Abstract
Despite many recent efforts, in silico identification of promoter regions is still in its infancy. However, the accurate identification and delineation of promoter regions is important for several reasons, such as improving genome annotation and devising experiments to study and understand transcriptional regulation. Current methods to identify the core region of promoters require large amounts of high-quality training data and often behave like black box models that output predictions that are difficult to interpret. Here, we present a novel approach for predicting promoters in whole-genome sequences by using large-scale structural properties of DNA. Our technique requires no training, is applicable to many eukaryotic genomes, and performs extremely well in comparison with the best available promoter prediction programs. Moreover, it is fast, simple in design, and has no size constraints, and the results are easily interpretable. We compared our approach with 14 current state-of-the-art implementations using human gene and transcription start site data and analyzed the ENCODE region in more detail. We also validated our method on 12 additional eukaryotic genomes, including vertebrates, invertebrates, plants, fungi, and protists.
Eukaryotic genomes are being sequenced at an ever-increasing pace. At the moment, nearly 50 complete genomes of eukaryotes are publicly available, and many more are in the pipeline to be sequenced in the next few years (Liolios et al. 2006). The proliferation of genome sequencing projects has driven the search for fast ways of sequence-based structural annotation, which involves the identification of genes and the modeling of their correct gene structure (Claverie et al. 1997; Mathé et al. 2002; Zhang 2002; Wang et al. 2004). Although great progress has been achieved in gene prediction, for instance by using comparative approaches (Wasserman et al. 2000; Liu et al. 2004; Jin et al. 2006; Wang and Zhang 2006), one of the more difficult tasks in the annotation of whole genomes remains the accurate identification and delineation of promoters (Fickett and Hatzigeorgiou 1997; Ohler 2000, 2001; Bajic et al. 2004, 2006a). Nevertheless, the prediction of the regions that control the transcriptional activation of genes is important for various reasons (Smale 2001; Butler and Kadonaga 2002; Bajic et al. 2004; Sonnenburg et al. 2006). On the one hand, promoter prediction can be used for the discovery of genes that are missed by gene predictors and/or for which experimental support (ESTs, cDNAs, etc.) is not available. On the other hand, the prediction of promoters is important for guiding further in silico searches and experimental work, for instance in narrowing down the regions that play the most important role in transcriptional regulation (Bajic et al. 2004, 2006a; Carninci et al. 2006; Solovyev et al. 2006).
The promoter is commonly referred to as the region upstream of a gene that contains the information permitting the proper activation or repression of the gene that it controls (Pedersen et al. 1999; Smale and Kadonaga 2003). The promoter region itself is typically divided into three parts: (1) the core promoter, which is the region that is responsible for the actual binding of the transcription apparatus and which is typically situated ∼35 bp upstream of the transcription start site (TSS); (2) the proximal promoter, a region containing several regulatory elements, which ranges up to a few hundred base pairs upstream of the TSS; and (3) the distal promoter, which can range several thousands of base pairs upstream of the TSS and contains additional regulatory elements called enhancers and silencers.
It has been known for quite some time that the properties of promoter regions are considerably different from those of other parts in the genome (Pedersen et al. 1998; Aerts et al. 2004; Florquin et al. 2005; Fukue et al. 2005; Tabach et al. 2007). Some features that have proven useful in the detection of promoters in vertebrate genomes are the so-called CpG islands close to the TSS (Delgado et al. 1998; Ioshikhes and Zhang 2000; Hannenhalli and Levy 2001), the presence of typical transcription factor binding sites (Solovyev and Shahmuradov 2003; Choi et al. 2004; Ohler 2006), and statistical properties of the core and proximal promoter (Down and Hubbard 2002; Bajic et al. 2006b; Fitzgerald et al. 2006). The similarities between orthologous promoters (Solovyev and Shahmuradov 2003; Jin et al. 2006) and information from mRNA transcripts (Liu and States 2002) have also been used to identify promoters. The more recent and sophisticated Promoter Prediction Programs (PPPs) look for these promoter-specific characteristics by using machine learning techniques such as discriminant analyses, Hidden Markov Models, and Artificial Neural Networks to predict and delineate promoters (for reviews, see Fickett and Hatzigeorgiou 1997; Rombauts et al. 2003; Bajic et al. 2004, 2006a; Sonnenburg et al. 2006). Programs and tools based on these techniques are difficult to train because they require a large amount of high-quality training data, preferably from an experimental setting (Munch and Krogh 2006). However, for most of the new genome projects there is only a limited amount of such data available. Another problem is that the outcome of programs based on these techniques is often difficult to interpret (Ratsch et al. 2006). Furthermore, all hitherto available programs are species-specific; i.e., they are trained on one species and are able to predict promoters only for that particular species. Another drawback of most PPPs is that they depend on specific motifs expected to be present in the core promoter. Indeed, some programs (Promoter2.0 [Knudsen 1999]; Eponine [Down and Hubbard 2002]; NNPP2.2 [Burden et al. 2004]) are based on the explicit presence of motifs such as the TATA box, which are very common in certain species, such as yeast (Struhl 1989), but much less common in mammals or plants (Suzuki et al. 2001; Butler and Kadonaga 2002; Fukue et al. 2004; Florquin et al. 2005). This hampers the ability of a single program to analyze different species with the same model, and does not facilitate the discovery of different types of promoters. Finally, there is the issue of scalability, as the best performing programs are unable to process large datasets (Ohler et al. 2002; Bajic et al. 2004, 2006a; Sonnenburg et al. 2006).
