Abstract
Using a combination of bacterial and phage-targeted metagenomics, we analyzed two geographically remote sludge bioreactors enriched in a single bacterial species Candidatus Accumulibacter phosphatis (CAP). We inferred unrestricted global movement of this species and identified aquatic ecosystems as the primary environmental reservoirs facilitating dispersal. Highly related and geographically remote CAP strains differed principally in genomic regions encoding phage defense mechanisms. We found that CAP populations were high density, clonal, and nonrecombining, providing natural targets for “kill-the-winner” phage predation. Community expression analysis demonstrated that phages were consistently active in the bioreactor community. Genomic signatures linking CAP to past phage exposures were observed mostly between local phage and host. We conclude that CAP strains disperse globally but must adapt to phage predation pressure locally.
Ecological theory is largely grounded on the study of macroscopic communities (Begon et al. 2006). Microbial communities are compelling alternative systems for testing ecological concepts because microorganisms have shorter generation times and can be studied under controlled conditions (Buckling and Rainey 2002; Jessup et al. 2004). However, microbial ecology has been limited by technological hurdles, namely the inability to characterize most microbial species because of a cultivation bottleneck and the inability to distinguish microorganisms at high resolution (species and strains) and track them in situ (Pace 1997).
Molecular methods developed over the past decade are addressing these limitations and allowing microbial ecology to mature as a discipline and, in the process, are challenging long-held assumptions about microbial populations. For example, multilocus sequence typing (MLST) (Maiden et al. 1998) has challenged the notion of general asexuality of microbial populations by demonstrating high rates of homologous recombination in some bacterial species (Feil et al. 2000). Another widely held belief, regarding the lack of geographic boundaries for microbial populations, has also been challenged by MLST-based studies of extremophiles (Papke et al. 2003; Whitaker et al. 2003).
Metagenomics, the application of shotgun sequencing to environmental samples, holds the promise of providing the least biased (culture-independent) and most comprehensive (genome-wide) resolution of sympatric populations (Whitaker and Banfield 2006). We analyzed metagenomic data from two Enhanced Biological Phosphorus Removal (EBPR) sludges dominated (up to 80% of the biomass) by an as-yet uncultured species, Candidatus Accumulibacter phosphatis (CAP) (Garcia Martin et al. 2006). The sludge samples were obtained from two geographically remote laboratory-scale bioreactors, one from Madison, Wisconsin, USA (US sludge), and the other from Brisbane, Queensland, Australia (OZ sludge). In addition, a phage-enriched sample of the US sludge was obtained for shotgun sequencing seven months after sampling for the bacterial metagenome. A microarray was prepared from both US data sets to examine gene expression of the bacterial and phage communities. Here we report global dispersal of, and local predation pressure on, the CAP populations revealed through comparative genomic analysis and expression data.
Results and Discussion
We began by searching for evidence of geographic isolation of the CAP populations by analyzing the phylogenetic distribution of 48 single-copy genes (Supplemental Table S1). Using single-copy genes ensures that any given strain is not represented by more than one sequence and therefore minimizes the possibility of misinterpreting paralogs as orthologs (Venter et al. 2004). PCR clone libraries were prepared for an additional gene, polyphosphate kinase (ppk), which has previously been used to strain-type CAP (McMahon et al. 2002). We determined the presence of multiple CAP strains in both the US and OZ sludge samples whose genes typically only diverged by up to 4% at the nucleotide level (Fig. 1). Additional distinct Accumulibacter populations were also identified with an average nucleotide sequence divergence of 15% from CAP (Fig. 1).
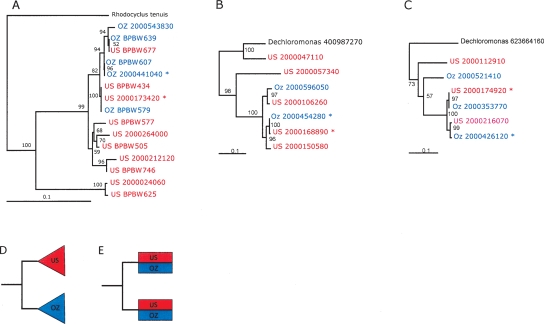
Figure 1.
