance plug-in of ImageJ and are represented as a box plot. The notch around the median represents ±5%. Outliers are defined as 1.5 times the Inter Quartile Range (IQR) (open circles). (B) (Left panel) Sketch of various telomere pairs represented as lines with TG repeats (black box), Y′ elements (open blue box), STR elements (open red box), and X core elements (green) indicated to the left. The shading between telomeres indicates >90% identity. (Right panel) Distance distributions between the telomeres in the left panel represented as box plots. The strains and number of cells analyzed per pair are as follows: (panel 1) GA-2686, n = 282, pair: 10L–10L; (panel 2) GA-2685, n = 315, pair: 9L–10L; (panel 3) GA-2753, n = 331, pair 14L–6L; (panel 4) GA-2691, n = 437, pair: 10L–16L; (panel 5) GA-2687, n = 367, pair: 9L–14R; (panel 6) GA-3268, n = 286, pair: 14R–10L; (panel 7) GA-2731, n = 91 pair: 14L–16L; (panel 8) GA-958, n = 231, pair: 6L–6R.
Confirming previous measurements made for Tel6R and Tel6L in another yeast background (W303; Bystricky et al. 2005), we found that these two telomeres were also closely juxtaposed in S288c cells, with a median 3D separation of 460 nm. In many cells, the two signals completely overlapped (Fig. 3B, bottom panel). To calibrate our distance measurements, we scored the distances between lacO and tetO arrays integrated next to each other on the same telomere using the same imaging and analysis method (Fig. 3B, top panel). The distance distribution for this physically linked pair was identical to that obtained for Tel6R and Tel6L (KS-test, P = 0.17), arguing that a mean center-to-center separation of 460 nm reflects the subdiffusive movement of two loci that are indeed physically adjacent. This value is not unexpected given the poor resolution in Z (0.5 μm), and the fact that a telomeric focus is 300–400 nm in diameter.
We next measured the separation between telomere pairs that share extensive sequence identity due to the presence of either PAU/VTH or COS family members. The two most homologous ends in this respect are Tel9L and Tel10L, which have >90% identity over 21 kb. Here, separation values peaked at 900 nm, with 50% of the values falling between 600 and 1200 nm (Fig. 3B, panel 2). For two highly homologous COS-containing ends, Tel6L and Tel14L, values centered around 1100 nm, and both sets of measurements argue against prolonged or stable colocalization in a telomeric focus. We nonetheless find it noteworthy that the distances separating the Tel9L–Tel10L pair are slightly smaller than those scored for Tel6L–Tel14L, or for two totally unrelated telomere pairs (Tel10L–Tel16L: Fig. 3B, panel 4, or Tel9L–Tel14R: Fig. 3B, panel 5). This difference cannot be attributed to the TAS, since all telomere pairs in panels 1–3 contain Y′, X, and STR motifs. Rather, the skewed Tel9L–Tel10L distribution may stem from infrequent events that bring the two ends together, even though they are not stably juxtaposed. This could either cause or result from the preferential recombination reported to occur between Y′ elements of these homologous ends (Louis et al. 1994). Despite this, our data argue that all these telomere pairs spend most of the time apart, allowing us to conclude that extensive subtelomeric homology does not promote telomere pairing in trans.
The Tel6R–Tel6L pair is characterized by having one telomere with a Y′ element and one telomere without it, which may also contribute to their juxtaposition. However, a similar alternating pattern of Y′ elements is found on the Tel9L–Tel14R pair and the Tel10L–Tel14R pair (Fig. 3, panels 5, 6), which show separations of ∼1000 nm. Furthermore, we see no correlation between telomere interaction and arm length when the tagged loci are on different chromosomes (Fig. 3B, panels 3, 7; see left column). Finally, in addition to the pairs reported above, five further telomere pairs were tested by visual inspection and nonautomated distance determination in the yeast background W303 (G. Van Houwe, data not shown). Thus, with over 12 telomere pairs examined, only Tel3R–Tel3L and Tel6R–Tel6L showed unusually close juxtaposition. From this extensive pairwise analysis of telomeres with extensive sequence identity, we concluded that neither similarity of subtelomeric sequence nor the presence of coordinately regulated gene families drives telomere interactions in yeast.
A sequence element in the Tel6R favors association with Tel6L
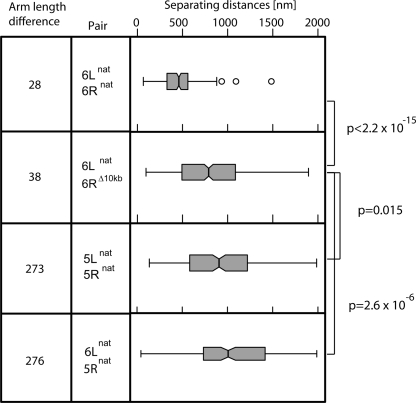
Given that only two sets of telomere ends showed significant interaction (the colinear ends of chromosomes 6 and 3), we considered two models that could explain their preferred interaction. One model argued that telomere juxtaposition arises from the fact that both chromosomes are short and metacentric. The second possibility invokes bridging factors that recognize a sequence element that is too short or too divergent to score in the homology scan used in Supplemental Figure 3. To test this latter hypothesis, we deleted the most distal 10 kb of Tel6R to create Tel6RΔ10kb and analyzed its separation from Tel6L in 3D (Fig. 4). The truncation shortens the chr 6R arm from 121 to 111 kb, a reduction of <10%. The protective TG1–3 cap remains and confers equal, if not more efficient, silencing and anchorage activity (Hediger et al. 2002). Nonetheless, removal of the Tel6R sequence led to a highly significant increase in the separation of Tel6RΔ10kb from Tel6L, shifting the median from 460 nm to 790 nm (Fig. 4; P < 2.2 × 10−15). The simplest explanation is that the 10-kb terminal truncation removes a positive determinant that favors interaction, which could map to either the subtelomeric X element or regions flanking a nonrepetitive ORF encoded in this region. Sequence analysis of these 10 kb revealed no highly conserved elements and few computationally determined transcription factor binding sites (Pachkov et al. 2007; Supplemental Fig. 7).