In light of all these caveats, we propose a simple technique for identifying and delineating (core) promoters that is based on the properties of long stretches of DNA. It has indeed been shown that sequence properties such as GC content and more general chemo-physical properties of the DNA, such as stabilizing energy of Z-DNA (Ho et al. 1990), DNA denaturation values (Blake and Delcourt 1998; Blake et al. 1999), protein-induced deformability (Olson et al. 1998), and duplex-free energy (Sugimoto et al. 1996), among others (for review, see Florquin et al. 2005), can be used to describe (core) promoters, and to discriminate between (core) promoter sequences and non (core) promoter sequences (Ohler et al. 2001; Florquin et al. 2005; Kanhere and Bansal 2005; Uren et al. 2006; Wang and Benham 2006). Because all these properties are calculated from conversion tables using di- or trinucleotides, one may argue that these properties are in fact exactly the same as the nucleotide sequence and do not offer any additional information. However, several studies have shown that this is not the case. Both Liao et al. (2000) and Baldi et al. (1998) have analyzed the correlation between the different properties, and their main conclusion was that the properties are largely independent. Moreover, Florquin et al. (2005) have clustered promoters based on these structural properties. The genes associated with the promoters in each cluster varied greatly for the different properties, which again indicates that the different properties contain complementary information. Bode et al. (2006) have shown that it is very hard to identify scaffold/matrix attachment regions (SMARs) from the sequence, as the important scaffold proteins recognize structural features instead of specific nucleotide sequences. Structural properties are known to have long-range interactions (up to 10 kb), so they can exhibit properties that are not visible in the sequence (Merling et al. 2003; Faiger et al. 2006). The Human Genomic Melting Map (Liu et al. 2007) shows a correlation between GC content and DNA denaturation, but due to the cooperative nature of DNA denaturation, this correlation is weaker on scales <500 bp.
We applied these different properties to the problem of promoter prediction and show that some properties give better performance than others, which again indicates differences in information content. In particular, we present the Easy Promoter Prediction Program (EP3), which uses GC content and large-scale structural features of DNA to identify and delineate promoter regions in whole-genome sequences. EP3 was applied to the human genome and compared with other state-of-the-art PPPs using two different evaluation techniques: One is commonly used for PPPs, while the other is based on a novel scheme also considering intergenic predictions that do not fall in a promoter. We evaluated the different ways of encoding the genomic DNA, and we also applied EP3 to a set of human noncoding RNA genes. Finally, we evaluated EP3 on 12 other eukaryotic genomes.
Results and Discussion
Properties of the core promoter region
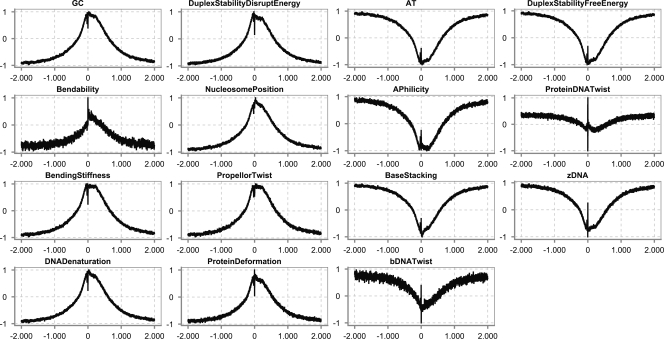
Figure 1 shows examples of numerical profiles for human promoter sequences. These profiles were obtained by lining up all the promoter sequences with the TSS at the same position. Next, we converted all sequences using the different structural properties of DNA into numeric sequences and, for each position, plotted the average over all numeric sequences. The X-axis is the position relative to the TSS, and the Y-axis is the normalized value of that property. It is clear that the human core promoter adopts a very specific intrinsic structure that stretches over quite long distances. Either a large peak is visible, representing highly stable regions in the DNA, or a cleft is visible, which represents highly unstable regions in the DNA. For instance, the GC profile shows a peak, while the AT profile shows a cleft. When zoomed in, it can be noticed that, within the broader region of stability, there is a small unstable region that corresponds to the TSS, and a second one that corresponds to the region where TBP binds (see below). The promoter prediction technique we present will identify those higher regions and are, thus, most likely to contain a TSS.
Figure 1.
Examples of numerical profiles representing structural properties of the DNA in human over long-range distances around the transcription start site (TSS) (2000 bp upstream and 2000 bp downstream). The numeric profiles contain the actual average values on each position and are not smoothed using a window. Structural properties can be divided into two broad categories: those for which the numerical profile shows a peak around the TSS (e.g., GC content, bendability, and DNA denaturation), and those for which the profile shows a cleft around the TSS (e.g., AT content, bDNA twist, and Duplex Stability-Free Energy). (X-axis) Position relative to the TSS; (Y-axis) normalized value for each of the different properties. The profiles are based on the properties listed in the first column of Table 2. The coordinates for the TSS were retrieved from Ensembl, and the sequences were extracted around this TSS.
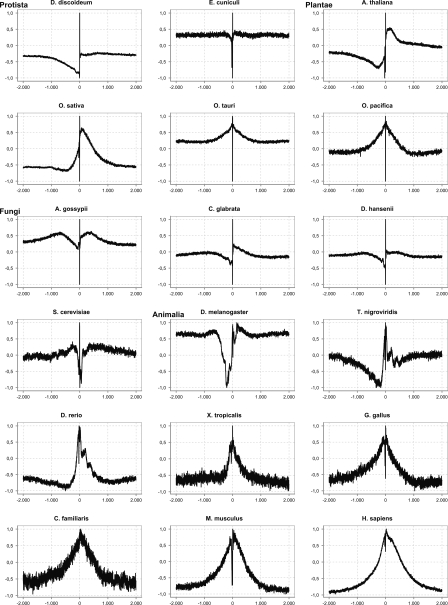
To see how general these properties of promoter sequences are, we created large-scale (2000 bp up- and downstream of the TSS) datasets for 16 different species. For each of these datasets, we analyzed the structural profiles. Over long-range distances, we observe several types of profiles (Fig. 2). The first one applies mostly to protists. In this type, the profile slowly decreases a few hundred base pairs upstream of the TSS, shows a transition of a strong peak and cleft on the TSS, and is flat again downstream of the TSS, which may indicate a relatively small promoter area in these organisms. The second type of profile is observed in mammals and shows a broad elevated region around the TSS, with again a strong local peak and drop on the TSS itself. Between these two extremes, we observe a more gradual transition in fungi and plants. The extremely high and low values that can be seen on the TSS are discussed in more detail in the next section.