Gene phylogenies reconstructed using nucleotide sequence show geographic intermingling of CAP strains. Sequences obtained from the US and OZ samples are shown in red and blue, respectively. Asterisks mark dominant strains. IMG (Markowitz et al. 2006) gene object identifiers (beginning with 2000) are given for genes derived from metagenomic data. Support for interior nodes are indicated by bootstrap resampling percentages. The following trees are shown. (A) Polyphosphate kinase (PCR-amplified clones begin with BPBW); (B) Ribosomal protein L9; (C) holiday junction resolvase, DNA-binding subunit RuvA. Schematics are provided for reference to show the expected tree topologies for endemic (D) and freely migrating (E) populations. Note that in the latter case, high recombination frequencies between local and introduced strains may mask dispersal patterns.
Contrary to recent findings in hot springs using high-resolution molecular methods (Papke et al. 2003; Whitaker et al. 2003), no phylogenetic separation based on geographic locale was observed. In all trees with adequate strain representation, the US and OZ CAP strains were intermingled. Furthermore, instances of identical US and OZ ppk genes were found (Fig. 1). This indicates global dispersal of CAP strains since the two sampled EBPR sludges have not been in direct contact and both laboratory-scale reactors were inoculated from local full-scale EBPR sludges that had been operating in EBPR mode for over 5 yr. To our knowledge, there was no intentional transfer of sludge between either the bioreactors or wastewater treatment plants from which they were derived.
To date, CAP has only been detected in activated sludges (Hesselmann et al. 1999; Crocetti et al. 2000; Zilles et al. 2002; Wong et al. 2005), which are sparse and tiny microbial reservoirs on a global scale and have only been in operation for about a century (Tchobanoglous et al. 2003). The relatively recent introduction of activated sludge systems suggests that CAP originated and therefore is able to survive in alternative environments. Indeed, we found that CAP has multiple genes encoding functions more likely to be used in oligotrophic habitats than in nutrient-rich activated sludge (Garcia Martin et al. 2006). These include complete pathways for nitrogen and carbon fixation, high-affinity phosphate transporters, and flagellar and chemotaxis genes (Garcia Martin et al. 2006). No CAP flagella have been observed in EBPR sludges in which this species forms large clusters of cells bound together by extracellular polymeric substances (EPS) (Crocetti et al. 2000).
To identify CAP habitats, we surveyed a range of environmental samples using Accumulibacter-specific PCR targeting the 16S rRNA and ppk genes that were subsequently confirmed by sequencing. Accumulibacter species were detected in both fresh and estuarine waters and associated sediments but were rarely observed in soil samples (Supplemental Table S2). We therefore suggest that CAP populations are distributed in the environment as sparse high-density point sources (EBPR sludges) linked by dispersal via widespread diffuse reservoirs (aquatic environments), conforming to the ecological definition of a metapopulation as a collection of contained populations connected by a small amount of gene flow (Hanski 1999).
The presence of multiple strains in each sludge sample allowed us to investigate CAP for evidence of homologous recombination between strains (Supplemental Fig. S1). Unlike recent studies in which microbial populations were found to be highly recombining (Tyson et al. 2004; Nesbo et al. 2006), CAP strains showed no compelling evidence for genomic mosaicism, or even modest levels of homologous recombination. This apparent asexuality would prevent homogenization of local and introduced strains and thereby highlight dispersal patterns (Fig. 1).