Figure 4.
Truncation of Tel6R impairs Tel6R–Tel6L interaction. Distance distribution box plots were performed and are represented as in Fig. 3 for the indicated pairs of telomeres. The relevant strains and numbers of cells analyzed are as follows: (panel 1) GA-958, n = 231, pair: 6L–6R; (panel 2) GA-3742, n = 368, pair: 6L–6RΔ10kb; (panel 3) GA-3708, n = 202, pair: 5L–5R natural; (panel 4) GA-957, n = 180, pair: 6L–5R. The indicated P-values compare the likelihood that the two indicated patterns are different, and was calculated in R using the two-sample Kolmogorov–Smirnov test.
Short telomeric sequences are insufficient to promote long-range interactions
We note that the median distance that separates the truncated Tel6RΔ10kb from Tel6L is still smaller than that separating the ends of the acrocentric chr 5 (Fig. 4, cf. Tel6Lnat–Tel6RΔ10kb and Tel5Lnat–Tel5Rnat). This argues for a combinatorial effect of linkage and/or arm length on telomere juxtaposition. To test the importance of either linkage (also called colinearity) or chromosome arm length, we developed a system that exchanges terminal sequences between two chromosome ends without otherwise altering the sequence of the chromosome. In this chromosome swap technique, an inducible, site-specific recombination event exchanges chromosome arms (Fig. 5). Since the target sites are tagged with individual loxP target sites, only the distal parts of chromosomes are exchanged. A detailed explanation is available in the Supplemental material and Supplemental Figure 5. Contour-clamped homogeneous electric field (CHEF) gel electrophoresis and Southern blot analysis confirmed in each case that a reciprocal swap occurred (Fig. 5D).
Figure 5.
Reciprocal exchange of telomeres and chromosome arms in yeast. (A) Depiction of native chr 5 (red) and chr 6 (blue). (B,C) Overview of the telomere swap (B) and chromosome arm swap (C) (see Supplemental Fig. 5 for detailed explanation). Black bars underneath the chromosomes indicate the annealing positions of the probes used for Southern hybridization. (D) Total intact genomic DNA in the form of chromosomes was isolated from the indicated strains before and after the exchange (swap). Chromosomes were separated by CHEF gel electrophoresis and stained with ethidium bromide. Southern blot analysis was performed with probes indicated in (A–C). (Lane 1) GA-3146, (lane 2) GA-1607, (lane 3) GA-1459, (lane 4) GA-1095, (lane 5) GA-3747, (lane 6) GA-3746, (lane 7) GA-3749, (lane 8) GA-3748, (lane 9) GA-3747, (lane 10) GA-3746, (lane 11) GA-3749, (lane 12) GA-3748.
Given that the loss of 10 kb from Tel6R affects its interaction with Tel6L, we next tested whether the terminal sequences of Tel5R can substitute for those removed in the Tel6R deletion. To achieve this, we exchanged the terminal 14 kb of Tel5R with those of Tel6R and monitored distances separating the newly formed Tel6R5Rsubtelo and Tel6L. As for the Tel6R deletion, the distances between Tel6L and Tel6R5Rsubtelo were significantly larger (median = 635 nm; Fig. 6A, panel 2) than those separating native chr 6 ends. Thus, restoration of length and the presence of the X element are insufficient for the optimal Tel6L–Tel6R interaction; instead, we conclude that a nonrepetitive element in the terminal 10 kb of chr 6R aids this selective interaction.
Figure 6.