Figure 2.
Examples of numerical profiles representing different species using the same property (inverted base stacking value) over long-range distances around the transcription start site (TSS) (2000 bp upstream and 2000 bp downstream). (X-axis) Position relative to the TSS; (Y-axis) normalized value of the property. The profile is calculated with a window size of three.
There seems to be a relationship between the genome size and the size of the peaks or clefts: In prokaryotes (not shown) and protists, these cover ∼100 bp, while they gradually increase to several hundreds of base pairs in fungi and up to ∼1000 bp in plants. In animals, smaller genomes such as that of Drosophila have a peak ranging from a few hundred base pairs on each side of the TSS; this gradually increases up to ∼2000 bp in mammals. In conclusion, the peak is most obvious in mammals, but is also clearly visible in fish, insects, fungi, and plants; it seems considerably smaller in protists.
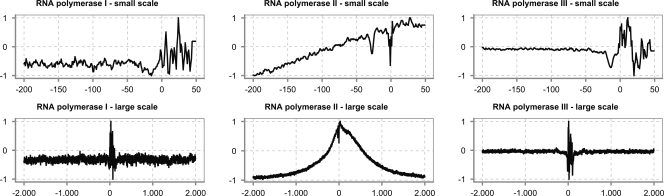
In humans, as well as in other eukaryotes, three types of RNA polymerases (RNAP) exist that are used to transcribe different types of genes. All three of them manage to identify the TSS in a whole-genome setting in their own way. Here, we focus on humans because this species is among the best documented ones. We retrieved the promoter sequences for several types of genes from Ensembl (see Methods) and constructed three different datasets for each of the promoter types: RNAP I, RNAP II, and RNAP III. For each of these datasets, we calculated a structural profile. Figure 3 shows the inverted base-stacking values over long-range distances around the TSS (2000 bp up- and downstream), as well as a zoomed-in view (top row) of a shorter region around the TSS (ranging from 200 bp upstream of the TSS to 50 bp downstream of the TSS) for sets of promoters recognized by the three different polymerases. Both on a large and a small scale, there are clear and important fluctuations in the profile around the TSS. One of the most striking features, at least for RNAP II promoters, is the presence of two clefts: one at the TSS and one near position −30. The latter one is related to the TATA binding protein that is important for transcription initiation.
Figure 3.
Structural profile of promoters for genes transcribed by different polymerases. (Top row) Numerical profiles over short-range distances around the TSS; (bottom row) profiles over long-range distances. The profile was calculated using the inverse of the base-stacking property. See text for details.
There is, however, a clear difference between RNAP II promoters on the one hand, and RNAP I and III promoters on the other hand. RNAP I and III promoters lack the large-scale stable region of RNAP II promoters and show local destabilization only around the TSS. These local instabilities around the TSS may be caused partly by the structural nature of the transcript originating from these genes. Drawing more definitive conclusions for the RNAP I and III promoters would require much more data than the few hundred sequences that are currently available in Ensembl. The lack of large-scale features is interesting as it may indicate that the region involved in regulation of these gene types is in general quite small. This might be in line with the fact that many genes transcribed by RNAP I and III promoters are produced in very large quantities and need less regulation. For instance, rRNA genes account for 80% of total steady-state cellular RNA and might thus require less sophisticated regulation (Jacob 1995). Another observation is that the regulation of r-protein genes is often post-transcriptional and therefore does not need extensive transcriptional regulation (Amaldi et al. 1989; Presutti et al. 1991). RNAP II promoters have an elevated region of nearly 2000 bp, which indicates a much larger area involved in transcriptional regulation of protein-coding genes.
Relationship between structural profiles and known core promoter elements
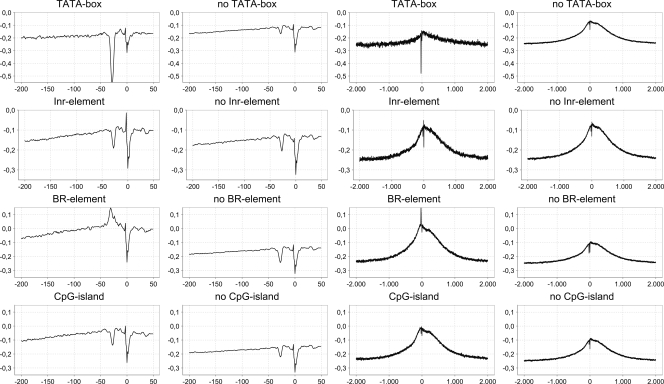
To investigate the relationship between the structural profile and the occurrence of known core promoter elements, we analyzed four elements (TATA, INR, BRE, and CpG islands) well known in animal genomes and the relation between the presence or absence of the element in the promoter sequence and the presence or absence of peaks and clefts in the structural profile. Again, we limited ourselves to humans because to detect motifs we require position weight matrices (PWM) or consensus sequences, which are not available for all organisms.