While most of the CAP strains were represented by unassembled reads or short contigs in the metagenomic data (indicating low abundance), one strain dominated each sludge, producing large contigs with high read depths, allowing assessment of within-strain heterogeneity. The dominant strain populations were found to be extremely homogeneous in the US and OZ sludges with an average of one confirmed single nucleotide polymorphism (SNP) per 163.2 and 65.6 kb, respectively (Supplemental Table S3). This indicates that both dominant CAP strains are virtually clonal.
The near clonality of the dominant strains, and their inability to recombine, means that the bulk of the biomass in each laboratory-scale EBPR sludge is composed of genetically identical cells. Such populations are natural targets for phage predation, via the so-called “kill-the-winner” phenomenon (Thingstad and Lignell 1997; Pernthaler 2005). Comparison of the dominant CAP strain genomes in the US and OZ sludges provided clues supporting this scenario. These dominant strains were highly similar, sharing over 95% nucleotide identity across most of the genome (Garcia Martin et al. 2006), implying that differences are the result of recent evolutionary dynamics. One striking difference was the variability of EPS gene cassettes (Garcia Martin et al. 2006). EPS can provide a first line of defense against phage predation by masking attachment sites on the cell surface. In response, lytic bacteriophages are known to encode strain-specific polysaccharases to degrade host EPS and allow access to the cell surface (Sutherland 2001). The observed redundancy and variability of EPS gene cassettes in CAP genomes may impede strain-specific targeting of EPS by phage.
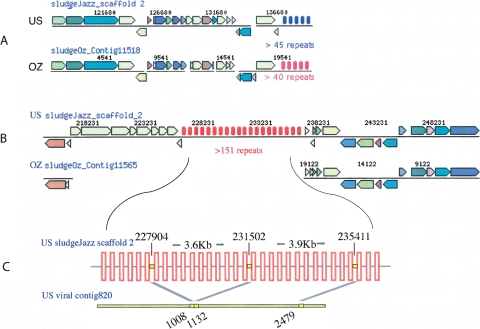
Another phage defense mechanism are clustered regularly interspaced short palindromic repeat (CRISPR) elements (Jansen et al. 2002; Barrangou et al. 2007). CRISPR elements are rapidly evolving clusters of short repeats regularly interspersed by unique sequences “spacers,” derived from foreign DNA entering the cell, including phages. It was recently demonstrated that spacers provide immunity to the phages from which they were derived (Barrangou et al. 2007). The bacterial metagenomes contained numerous CRISPR elements of which five could be unambiguously assigned to CAP strains (Supplemental Table S4). Both substitutions and insertions of CRISPR elements were observed between CAP strains (Fig. 2A,B; Supplemental Table S4). Only one type of repeat sequence and no spacers were common to the two data sets, suggesting exposure to different local phage populations.
Figure 2.
Sample alignments of homologous regions in the dominant US and OZ strains showing substitution (A) and insertion (B) of CRISPR elements. CRISPR repeats are indicated by sets of vertical bars with colors denoting different repeat sequences. Total number of repeats for each CRISPR element is unknown because of incomplete sequence information in the draft assembly; therefore, a minimum estimate is given. (C) Schematic magnification of dominant CAP CRISPR element and a contig from the phage virion metagenome revealing a phage that has previously infected CAP. All spacers targeting the phage had the same orientation. The starting positions of each spacer and the matching segments in the phage are indicated. The drawing is not to scale, and the actual number of repeats is significantly higher.
CRISPR elements and EPS gene clusters were among the most notable differences between closely related strains of Streptococcus thermophilus, which is used in coculture with Lactobacillus species for industrial yogurt and cheese production (Bolotin et al. 2004). Therefore, rapid acquisition and substitution of EPS gene cassettes and CRISPR elements may be a widespread response in bacteria to the pressure of phage predation in low complexity engineered ecosystems.