Equal chromosomal arm length and colinearity promote telomere interaction. (A) Distance distributions were determined between the indicated lacO and tetO tags inserted in isogenic strains. The results are represented as box plots as in Fig. 3. A representation of the analyzed chromosome is indicated as folded in a Rabl configuration. The relevant pairs of loci analyzed are as follows: (1) Tel6L–Tel6R (GA-958, n = 231); (2) Tel6L–Tel6R5Rsubtelo (GA-1094, n = 194); (3) Tel6L–Tel5R6Rsubtelo (GA-1095, n = 265); (4) Tel6L–Tel5R (GA-957, n = 180). (B) Superposition of a brightfield image with a maximal projection of fluorescence image stacks is shown for anaphase cells bearing differentially tagged telomeres as indicated. Bar, 2 μm. (Upper left panel) A cell bearing lacO and tetO tags at Tel6L and Tel6R, respectively (GA-958), (upper right panel) Tel5L and Tel5R (GA-3746), (lower right panel) Tel6L and Tel5R (GA-957), (lower left panel) probable configuration of chr 6 in anaphase. (C) Quantitative analysis of Tel6L–Tel 6R (GA-958, n = 36), Tel5L–Tel5R (GA-3746, n = 33), and Tel6L–Tel5R (GA-957, n = 30) in anaphase and telophase cells. The two-sample Kolmogorov–Smirnov test confirmed that the differences between the distributions are significant. (D) As in A, but box plots are shown for the following strains resulting from chromosome arm exchange: (1) Tel5L–Tel5R (GA-3746, n = 307); (2) Tel5L–ARS514 (GA-3747, n = 320); (3) Tel5L–ARS5146Rsubtelo (GA-3749, n = 211); (4) Tel5L–Tel6R5Rarm (GA-3748, n = 296); (5) Tel5L–ARS514 (GA-3689) in cells expressing either LexA-Ku80 or LexA-Sir4, which then bind ARS514 bearing 8× LexA binding sites.
Given the importance of the distal 10 kb of Tel6R, we next examined whether it was sufficient to promote a stable interaction with Tel6L in trans. We therefore measured the separation of the recipient in the telomere swap, Tel5R6Rsubtelo, from Tel6L. However, the presence of sequences from Tel6R on chr 5 had no effect on the relative position of Tel6R and Tel5R (Fig. 6A, panels 3,4; KS-test, P = 0.18). We conclude that the positive element at Tel6R cannot promote interaction with an unlinked telomere (i.e., Tel6L–Tel5R6Rsubtelo).
This failure might be due to a need for centromere-proximal sequences from chr 6R that cooperate with the distal element to promote interaction with Tel6L in trans. We could test this by analyzing the distance distribution of Tel6L–Tel6R in a diploid strain in which the two fluorescent tags were integrated either on the same chromosome (in cis) or one on each of the two chr 6 homologues (in trans, Supplemental Fig. 4). When the two tags were on opposite ends of distinct homologs (i.e., in trans), we found that preferred juxtaposition was lost (median = 1065 nm, Supplemental Fig. 4). In contrast, the distance distribution for the two tags placed on opposite ends of the same chromosome in a diploid was indistinguishable from that in a haploid (median = 490 nm and 460 nm, respectively). This suggests that the distal 10 kb of Tel6R is not sufficient to mediate interaction with Tel6L when the two are unlinked, and that its inability to function on Tel5R is not due to the absence of more centromere proximal sequences. We conclude therefore that the contribution of a subtelomeric Tel6R sequence to interaction with Tel6L requires a predisposition for juxtaposition conferred by colinearity.
Rabl folding and chromosome arm length influence telomere interactions
Chromosome 6 is the third smallest chromosome in yeast and is metacentric, with arm lengths of 121 kb and 148 kb. We note that 30 kb of folded chromosomal DNA covers a distance of ≅160–180 nm in living cells (Bystricky et al. 2004), which is significantly less than the diameter of the telomeric focus. Thus, if chr 6 were to fold centrally, Tel6R and Tel6L would be equidistant from the centromere. This conformation occurs in living cells during anaphase, when chromosomes are pulled by their centromeres to opposite ends of the nucleus. In this so-called Rabl configuration, sites that are equidistant in kilobases from the centromere could be near each other in 3D space. This phenomenon, originally described in amphibians (Rabl 1885), was confirmed for yeast by monitoring the distance between tagged Tel6R and Tel6L in late anaphase and telophase cells (Fig. 6B,C). For acrocentric chromosomes such as chr 5, telomeres were not juxtaposed in anaphase (median separation = 670 nm; Fig. 6C), although this value was significantly smaller than the separation of unlinked telomeres (Tel6L–Tel5R) or even the separation of Tel5R–Tel5L in interphase. These considerations suggest that particularly in anaphase, the linear distance of a sequence from the centromere may influence juxtaposition both during segregation and in the ensuing G1-phase nucleus. A rigorous test of this hypothesis could be performed by exploiting the chromosome swap technique. We set out to modulate the length of the long arm of chr 5, creating a metacentric chromosome from the natural acrocentric chr 5 (Fig. 5C). If equal chromosome arm lengths are important, this alteration would be expected to reduce the separation of Tel5R and Tel5L in interphase cells.
We first analyzed the distance between Tel5L and a fluorescent tag inserted at ARS514, an origin of replication on the right arm of chr 5 equidistant with Tel5L from the centromere (Fig. 6D). The box plot for the distances measured between Tel5L and ARS514 shows significantly smaller distances than those separating Tel5L and Tel5R (KS-test, P = 6 × 10−7). While this is consistent with our hypothesis that the polarized anaphase organization has an effect on long-range interactions, these sites were nonetheless not as close as Tel6R–Tel6L.