The first element we considered is the TATA box, also known as the Hogness box, which binds the TATA binding protein (TBP) and is often involved in transcription initiation (Smale and Kadonaga 2003). The structural profile for the promoters containing the TATA motif is shown in Figure 4 (top row). As can be noticed, the numerical profile of promoters that do contain the TATA box lies lower than those that do not. Furthermore, the cleft around position −30 is much deeper in TATA-box-containing promoters, implying that the DNA denatures more easily on that position, which conforms to the knowledge that the TBP is involved in the denaturation of DNA. Although it is clear that promoters containing the TATA motif have a much deeper cleft than those without, we must note that the cleft is visible as well. This indicates that the TBP, which is important for transcription to start, requires a region that denatures more easily, whether it contains the motif or not, to allow efficient binding (Comai et al. 1992; Cormack and Struhl 1992; White and Jackson 1992). Therefore, we can conclude that the TATA motif is responsible for the instability of this region, but other combinations of nucleotides also can result in a region that is unstable enough. Also, on a larger scale (Fig. 4, right side), we see that the profile is slightly lower for promoters containing TATA than for non-TATA promoters.
Figure 4.
Structural profiles of RNAP II promoters containing known motifs or elements versus promoters for which the presence of motifs cannot be demonstrated. (Left two columns) Short-range distances around the TSS; (right two columns) long-range distances around the TSS. The profile was calculated using the inverse of the base-stacking property. (Inr) Initiator element, (BR) TFIIB recognition element.
A second well-known core promoter element is the Initiator element (INR) that is found mostly on position 0; i.e., the TSS itself. In Figure 4 (second row), the structural profiles for promoters with and without the INR motif are displayed. Overall, the two profiles are very similar, except for one remarkable difference visible on the small-scale plot: On position −1 there is a significant peak (up to −0.05) upstream of the TSS cleft in the INR-containing promoters, while this peak is missing in the promoters that lack INR. This seems to indicate that this peak, which increases the stability–instability transition, plays a role in the accurate transcription initiation because the INR motif is known to be important for accurate transcription initiation (Lo and Smale 1996). On a larger scale, we do not observe a significant difference between INR-containing promoters and those that do not.
The TFIIB recognition element (BRE) consists of two parts, BREu, which is located upstream of the position of the TATA box (Lagrange et al. 1998), and BREd, which is located downstream of the TATA motif (Deng and Roberts 2005). The profile of BRE promoters is plotted in the third row of Figure 4. It is clear that the profile of promoters that contain a BRE motif is higher than that of promoters lacking the BRE motif. This is not surprising because the BRE motif consists entirely of G and C nucleotides, which results in higher inverted base-stacking values. The local profile for BRE-containing promoters shows a rather stable area from position −40 to −20. These features fit well with the observation that TFIIB (GTF2B) interacts with the major groove upstream of the TATA box and with the minor groove downstream of the TATA box (Nikolov et al. 1995; Lagrange et al. 1998; Deng and Roberts 2005, 2006). Both interactions leave their tracks on the structural profile. The interaction with the major groove is visible as a peak at position −35, while the interaction with the minor groove is visible as a peak at position −25. These two peaks form a cleft at position −30 that will be used by TBP to bind. The BRE motif is more prevalent among promoters without the TATA motif, and the motif might be a functional substitute for the TATA box (Deng and Roberts 2006). On a larger scale, we see that the profile of promoters lacking the BRE motif has a much lower amplitude than that of promoters containing the BRE motif.
A last well-known characteristic of core promoters is the abundance of CpG islands near protein-coding genes in mammals (Bird 2002). CpG islands are regions that are mainly associated with mammalian promoters, but are also observed in plants (Rombauts et al. 2003). They are related to housekeeping genes (Baek et al. 2007), dynamic usage of TSSs (Kawaji et al. 2006), and bidirectional promoter activity (Trinklein et al. 2004). The CpG dinucleotide is underrepresented in the genome because the cytosine can be converted into thymine after methylation. Only DNA stretches that are under evolutionary stress, such as promoters, are preserved, and these regions are rich in CpG dinucleotides that are unmethylated. The unmethylated CpG-rich regions are often associated with promoters of protein-coding genes (Bird 2002). These islands are typically found upstream of the core promoter, and, when present, these promoters usually lack a TATA box and often have alternative start sites. The local profile does not vary much between promoters having CpG islands and the others, with only an expected shift toward higher stability values due to GC richness. On a larger scale, as with BRE elements, the profile of promoters lacking CpG islands has a much lower amplitude than that of promoters with CpG islands. This may indicate that the existence of either feature is correlated with long-range regulation, which would require further investigation.
Promoter prediction and comparison to the state of the art
We have shown that in eukaryotes the region near the TSS has a very distinct structural profile, while this is absent in the remainder of intergenic sequences and in coding sequences (Fig. 1). The peak in this profile, for instance using base stacking values, is a well-positioned property of the eukaryotic promoter that stretches over several hundreds of base pairs with its top located on the TSS, which can be used to identify promoters. As computation of this profile for individual sequences involves smoothing over a few hundred base pairs, this feature is too coarse to predict the actual TSS; it is nevertheless very well suited to predict the core promoter region. Furthermore, due to the fact that it is such a large-scale property, there are very few false positives when scanning the genome. Using large-scale structural features is new to the field of promoter prediction, because previous studies usually analyzed no more than 200 bp around the TSS (Ohler and Niemann 2001; Fukue et al. 2004, 2005; Florquin et al. 2005; Kanhere and Bansal 2005).
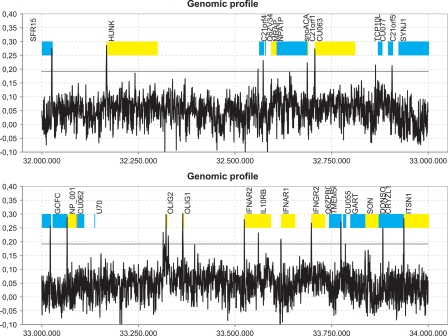
In applying EP3 to the human genome, we observed clear peaks near the start of most genes, which is exemplified in Figure 5 for a small region of the human genome. This figure shows a plot of the structural profile for 2 Mbp on the human chromosome 21 using DNA base-stacking values and a window size of 400 bp (see Methods). To demonstrate the predictive strength of these structural profiles, we have added the gene annotation retrieved from Ensembl to the predictions. This figure clearly shows that most genes do have a peak near the 5′ end, and that there are virtually no peaks outside this region (false positives), indicating that the large-scale structural properties we observe are distinct enough for EP3 to efficiently predict core promoter regions. It should be noted that, while this single property proves to work surprisingly well, future research may focus on combining multiple properties to describe the promoter region even better, as the overlap between the predictions obtained through different properties may vary significantly (Supplemental Table 1). For example, the overlap between Bendability and Duplex Stability-Free energy is only 1%, while the overlap between predictions from Duplex Stability-Free energy and DNA denaturation is ≥80%. The average overlap between a pair of properties is 44% (SD = 29%). Improving the models for the different structural properties may also increase the overall performance.