To test the hypothesis that phages are playing a major role in structuring CAP populations in EBPR, we sampled the phage virion metagenome of the US sludge 7 mo after sampling the bacterial metagenome. Eleven US CRISPR spacers, eight of which belonged to the dominant CAP strain, had matches to phage genome fragments (Supplemental Table S5), with some phages being targeted by multiple spacers (Fig. 2C) and some spacers targeting multiple related phages. This provides a direct link between the uncultivated bacterial host and phage virions and confirms that the CAP population had previously been infected by these phages. Two CRISPR spacers found only in the dominant OZ CAP strain had matches to the US phage community, supporting geographic dispersal of the host and/or phage.
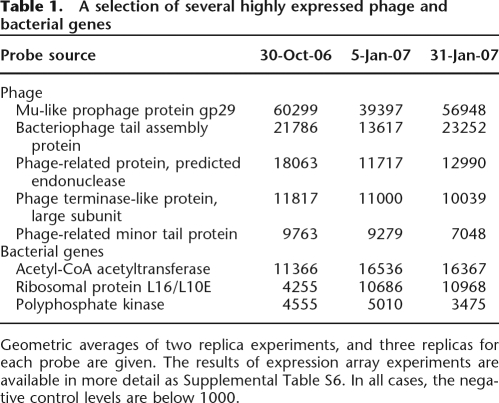
To confirm that phages are active in the sludge ecosystem, we monitored the US sludge at three time points spanning 3 mo using expression arrays targeting both phage and bacterial genes obtained from the metagenomic data sets. We found that large numbers of genes originating from the phage virion metagenome and some genes in the bacterial metagenome of putative prophage origin were highly expressed (Table 1 and Supplemental Table S6). These included many hypothetical proteins but also proteins associated with phage tail assembly, a phage-specific endonuclease and terminase (Supplemental Table S6), suggesting that phages are continuously active in the sludge. Since the microarray was based on phage virion genes sampled almost 2 yr prior to the expression analysis, some phages must persist for long periods in the sludge. These data imply that the bacterial community is under persistent local predation pressure by phages and live in a volatile but relatively stable coexistence.
Table 1.
A selection of several highly expressed phage and bacterial genes
Geometric averages of two replica experiments, and three replicas for each probe are given. The results of expression array experiments are available in more detail as Supplemental Table S6. In all cases, the negative control levels are below 1000.
In summary, we have shown that (1) CAP is globally dispersed, (2) highly related and geographically remote CAP strains differ principally in genomic regions encoding phage defense mechanisms, (3) high-density, clonal, nonrecombining CAP populations in EBPR bioreactors are natural targets of “kill-the-winner” phage predation, (4) phages are consistently active in EBPR bioreactor communities, and (5) signatures of past phage infections in CAP are observed mostly between local phage and host. We therefore conclude that CAP strains disperse globally but must adapt to local persistent phage predation pressure. The present study illustrates the value of combining high-throughput sequence and gene expression data from the bacterial and viral fractions of an ecosystem to elucidate population structure, biogeography, and host–parasite interdependencies.
Methods
Metagenomic sequencing
Sludge samples for the US and Australian (OZ) bacterial metagenomes were obtained on July 3 and August 18, 2004, respectively. Sequencing, assembly, and gene prediction of the bacterial metagenomes are described elsewhere (Garcia Martin et al. 2006). To obtain a phage virion metagenome, a sludge sample from the US bioreactor was taken on February 7, 2005, 7 mo after sampling for the bacterial metagenome. Virion purification techniques, construction of shotgun libraries, sequencing, assembly, and gene calling are described in the Supplemental Research Data.
Bioinformatic analyses
Single-copy gene analysis was performed to infer biogeographical patterns by (1) selecting 47 conserved single-copy gene families in isolate genomes in the Integrated Microbial Genomes (IMG) database (Markowitz et al. 2006) using PFAM (Bateman et al. 2004) profile searches with rps-BLAST (Altschul et al. 1997), (2) identifying members of these families in the bacterial sludge metagenomes, (3) aligning each family with ClustalX (Thompson et al. 1994), and (4) generating neighbor-joining trees using ClustalX. See Supplemental Research Data for details.