We next tested whether we could improve the interaction by inserting a telomere at the ARS514 site. This manipulation would recreate the overall chromosomal architecture of chr 6 within chr 5. To achieve this without losing essential information on the distal part of chr 5R, we swapped the entire ARS514-distal 5R arm with the terminal 10-kb fragment of Tel6R and monitored the distances separating the existing centromere-proximal fluorescent tags in 3D. The truncation of chr 5R improved interaction between the ARS514 tag and Tel5L, giving a median separation of 642 nm, which is statistically indistinguishable from the distance separating Tel6L from Tel6R5Rsubtelo (Fig. 6A, panel 2; KS-test P = 0.212). Remarkably, the distances separating the ends of this new metacentric chr 5 (chr 5 with chr 6-like architecture) became smaller than those separating Tel6L from truncated Tel6RΔ10kb (Fig. 4). In other words, by rendering chr 5 metacentric with the terminal sequences of Tel6R at one end, we obtained the same degree of juxtaposition as we detected for Tel6L–Tel6R5Rsubtelo. This juxtaposition is closer than that of Tel6L–Tel6RΔ10kb and suggests that long-range interactions require that the interacting sites are colinear and roughly equidistant from the centromere (±30 kb), even though interaction can also be aided by subtelomeric sequences.
Finally, we examined why ARS514 itself fails to interact with Tel5L. We note that terminal TG1–3 arrays bind Rap1 and lead to the accumulation of both Sir4 and yKu at telomeres (for review, see Gasser and Cockell 2001). Given that yKu or Sir4 interact with each other, dimerize with other Sir factors, and bind NE anchors (Hediger et al. 2002; Taddei et al. 2004a), it seemed plausible that their presence alone might be able to favor long-range interaction at sites equidistant from a given centromere. To test whether either Sir protein or yKu binding is sufficient to improve the juxtaposition of ARS514 to Tel5L without creation of a telomere, we integrated an array of lexA sites near the lacO insert at ARS514 and monitored distances separating this internal tag from Tel5L in the presence of either LexA-Ku80 or LexA-Sir4C (Fig. 6D, panel 5). The targeted binding of either fusion protein did not improve interaction with Tel5L, even though both constructs are able to mediate the stable relocation of internal sequences to the NE (Supplemental Fig. 6). Other evidence further rules out a correlation of efficient perinuclear attachment with telomere pairing, since Tel6R and Tel6R5Rsubtelo bind the NE with the same efficiency (Supplemental Fig. 6) yet pair with Tel6L with significantly different efficiency (Fig. 6A, cf. panels 1 and 2). These results argue that neither the efficiency of Sir factor or yKu binding nor the NE attachment they mediate is relevant for interactions in trans.
Our data strongly suggest that colinearity is a factor, but is not sufficient, to promote telomere–telomere interaction. We further explored this effect on the native acrocentric chr 5, by comparing the separation of its telomeres when they were either colinear or when the long arm of chr 5R was transferred to Tel6R. We found that the distances separating Tel5R from Tel5L were ∼1000 nm, whether or not the two were colinear (Fig. 6D, panels 1 and 4). This distribution was the same as that of two unlinked telomeres, Tel6L and Tel5R (Fig. 6A, panel 4). Because equilibrating the length of the two chr 5 arms significantly improved the juxtaposition of chr 5 telomeres, we conclude that metacentric structure is a necessary prerequisite for significant telomere interactions. Similar arm length without colinearity, however, is not sufficient, because no significant pairing could be detected between Tel6L–Tel6R when tags were placed on unlinked homologs in a diploid strain (Supplemental Fig. 4).
Discussion
The optimization of techniques for altering gross chromosomal organization and for monitoring spatial separation of loci in living cells has allowed us to examine what promotes long-range interactions between chromosomal sites in interphase nuclei. The 3D distance-measuring tool is applicable both to live imaging of CFP, YFP, GFP, or RFP-tagged foci in living cells, as well as two-color FISH or multicolor immunofluorescence labeling. Its application is highly appropriate for, but not restricted to, questions of nuclear organization. The second tool is an elegant chromosome swap technique that achieves a reciprocal exchange of chromosome arms, allowing one to reorganize eukaryotic chromosomes without a net loss of sequence. This has allowed us to examine the factors that influence telomere–telomere interaction in budding yeast in an unprecedented manner. Although an independent methodology for sequence exchange has been published (Delneri et al. 2003), the current protocol introduces a selection event that can be readily adapted to use in other organisms. In this current study, chromosome arm swapping has allowed us to show that metacentric chromosomal organization influences telomere–telomere interactions in yeast.
Short sequences, colinearity, and equal arm length drive telomere clustering
An analysis of subtelomeric sequences of yeast chromosomes indicated that they fall into seven subgroups reflecting the presence of subtelomeric gene families (Supplemental Fig. 3; see also www.nottingham.ac.uk/genetics/louis). Given that this number agreed roughly with the number of telomeric foci found in interphase, we went on to test the importance of sequence conservation for telomere–telomere pairing. We found no significant effect: Yeast chromosome ends bearing homologous genes, such as the co-regulated COS or PAU genes, were not found in a common focus. Even Tel9L and Tel10L, which have preferential Y′ exchange through recombination (Louis et al. 1994) and share >21 kb of sequence identity, do not colocalize. While interaction is obviously essential for homologous recombination, our data further suggest that stable juxtaposition is unlikely to be the rate-limiting factor in recombination between chromosomes in vegetatively growing yeast cells (Lee et al. 1999).