Figure 5.
Structural profile (blue) of human chromosome 21 between position 32,000,000 bp and 33,000,000 bp. The profile, based on inverted base-stacking values, was made using a window size of 400 bp and using nonoverlapping windows. Experimentally annotated genes from Ensembl are indicated: (yellow bands) positive strand; (blue bands) negative strand. Peaks in the profile that exceed the classification threshold (T = 0.19, horizontal gray line) are predicted as promoter regions. (X-axis) Position in the genomic sequence, (Y-axis) inverse of the value for base-stacking energy. The labels on the plot show the gene names as they appear in Ensembl.
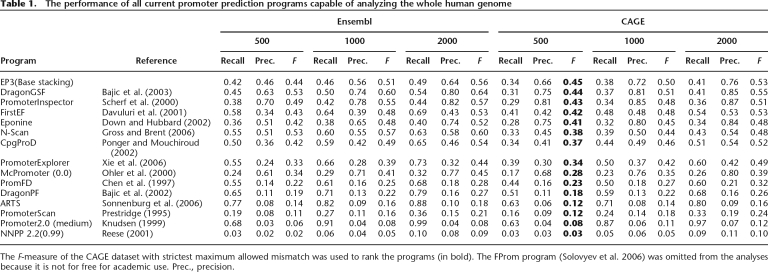
Next, we compared our program with a broad range of programs that are currently available for human promoter predictions (Table 1). We used both the Ensembl gene annotation and the CAGE transcription start data and tested GC content as the simplest feature and compared it with the other structural features. All programs were run using default settings unless indicated otherwise. For those programs that depend on a parameter, we included the best result in the table. For EP3, we included the values for the base-stacking property, as these gave the best results (Table 2). To rank the different programs, we used the F-measure of the program with a maximum allowed distance of 500 bp with the true TSS compared with the CAGE dataset. This is in contrast to other recent reviews and promoter prediction papers that used less strict validation (Bajic et al. 2004, 2006a; Sonnenburg et al. 2006; Xie et al. 2006). We included the less strict mismatch distances (1000 and 2000 bp) for reference with the other papers that allowed a distance of 1000 bp (Bajic et al. 2006a) or 2000 bp (Bajic et al. 2004; Sonnenburg et al. 2006). When using larger values, more distant predictions will also be considered to be true positive (TP). This is not desirable, as predictions should be as close as possible to the TSS. The F-measure clearly decreases when the evaluation becomes stricter, indicating that all programs output predictions that are quite distant from the actual TSS.
Table 1.
The performance of all current promoter prediction programs capable of analyzing the whole human genome
The F-measure of the CAGE dataset with strictest maximum allowed mismatch was used to rank the programs (in bold). The FProm program (Solovyev et al. 2006) was omitted from the analyses because it is not for free for academic use. Prec., precision.
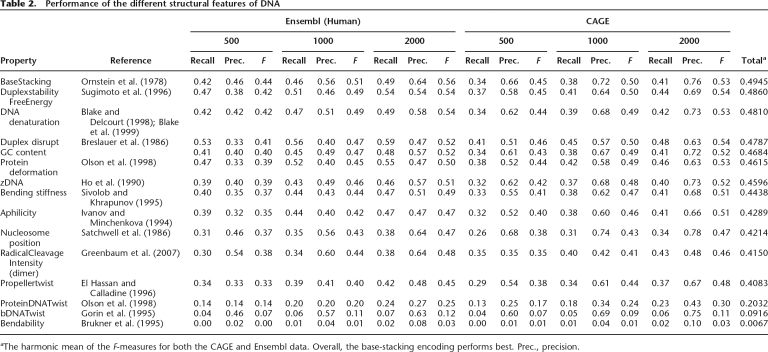
Table 2.
Performance of the different structural features of DNA
aThe harmonic mean of the F-measures for both the CAGE and Ensembl data. Overall, the base-stacking encoding performs best. Prec., precision.
For NNPP2.2, McPromoter, and Promoter2.0, we considered different parameter settings. We included only the setting that resulted in the highest F-measure. We considered two different techniques to calculate the number of TPs, false positives (FPs), and false negatives (FNs) (see Methods). The first one is based simply on gene annotation and was used before in several studies on promoter predictions (Bajic et al. 2004, 2006a; Sonnenburg et al. 2006; Xie et al. 2006). The second one is an alternative technique we developed that also includes intergenic predictions but requires transcriptional data, such as the CAGE dataset.
For the Ensembl annotation, we see that Dragon Gene Start Finder (GSF) (F = 0.53) and PromoterInspector (F = 0.49) perform better than EP3 (F = 44). However, when using the CAGE annotation, EP3 performs slightly better (F = 0.45) than DragonGSF (F = 0.44) and PromoterInspector (F = 0.43). Although these differences seem small, it still concerns several hundreds of genes or TSSs. For instance, a 1% lower recall means 200 genes in the Ensembl dataset that are missed by that program. One percent less precision means 200 extra false predictions, so even the small differences are significant. In the case of the CAGE dataset, 1% difference corresponds to a difference of ∼1250 TSSs. Combining this into the F-measure means that a program that has 1% lower value misses 1250 TSSs and predicts an additional 1250 wrong ones.