To refine the resolution of the single-copy gene analysis, we PCR-amplified the ppk gene from the sludge biomass and environmental samples. The amplification product was cloned into Escherichia coli, and 96 clones were picked from each library for sequencing. See Supplemental Research Data for further details.
Quantification of SNP frequency was done using CONSED program (Gordon et al. 1998) on the largest 10 contigs, and reported polymorphisms were manually rechecked. The screen for homologous recombination was done using SNP-VISTA (Shah et al. 2005). CRISPR elements were identified using piler-cr (Edgar 2007), and BLASTN (Altschul et al. 1997) was used to link between CRISPR spacers and genomic regions. See Supplemental Research Data for details.
Microarrays
Combimatrix CustomArray 12K microarrays were constructed from predicted genes from both bacterial and phage metagenomes. Samples from US sludge were extracted on October 30, 2006; January 5, 2007; and January 31, 2007. RNA was purified, labeled, and hybridized to the arrays. For each probe, we calculated the geometric average of all replicates, with the exclusion of dubious spots (Supplemental Table S6). See Supplemental Research Data for further details.
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6835308
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Boyaval P., Moineau S., Romero D.A., Horvath P., Moineau S., Romero D.A., Horvath P., Romero D.A., Horvath P., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Marshall M., Moxon S., Sonnhammer E.L., Moxon S., Sonnhammer E.L., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon M., Townsend C.R., Harper J.L., Townsend C.R., Harper J.L., Harper J.L. Ecology: From individuals to ecosystems. Blackwell; Malden, MA: 2006. [Google Scholar]
- Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Quinquis B., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Goltsman E., Mazur M., Pusch G.D., Mazur M., Pusch G.D., Pusch G.D., et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A., Rainey P.B., Rainey P.B. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- Crocetti G.R., Hugenholtz P., Bond P.L., Schuler A., Keller J., Jenkins D., Blackall L.L., Hugenholtz P., Bond P.L., Schuler A., Keller J., Jenkins D., Blackall L.L., Bond P.L., Schuler A., Keller J., Jenkins D., Blackall L.L., Schuler A., Keller J., Jenkins D., Blackall L.L., Keller J., Jenkins D., Blackall L.L., Jenkins D., Blackall L.L., Blackall L.L. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E.J., Enright M.C., Spratt B.G., Enright M.C., Spratt B.G., Spratt B.G. Estimating the relative contributions of mutation and recombination to clonal diversification: A comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 2000;151:465–469. doi: 10.1016/s0923-2508(00)00168-6. [DOI] [PubMed] [Google Scholar]
- Garcia Martin H., Ivanova N., Kunin V., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Ivanova N., Kunin V., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Kunin V., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Yeates C., He S., Salamov A.A., Szeto E., He S., Salamov A.A., Szeto E., Salamov A.A., Szeto E., Szeto E., et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hanski I. Metapopulation ecology. Oxford University Press; Oxford, UK: 1999. [Google Scholar]
- Hesselmann R.P., Werlen C., Hahn D., van der Meer J.R., Zehnder A.J., Werlen C., Hahn D., van der Meer J.R., Zehnder A.J., Hahn D., van der Meer J.R., Zehnder A.J., van der Meer J.R., Zehnder A.J., Zehnder A.J. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- Jansen R., Embden J.D., Gaastra W., Schouls L.M., Embden J.D., Gaastra W., Schouls L.M., Gaastra W., Schouls L.M., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jessup C.M., Kassen R., Forde S.E., Kerr B., Buckling A., Rainey P.B., Bohannan B.J.M., Kassen R., Forde S.E., Kerr B., Buckling A., Rainey P.B., Bohannan B.J.M., Forde S.E., Kerr B., Buckling A., Rainey P.B., Bohannan B.J.M., Kerr B., Buckling A., Rainey P.B., Bohannan B.J.M., Buckling A., Rainey P.B., Bohannan B.J.M., Rainey P.B., Bohannan B.J.M., Bohannan B.J.M. Big questions, small worlds: Microbial model systems in ecology. Trends Ecol. Evol. 2004;19:189–197. doi: 10.1016/j.tree.