We confirmed past work that reported that the two telomeres of chr 6 associate to a high degree. It had been proposed that short metacentric chromosome structure would favor telomere interaction (Bystricky et al. 2005), a hypothesis that could be tested with our chromosome swap technique. We converted the acrocentric chr 5 into a metacentric chromosome by swapping the 5R and 6R arms. We found that this conversion led to a significantly closer juxtaposition of the ends of chromosome 5. The two ends were not as close as the two natural ends of chr 6: Contact between Tel6R and Tel6L is enhanced by a 10-kb subtelomeric region on Tel6R. Importantly, however, the Tel6R sequence promotes interaction only when two telomeres are physically connected. In conclusion, our experiments argue that colinearity is a prerequisite for what we presume to be a factor-mediated interaction between two telomeres.
Rabl organization in anaphase contributes to long-range interphase interactions
How can the colinearity of the extreme ends of a chromosome and distance from the centromere facilitate the pairing of telomeres in interphase? In both open and closed mitoses, centromeres are actively and coordinately pulled toward the spindle pole body or centrosome by the mitotic spindle, while the rest of the chromosome and the telomeres follow passively (Fig. 7). This anaphase pattern was first described in 1885 by Carl Rabl, who observed it in cells from an axolotl-like amphibian (Rabl 1885). Given a reasonably uniform compaction ratio for yeast mitotic chromatin (Lavoie et al. 2002), it follows from the Rabl configuration that sequences on opposite arms of the same chromosome equidistant from the centromere will be spatially juxtaposed in late metaphase (Fig. 7). This we confirmed by measurements in living yeast cells in anaphase (Fig. 6C). Because telomeres are released from the NE in mitosis and re-anchor in G1 (Laroche et al. 2000; Smith et al. 2003), it appears that the critical time for reestablishing telomere–telomere interactions is the transition from telophase to G1.
Figure 7.
The Rabl configuration in telophase can influence telomere juxtaposition. Shown is a model suggesting how the Rabl organization of chromosomes in late anaphase and telophase might influence telomere–telomere clustering into perinuclear foci in interphase. The drawing indicates how arm length and metacentricity lead to late anaphase juxtaposition. An unknown sequence- or structure-recognizing factor may further promote interaction between Tel6R and Tel6L.
The Rabl organization of yeast chromosomes occurs not only in anaphase but also in interphase cells (Bystricky et al. 2004, 2005). This configuration, combined with metacentric chromosome structure, ensures that telomere–telomere interactions can be initiated in telophase and stabilized in early G1 phase. We find that interactions between the left and right telomeres of the same chromosome are favored (Fig. 6B,C). The obvious candidates for stabilizing interactions (Sir proteins or yKu) are present on all telomeres and therefore cannot be responsible for selective interaction. The origin recognition complex (ORC) is also bound to TAS and proARS sites in subtelomeric regions and contributes to interactions between sister chromatids (Shimada and Gasser 2007). However, we find that the nucleation of silent chromatin is unable to promote interaction of the internal ARS514 with Tel5L (Fig. 6D, panel 5), suggesting that none of these elements is sufficient to induce telomere juxtaposition in trans. Rather, the coherence of the linear DNA polymer, which impairs random motion to define a spatial “chromosome territory” (Cremer and Cremer 2001), and the polarized Rabl configuration at the end of mitosis, function together to determine which telomeres cluster in yeast.
Superimposed on these fundamental factors, we have identified a small region in the distal sequences of Tel6R that enhances the interaction of both Tel6R with Tel6L, and when transferred to chr 5R, the interaction of Tel5Rsubtelo6R with Tel5L. We have examined the distal Tel6R sequence for unique or unusual characteristics. Genome-wide mapping studies have shown that Rap1, Sir protein, ORC, Htz1, and Cohesin distributions on both telomeres of chr 6 are indistinguishable from those at other telomeres (Lieb et al. 2001; Wyrick et al. 2001; Glynn et al. 2004; Raisner et al. 2005). While the deletion of YKU70 had minor effects on Tel6R–Tel6L interaction (Gehlen et al. 2006), we note that yKu binds all yeast telomeres, arguing that at the very least its influence must depend on another Tel6R-specific factor.
Because there are two hypothetical ORFs in the 10-kb region of contact-promoting Tel6R sequence, we speculated that Tel6L–Tel6R interaction may reflect the presence of binding sites for bivalent transcription factors (TF) at both chromosomal ends. Recently, van Nimwegen and colleagues computationally predicted binding sites for 75 different TFs over the entire S. cerevisiae genome, including the subtelomeric regions (Pachkov et al. 2007). We find several TF binding sites shared between Tel6L and Tel6R (Supplemental Fig. 7) that are absent from Tel5R (e.g., Thi2, Rpn4, Ste12, and Tec1). However, there is no evidence that these factors or their ligands dimerize. Based on predicted TF distributions, we entertain an alternative model for the effect associated with the deletion of the distal 10 kb of Tel6R. We propose that telomere interaction arises by default through the combined effect of anaphase juxtaposition and bivalent telomere factors (e.g., Sir4, yKu, Sir3, Rif1/2), while subtelomeric TF could attenuate such pairing. In this scenario, the difference between the interaction of Tel6L with Tel6R and with the truncated Tel6RΔ10kb might reflect the fact that truncation of Tel6R juxtaposes new TF consenses near the terminal complex, antagonizing telomere interactions (Supplemental Fig. 7, bottom panel). Since the positioning of such factors is still theoretical, a test of this model requires biochemical analysis of TF binding site occupancy.