In conclusion, the best performing programs are EP3, DragonGSF, PromoterInspector, Eponine, and FirstEF, which have an F-measure > 0.4. Several programs (CpGProD, PromoterExplorer, N-Scan, and McPromoter) still perform quite well and have an F-measure > 0.25. The rest of the programs (PromFD, ARTS, DragonPF, PromoterScan, NNPP2.2, and Promoter2.0) do not perform very well on the complete human genome (F < 0.25). In all cases, these low F-measures are caused by a very low precision, which is obtained when the PPP outputs many FPs, a problem reminiscent of promoter prediction since the beginning.
The performance of PPPs on the Ensembl dataset is generally higher than that on the CAGE dataset, probably because of the different counting schemes for Ensembl data and the CAGE dataset, whereas the scheme for Ensembl data ignores all intergenic predictions. Some of the other programs may also perform better on the Ensembl data than EP3, because these programs have been trained on promoters of protein-coding genes and are therefore more gene-centric. For the top-performing programs, the balance between the recall (sensitivity) and precision (specificity) is mostly favoring the precision, with the exception of FirstEF, which is perfectly balanced on the CAGE dataset. The other programs often have high recall values, but at the cost of very low precision values.
Performance on the ENCODE region
The ENCODE project aims to carefully annotate all functional elements in a small portion (1%) of the human genome. We used EP3 to make predictions on the 44 regions that cover ∼30 Mb and compared the prediction with three different datasets. First, we compared our predictions to a set of known functional promoters (Cooper et al. 2006). The dataset is only partial for the ENCODE region because Cooper et al. tested only 642 putative promoters of the 921 they predicted. These partial data are insufficient to assess the precision of EP3. Of the 642 promoters tested, 387 were discovered to be functional. EP3 predicts 24% (recall) of the promoters that are marked as functional by Cooper et al. when using a maximum distance of 500 bp. This low recall rate is rather surprising, as the performance on the GENCODE and CAGE data is very good (see below). But when we look in detail, a much higher recall rate (40%) is obtained for the genes expressed in all 16 cell lines, which indicates that EP3 is biased toward broadly expressed genes. Of the 257 nonfunctional promoters, 18 (7%) are predicted by EP3.
Next, we compared the predictions of EP3 on the ENCODE region with the gene annotation from the GENCODE project (Harrow et al. 2006) and with the CAGE data from Riken. The GENCODE annotation was compared with the predictions using the classic method for calculating the performance, as it is a gene annotation similar to the one of Ensembl. We obtained a recall of 0.46, a precision of 0.72, and an F-measure of 0.56. The performance on the CAGE data was calculated with the novel method presented here and has 0.61 recall, 0.87 precision, and 0.72 for the F-measure. Both performances were calculated using a maximum distance of only 500 bp. As expected from previous analyses of the ENCODE region (Bajic et al. 2006a), we see that the performance of our program is better on this region than on the rest of the genome, which indicates that some of the FP in the genome setting are actually missed genes or missed TSSs.
To test this last claim, we retrieved datasets from the ENCODE project for Affymetrix Transcribed Fragments, Yale Transcriptionally Active Regions (TARs), and novel TARs from the DART system (Rozowsky et al. 2007). We combined these three sets with the CAGE data from Riken (single CAGE tags included). This set is called the Evidence for Transcriptional Activity set (EFTA). When comparing the predictions of EP3, we found that of all predictions made by EP3, 87% have a hit with EFTA within 125 bp, 95% have a hit within 500 bp, and 98% have a hit within 2000 bp within EFTA. When excluding the single CAGE tags, the rates drop to 80%, 92%, and 97%, respectively. These numbers indicate that EP3 has a very strong specificity and that many of the so-called FPs discussed above are in fact associated with transcriptionally active regions.
Furthermore, we compared the predictions of EP3 with two sets of DNase hypersensitivity sites (DHSS) retrieved from the ENCODE project. For the first set (encodeNhgriDnaseHsMpssCd4, seven cell types), 50% of the DHSS are near a prediction of EP3, and for the second set (encodeRegulomeDnaseGM06990Sites, one cell type), 28% of the DHSS are near an EP3 prediction. For both sets, the recall rate is lower than that on the CAGE set, but this was to be expected, as the DNase dataset covers only a limited number of cell types.
Performance on different eukaryotic genomes
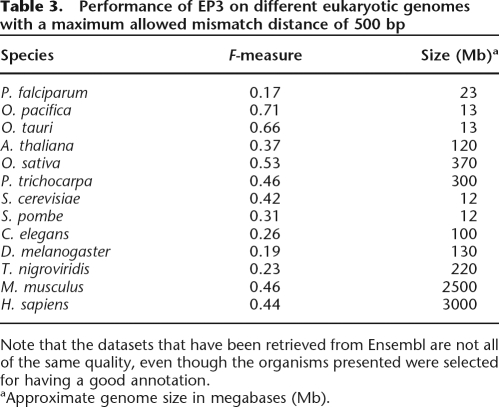
In the previous sections, we demonstrated the performance of EP3 on the human genome. We have also tested its performance on a wide range of other eukaryotes, including animals (Mus musculus, Tetraodon nigroviridis, Drosophila melanogaster), fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe), algae (Ostreococcus tauri, Ostreococcus pacifica), higher plants (Arabidopsis thaliana, Oryza sativa, Populus trichocarpa), and a protist (Plasmodium falciparum). Only data for human and mouse are available from the CAGE technique; therefore, we limited the analyses for the other eukaryotes to the data available from Ensembl. Table 3 shows the result when we used EP3 to predict promoter regions in other eukaryotes. The F-measure ranges from 0.17 to 0.71 on the different species, with P. falciparum (F = 0.17) and D. melanogaster (F = 0.19) on the low end of the scale, and O. pacifica (F = 0.71) and O. tauri (F = 0.66) giving the best results. We evaluated the performance of EP3 only; the other programs are not suitable for all other genomes because they are specifically trained for a single species. EP3 obtains a good score for some species, while for other species the score is worse. The F-score is a bit higher for mouse than for human, which indicates that the program performs well for mammals. Within the green lineage (green algae and land plants), there seem to be two groups, based on the genome size. The performance for the two algae (first group) is excellent (F > 0.65), which is probably partly due to the very small genome size and the still large gene space (∼8000 genes). Due to the window approach to assess TPs and FPs, most predictions will be a TP because of the small genome. The second group comprised of rice, Arabidopsis, and poplar has larger genomes, and the performance of EP3 is comparable to that on mammals. The performance for the two yeasts is lower than that on the two Ostreococcus genomes, which have roughly the same genome size. This is probably due to the less obvious profile in yeasts compared with the one in algae (see Fig. 2). The performance of EP3 on Drosophila and Plasmodium is weak. In the case of the fruit fly, this is likely due to its very different structural profile, as observed in Figure 2, while for Plasmodium the low performance is probably caused by the rather flat profile observed in protists that makes it very difficult for EP3 to distinguish promoter regions from other parts of the genome. Therefore, for some species, for example D. melanogaster, specifically trained programs might perform better (Ohler 2006).