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Maiden M.C., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhou J., Zurth K., Caugant D.A., Zurth K., Caugant D.A., Caugant D.A., et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V.M., Korzeniewski F., Palaniappan K., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Korzeniewski F., Palaniappan K., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Palaniappan K., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Dubchak I., Hugenholtz P., Anderson I., Hugenholtz P., Anderson I., Anderson I., et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K.D., Dojka M.A., Pace N.R., Jenkins D., Keasling J.D., Dojka M.A., Pace N.R., Jenkins D., Keasling J.D., Pace N.R., Jenkins D., Keasling J.D., Jenkins D., Keasling J.D., Keasling J.D. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microbiol. 2002;68:4971–4978. doi: 10.1128/AEM.68.10.4971-4978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø C.L., Dlutek M., Doolittle W.F., Dlutek M., Doolittle W.F., Doolittle W.F. Recombination in Thermotoga: Implications for species concepts and biogeography. Genetics. 2006;172:759–769. doi: 10.1534/genetics.105.049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Papke R.T., Ramsing N.B., Bateson M.M., Ward D.M., Ramsing N.B., Bateson M.M., Ward D.M., Bateson M.M., Ward D.M., Ward D.M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Shah N., Teplitsky M.V., Minovitsky S., Pennacchio L.A., Hugenholtz P., Hamann B., Dubchak I., Teplitsky M.V., Minovitsky S., Pennacchio L.A., Hugenholtz P., Hamann B., Dubchak I., Minovitsky S., Pennacchio L.A., Hugenholtz P., Hamann B., Dubchak I., Pennacchio L.A., Hugenholtz P., Hamann B., Dubchak I., Hugenholtz P., Hamann B., Dubchak I., Hamann B., Dubchak I., Dubchak I. SNP-VISTA: An interactive SNP visualization tool. BMC Bioinformatics. 2005;6:292. doi: 10.1186/1471-2105-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. Biofilm exopolysaccharides: A strong and sticky framework. Microbiol. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- Tchobanoglous G., Burton F.L., Stensel H.D., Burton F.L., Stensel H.D., Stensel H.D. Wastewater engineering: Treatment and reuse. Metcalf & Eddy–McGraw-Hill; Boston: 2003. [Google Scholar]
- Thingstad T., Lignell R., Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 1997;13:19–27. [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Rubin E.M., Rokhsar D.S., Banfield J.F., Rokhsar D.S., Banfield J.F., Banfield J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Wu D., Paulsen I., Nelson K.E., Nelson W., Paulsen I., Nelson K.E., Nelson W., Nelson K.E., Nelson W., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Whitaker R.J., Banfield J.F., Banfield J.F. Population genomics in natural microbial communities. Trends Ecol. Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Whitaker R.J., Grogan D.W., Taylor J.W., Grogan D.W., Taylor J.W., Taylor J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Wong M.T., Mino T., Seviour R.J., Onuki M., Liu W.T., Mino T., Seviour R.J., Onuki M., Liu W.T., Seviour R.J., Onuki M., Liu W.T., Onuki M., Liu W.T., Liu W.T. In situ identification and characterization of the microbial community structure of full-scale enhanced biological phosphorous removal plants in Japan. Water Res. 2005;39:2901–2914. doi: 10.1016/j.watres.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Zilles J.L., Peccia J., Kim M.W., Hung C.H., Noguera D.R., Peccia J., Kim M.W., Hung C.H., Noguera D.R., Kim M.W., Hung C.H., Noguera D.R., Hung C.H., Noguera D.R., Noguera D.R. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 2002;68:2763–2769. doi: 10.1128/AEM.68.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]