The regularity of telomeric foci composition
Among more than 12 pairs of differentially tagged yeast telomeres, our laboratory has found a reproducible juxtaposition for only three pairs: Tel6R–Tel6L, Tel3R–Tel3L, and now for the ends of a metacentric (modified) chr 5. From this sample, we deduce that the composition of telomeric foci is largely stochastic, reflecting the chance juxtaposition of chromosome ends in anaphase, rather than a tightly orchestrated distribution. We propose that the formation of telomeric foci results from the combined effects of chromosome arm length and Rabl organization, which can be modulated by sequence-specific factors (see above). Given that there is significantly limited mobility of yeast telomeres in interphase cells (Heun et al. 2001; Hediger et al. 2002), the telomeres that end up proximal to each other in telophase are likely to remain close to each other in nuclei throughout G1 and S phase.
Very little is known about the mechanisms that drive heterochromatin clustering in other species. It was reported for Plasmodia that the loss of subtelomeric repeats affects interactions in trans (Figueiredo et al. 2002), yet the factors involved are unknown. In Schizosaccharomyces pombe, the RNAi machinery (ago1, dcr1, and rdp1) is crucial for the maintenance of telomere clustering (Hall et al. 2003), yet again, bridging molecules are unknown. Nonetheless, the loss of RNAi led to the loss of histone H3K9 methylation and displacement of Swi6 (HP1), yet did not disrupt telomeric repression. Furthermore, in both Drosophila and mammalian cells, the down-regulation or displacement of HP1 failed to disrupt the chromocenter, which derives from long-range interactions of pericentric heterochromatin (Maison et al. 2002; Peters et al. 2002). Based on our studies and on observations in other organisms, we propose that the global arrangement of chromosomes in anaphase, rather than local chromatin structure, has the dominant impact on long-range interactions in the subsequent interphase nucleus. Interactions seem to arise in part due to stochastic juxtaposition, yet lead to the variegated repression mediated by heterochromatin. Adaptations of the chromosome swap protocol presented here may facilitate the evaluation of such parameters in other species.
Methods
Live fluorescence microscopy
For live imaging, cultures were grown exponentially in synthetic medium to a concentration of <1 × 105 cells/mL. Live microscopy was performed at 30°C on cells spread on agarose patches containing synthetic complete medium with 4% glucose. Strains bearing integrated and fully functional fusions of Rap1-GFP and Sir3-GFP were imaged using a Ludin Chamber on a Metamorph-driven spinning disk confocal unit (Yokogawa, CSU22) built on a Zeiss AXIO microscope equipped with two Cascade II cameras. Image stacks of 21 focal planes were captured with a z-step size of 300 nm at 20-sec intervals for Rap1-GFP and at 30-sec intervals for Sir3-GFP, over periods up to 90 min. Image stacks were deconvolved using the Huygens software. Images for telomere–telomere distance measurements were captured on a Metamorph-driven Olympus IX70 wide-field microscope equipped with a Coolsnap HQ camera (Roper Scientific Photometrics). Alternating the wavelength between 437 nm (CFP) and 517 nm (YFP) at every image plane, stacks of 21 images were acquired with a step size of 0.2 μm. A 100×/1.4 Oil Plan-Apochromat objective from Zeiss was used.
Telomere to telomere distance measurement
A novel software package, called SpotDistance, implemented as a Java plug-in for the public-domain software ImageJ, was developed to measure distances between two target telomeres in an automated manner. It is freely available to the research community at http://bigwww.epfl.ch/spotdistance/. The plug-in places two input z-stacks, YFP-source and CFP-source images, into the red and green channels, respectively. For details of its functionality see Supplemental Figure 1. For graphic display, the measured distances were loaded into R software (www.r-project.org), and the distance distributions for each pair were displayed as a box plot. Different distance distributions were scored for significance using the two-sample Kolmogorov–Smirnov test.
Contour-clamped homogeneous electric field (CHEF) electrophoresis
Pulsed-field electrophoresis was conducted using the CHEF-DRII System from Biorad. Preparation of agarose-embedded whole chromosomes was done according the Biorad user’s manual. The gel was run at the following conditions: 5 V/cm2, 1× TAE, 10°C, block 1: 60-sec switch time for 15 hr, block 2: 90-sec switch time for 7 hr. Southern hybridization was performed using standard procedures with randomly primed probes that were amplified by PCR from genomic DNA.