Table 3.
Performance of EP3 on different eukaryotic genomes with a maximum allowed mismatch distance of 500 bp
Note that the datasets that have been retrieved from Ensembl are not all of the same quality, even though the organisms presented were selected for having a good annotation.
aApproximate genome size in megabases (Mb).
Recognizing different promoter types
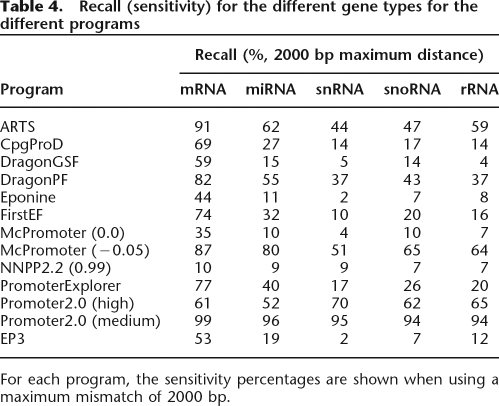
Besides genes that code for proteins, there are also genes that are transcribed but for which the RNA is not translated into proteins, so-called noncoding RNAs. These genes produce transcripts that function directly as structural, catalytic, or regulatory RNAs. Recent screens for such genes revealed a surprisingly large number of them, with prominent roles such as guiding the post-transcriptional regulation of protein-coding genes (Eddy 2001; Bartel 2004). Previous studies on promoter prediction focused mainly on a single type of promoter, most often the promoter of protein-coding genes, which are transcribed by RNAP II. Using the Ensembl annotation for humans, we show that our approach is also suited to predict other types of promoters. Although the program works best to identify and delineate promoters of protein-coding genes, it can also be used to detect promoters of snRNA, rRNA, miRNAs, snoRNA, and tRNA genes. Other types of noncoding genes such as scRNA and mitochondrial rRNA were not considered because of the lack of data in the Ensembl database. Table 4 shows the different recall (sensitivity) rates for the different types of genes in this study (snRNA, rRNA, miRNA, snoRNA, and tRNA). The precision cannot be calculated for these analyses because the program will always predict all types of promoters and it will never give predictions specific for a single type of promoter. Therefore, we focus only on how many known noncoding RNA promoters we can identify with our approach. Although EP3 can identify non-protein-coding genes, other programs have higher recall rates. However, the programs with high recall rates are also the ones that performed worse when we applied them to humans. From Table 1, we see that the programs that have high recall and low precision have the lowest F-measure, those also being the programs that have the highest recall for the non-protein-coding genes. Although EP3 is thus also capable of predicting the promoters of noncoding genes, its performance is significantly lower, most probably because the peak in the profile is much smaller (see Fig. 2). Nevertheless, compared with the other top-performing programs on the whole genome (DragonGSF and PromoterInspector), EP3 has very similar recall rates. From this analysis, it is clear that general-purpose PPPs such as those listed in Table 1 are best suited for the prediction of protein-coding gene promoters. For miRNA and tRNA promoters, there are probably better approaches using specifically trained tools for identification of these types of promoters (Lowe and Eddy 1997; Zhou et al. 2007). Finally, EP3 is slightly biased toward GC-rich promoters because the structural feature is most outspoken in these promoters. This bias toward GC-rich promoters is also present in all top-performing PPPs from Table 1 (Scherf et al. 2001; Bajic et al. 2004) and indicates that CpG-island-associated housekeeping genes are favored in the predictions.
Table 4.
Recall (sensitivity) for the different gene types for the different programs
For each program, the sensitivity percentages are shown when using a maximum mismatch of 2000 bp.
Conclusion
The evaluation of PPPs in a whole-genome context is crucial to understand the true performance of the program. Evaluation on a small test set, such as the Eukaryotic Promoter Database (Schmid et al. 2006), does not provide sufficient insight into the real performance of the program when used in actual genome annotation projects. The recall and precision values we found in our analysis are lower than those reported in the original papers, where in most cases the evaluation was done on a (much) smaller dataset. Even the evaluation of a complete chromosome is not sufficient, as there are huge differences in nucleotide content and gene density. If possible, it is advisable to use transcription data, such as the CAGE data, to assess the performance. Transcription data are superior to the gene annotation and its associated way of counting TPs; the gene annotation may not give a complete picture on the performance because it completely ignores intergenic predictions. In short, one should assess a promoter predictor on the whole genome, preferably validating with TSS data.