Acknowledgments
We thank Thierry Laroche and Jens Rietdorf from the imaging platform at FMI for assistance, and Peter Meister, Angela Taddei, and members of the Gasser laboratory for discussions and advice. Our research is supported by the Swiss National Science Foundation, the NCCR program “Frontiers in Genetics,” and the Novartis Research Foundation. M.R.G. was supported by the Novartis Foundation during a sabbatical in the Gasser laboratory. M.V.P. thanks the Spanish Ministry of Science and Education for financial support.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6687808
References
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R., Neiman A.M., Zappulla D.C., Sternglanz R., Zappulla D.C., Sternglanz R., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Simpson M., West A., Felsenfeld G., West A., Felsenfeld G., Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. (Suppl. 4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Heun P., Gehlen L., Langowski J., Gasser S.M., Heun P., Gehlen L., Langowski J., Gasser S.M., Gehlen L., Langowski J., Gasser S.M., Langowski J., Gasser S.M., Gasser S.M. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Laroche T., van Houwe G., Blaszczyk M., Gasser S.M., Laroche T., van Houwe G., Blaszczyk M., Gasser S.M., van Houwe G., Blaszczyk M., Gasser S.M., Blaszczyk M., Gasser S.M., Gasser S.M. Chromosome looping in yeast: Telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S.E., Drewell R.A., Drewell R.A. Chromatin looping mediates boundary element promoter interactions. Bioessays. 2007;29:7–10. doi: 10.1002/bies.20520. [DOI] [PubMed] [Google Scholar]
- Chan C.S., Tye B.K., Tye B.K. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer C., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N., Rippe K., Dekker M., Kleckner N., Dekker M., Kleckner N., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Delneri D., Colson I., Grammenoudi S., Roberts I.N., Louis E.J., Oliver S.G., Colson I., Grammenoudi S., Roberts I.N., Louis E.J., Oliver S.G., Grammenoudi S., Roberts I.N., Louis E.J., Oliver S.G., Roberts I.N., Louis E.J., Oliver S.G., Louis E.J., Oliver S.G., Oliver S.G. Engineering evoluation to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C., Therizols P., Lafontaine I., Dujon B., Fairhead C., Lafontaine I., Dujon B., Fairhead C., Dujon B., Fairhead C., Fairhead C. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- Figueiredo L.M., Freitas-Junior L.H., Bottius E., Olivo-Marin J.C., Scherf A., Freitas-Junior L.H., Bottius E., Olivo-Marin J.C., Scherf A., Bottius E., Olivo-Marin J.C., Scherf A., Olivo-Marin J.C., Scherf A., Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.G., Merkenschlager M., Merkenschlager M. Gene silencing, cell fate and nuclear organisation. Curr. Opin. Genet. Dev. 2002;12:193–197. doi: 10.1016/s0959-437x(02)00286-1. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Cockell M.M., Cockell M.M. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Hediger F., Taddei A., Neumann F.R., Gartenberg M.R., Hediger F., Taddei A., Neumann F.R., Gartenberg M.R., Taddei A., Neumann F.R., Gartenberg M.R., Neumann F.R., Gartenberg M.R., Gartenberg M.R. The function of telomere clustering in yeast: The circe effect. Cold Spring Harb. Symp. Quant. Biol. 2004;69:327–337. doi: 10.1101/sqb.2004.69.327. [DOI] [PubMed] [Google Scholar]
- Gehlen L.R., Rosa A., Klenin K., Langowski J., Gasser S.M., Bystricky K., Rosa A., Klenin K., Langowski J., Gasser S.M., Bystricky K., Klenin K., Langowski J., Gasser S.M., Bystricky K., Langowski J., Gasser S.M., Bystricky K., Gasser S.M., Bystricky K., Bystricky K. Spatially confined polymer chains: Implications of chromatin fibre flexibility and peripheral anchoring on telomere–telomere interaction. J. Phys. Condens. Matter. 2006;18:245–252. [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Koshland D.E., DeRisi J.L., Gerton J.L., DeRisi J.L., Gerton J.L., Gerton J.L. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M., Formenton A., Maillet L., Scherthan H., Gasser S.M., Maillet L., Scherthan H., Gasser S.M., Scherthan H., Gasser S.M., Gasser S.M. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type S. cerevisiae. J. Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Noma K., Grewal S.I., Noma K., Grewal S.I., Grewal S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G.R., Bumgarner S., Styles C., Fink G.R., Styles C., Fink G.R., Fink G.R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M., Van Houwe G., Dubrana K., Gasser S.M., Dubrana K., Gasser S.M., Gasser S.M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Heun P., Laroche T., Shimada K., Furrer P., Gasser S.M., Laroche T., Shimada K., Furrer P., Gasser S.M., Shimada K., Furrer P., Gasser S.M., Furrer P., Gasser S.M., Gasser S.M. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Robertson E.D., Donaldson A.D., Robertson E.D., Donaldson A.D., Donaldson A.D. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J. 2006;25:1505–1514. doi: 10.1038/sj.emboj.7601038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov A.N. Foundations of the theory of probability. Chelsea Publishing Company; New York: 1956. [Google Scholar]
- Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M., Pryde F.E., Louis E.J., Gasser S.M., Louis E.J., Gasser S.M., Gasser S.M. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Tsai-Pflugfelder M., Gasser S.M., Martin S.G., Tsai-Pflugfelder M., Gasser S.M., Tsai-Pflugfelder M., Gasser S.M., Gasser S.M. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 2000;129:159–174. doi: 10.1006/jsbi.2000.4240. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Hogan E., Koshland D., Hogan E., Koshland D., Koshland D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Paques F., Sylvan J., Haber J.E., Paques F., Sylvan J., Haber J.E., Sylvan J., Haber J.E., Haber J.E. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Lieb J.D., Liu X., Botstein D., Brown P.O., Liu X., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Louis E.J., Haber J.E., Haber J.E. The structure and evolution of subtelomeric Y′ repeats in S. cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E.J., Naumova E.S., Lee A., Naumov G., Haber J.E., Naumova E.S., Lee A., Naumov G., Haber J.E., Lee A., Naumov G., Haber J.E., Naumov G., Haber J.E., Haber J.E. The chromosome end in yeast: Its mosaic nature and influence on recombinational dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M., Gotta M., Marcand S., Gilson E., Gasser S.M., Marcand S., Gilson E., Gasser S.M., Gilson E., Gasser S.M., Gasser S.M. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Taddei A., Lachner M., Jenuwein T., Almouzni G., Lachner M., Jenuwein T., Almouzni G., Jenuwein T., Almouzni G., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., Ciosk R., Nasmyth K., Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Mitchell J.A., Lopes S., Reik W., Lopes S., Reik W., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Ozcan S., Johnston M., Johnston M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachkov M., Erb I., Molina N., van Nimwegen E., Erb I., Molina N., van Nimwegen E., Molina N., van Nimwegen E., van Nimwegen E. SwissRegulon: A database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M., Gilson E., Axelrod A., Pillus L., Gasser S.M., Axelrod A., Pillus L., Gasser S.M., Pillus L., Gasser S.M., Gasser S.M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Mermoud J.E., O’Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T., Mermoud J.E., O’Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T., O’Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T., Pagani M., Schweizer D., Brockdorff N., Jenuwein T., Schweizer D., Brockdorff N., Jenuwein T., Brockdorff N., Jenuwein T., Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Poirey R., Despons L., Leh V., Lafuente M.J., Potier S., Souciet J.L., Jauniaux J.C., Despons L., Leh V., Lafuente M.J., Potier S., Souciet J.L., Jauniaux J.C., Leh V., Lafuente M.J., Potier S., Souciet J.L., Jauniaux J.C., Lafuente M.J., Potier S., Souciet J.L., Jauniaux J.C., Potier S., Souciet J.L., Jauniaux J.C., Souciet J.L., Jauniaux J.C., Jauniaux J.C. Functional analysis of the S. cerevisiae DUP240 multigene family reveals membrane-associated proteins that are not essential for cell viability. Microbiol. 2002;148:2111–2123. doi: 10.1099/00221287-148-7-2111. [DOI] [PubMed] [Google Scholar]
- Rabl C. Über Zellteilung. Morphologisches Jahrbuch. 1885;10:214–330. [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Schreiber S.L., Rando O.J., Madhani H.D., Rando O.J., Madhani H.D., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Willhelm C., Sudlow G., Murray A., Belmont A.S., Sudlow G., Murray A., Belmont A.S., Murray A., Belmont A.S., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Figueiredo L.M., Freitas-Junior L.H., Figueiredo L.M., Freitas-Junior L.H., Freitas-Junior L.H. Plasmodium telomeres: A pathogen’s perspective. Curr. Opin. Microbiol. 2001;4:409–414. doi: 10.1016/s1369-5274(00)00227-7. [DOI] [PubMed] [Google Scholar]
- Shimada K., Gasser S.M., Gasser S.M. The origin recognition complex functions in sister-chromatid cohesion in S. cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- Smith C.D., Smith D.L., DeRisi J.L., Blackburn E.H., Smith D.L., DeRisi J.L., Blackburn E.H., DeRisi J.L., Blackburn E.H., Blackburn E.H. Telomeric protein distributions and remodeling through the cell cycle in S. cerevisiae. Mol. Biol. Cell. 2003;14:556–570. doi: 10.1091/mbc.E02-08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M., Hediger F., Neumann F.R., Bauer C., Gasser S.M., Neumann F.R., Bauer C., Gasser S.M., Bauer C., Gasser S.M., Gasser S.M. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004a;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Gasser S.M., Hediger F., Neumann F.R., Gasser S.M., Neumann F.R., Gasser S.M., Gasser S.M. The function of nuclear architecture: A genetic approach. Annu. Rev. Genet. 2004b;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E., Olivo-Marin J.C., Dujon B., Fabre E., Dujon B., Fabre E., Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakainen H., Naumov G., Naumova E., Korhola M., Naumov G., Naumova E., Korhola M., Naumova E., Korhola M., Korhola M. Physical mapping of the MEL gene family in S. cerevisiae. Curr. Genet. 1993;24:461–464. doi: 10.1007/BF00351706. [DOI] [PubMed] [Google Scholar]
- Viswanathan M., Muthukumar G., Cong Y.S., Lenard J., Muthukumar G., Cong Y.S., Lenard J., Cong Y.S., Lenard J., Lenard J. Seripauperins of S. cerevisiae: A new multigene family encoding serine-poor relatives of serine-rich proteins. Gene. 1994;148:149–153. doi: 10.1016/0378-1119(94)90249-6. [DOI] [PubMed] [Google Scholar]
- Wyrick J.J., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Young R.A., Bell S.P., Aparicio O.M., Bell S.P., Aparicio O.M., Aparicio O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]