While EP3 does not outperform its peers by much, the program has several additional advantages compared with other PPPs. EP3 requires no training or parameter tuning, unlike other programs that need extensive amounts of experimentally determined data for the training of their model (Ohler et al. 2000; Scherf et al. 2000; Davuluri et al. 2001; Down and Hubbard 2002; Bajic et al. 2003). When working on a genomic scale, speed and memory requirements also are of importance. EP3 is very fast (for instance, it takes <1 h to annotate the complete human genome), requires little memory, and can thus be run on a home computer; in contrast, some programs require a computer cluster of 80 machines for nearly a week to process the human genome (data not shown). Besides performing very well, especially in light of its simplicity, EP3 can handle many eukaryotic genomes without modifications, as was shown here.
Up to now, most PPPs have used biologically driven small-scale features to build a model for promoter recognition (Fickett and Hatzigeorgiou 1997; Hannenhalli and Levy 2001; Werner 2003). These features are different for each species, and thus the current programs need to be retrained for each species. Here, we have presented a simple approach that employs more global structural features of the DNA sequence in promoter and nonpromoter regions. Our technique does not use any complex machine learning algorithms, for which it is often impossible to infer any new knowledge from the model itself. On the contrary, our method requires no training whatsoever and can be applied to any eukaryotic genome. Numerical profiles representing sequence-dependent properties investigated on a large scale and a very large number of genes show a remarkable feature of promoters. To the best of our knowledge, this is the first time that this feature is described for multiple eukaryotes. The feature associated with promoters is an extended region where the DNA is more stable. This large-scale feature is not present in other types of genomic sequence. While it is most outspoken in vertebrate promoters, it is also present in other eukaryotic promoters, so it seems to be universal and largely independent of the presence or absence of binding sites. As a result, EP3 is capable of performing very well on several eukaryotic species, without the need for any training, which is a unique achievement among promoter prediction software, allowing true “ab initio” promoter prediction. Finally, analysis of the well documented ENCODE region shows that most predictions are indeed associated with transcriptionally active regions.
Methods
Datasets
The CAGE datasets for human have been retrieved from the FANTOM3 project (http://fantom.gsc.riken.go.jp/). The database was compiled by Carninci et al. (2006) and was obtained through the CAGE technique, which identifies all possible TSSs with very high accuracy (Shiraki et al. 2003). This dataset covers the whole human genome and can thus be used for validation purposes. Only tag clusters with at least two mapped tags on the same genomic location were considered to be real TSSs, which should filter out most FPs from the CAGE technique. This filtering resulted in 123,400 unique start sites for human. The whole genome sequences for human (hg17) and mouse (mm5) were retrieved from the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) (Kuhn et al. 2007).
We retrieved the following genes in human using the BioMart tool at the Ensembl website, release 37 (http://www.ensembl.org): (1) 20,297 protein-coding, (2) 1382 small nuclear RNA (snRNA), (3) 330 ribosomal RNA (rRNA), (4) 326 microRNA (miRNA), and (5) 642 small nucleolar RNA (snoRNA) (Hubbard et al. 2007). The dataset for transfer RNA (tRNA) was downloaded from the tRNAscan-SE homepage (http://lowelab.ucsc.edu/GtRNAdb/Hsapi/) (Lowe and Eddy 1997). The datasets with annotations for different species (Table 3) were retrieved from two different sources: The protein-coding genes for human, mouse, Tetraodon, fruit fly, and yeast were retrieved using BioMart, while the genomic sequences were retrieved from the Ensembl ftp server (ftp://ftp.ensembl.org/pub/). The sequences and annotation for the five plant species were retrieved from in-house data (http://bioinformatics.psb.ugent.be). The genome and annotation for P. falciparum were retrieved from the Sanger database (http://www.sanger.ac.uk/).
The data for the ENCODE analysis have been downloaded from the UCSC ENCODE repository (http://genome.ucsc.edu/ENCODE/). The data for the functional promoters in the encode region were retrieved from Supplemental Table D (Cooper et al. 2006). The data for the novel transcriptionally active regions were retrieved from the DART database (http://dart.gersteinlab.org/ENCODE/GT/).
Calculating structural profiles
Profiles from DNA sequences are calculated as follows: First, the nucleotide sequence is converted into a sequence of numbers (i.e., a numerical profile). This is done by replacing each dinucleotide (or trinucleotide, depending on the physico-chemical feature used) with its corresponding structural value, which is obtained from experimentally validated conversion tables. Florquin et al. (2005) provide references to the protocols to obtain these conversion tables. These contain values, for example, for stabilizing energy of Z-DNA (Ho et al. 1990), DNA denaturation (Blake and Delcourt 1998; Blake et al. 1999), protein-induced deformability (Olson et al. 1998), and duplex-free energy (Sugimoto et al. 1996). Next, we average over several values, and the number of values we use for computing this average is called the window size, in our case 400 bp. The value 400 was chosen because it gave the best performance in a whole-genome context (see Results). If a value in the profile exceeds a certain threshold, this points to a putative promoter region (see below). For properties that show a cleft instead of a peak (see below), we took the inverse value to make sure we also obtained peaks. This was done to be certain we could use the same approach for all structural features.
Prediction algorithm
The algorithm we use (EP3) to identify promoters has two internal parameters, one for the length of the window and one for the deviation from the average. Both have been determined empirically. The optimal length of the window is 400 bp. We determined this by trying values between 50 and 2000 and seeing which one performed best. Smaller windows result in more predictions, but the number of false predictions increases faster than the rate of true predictions. Larger values result in fewer predictions but higher precision. The F-measure indicates that 400 bp is optimal for all organisms. This value appears to be independent of the size of the genome because the organisms we analyzed ranged in size from 12 Mbp to 3 Gbp, and 400 bp was always the optimal value. Because the overall profile of a genome can vary significantly between different species due to different GC content, the threshold should be determined dynamically for each genome. As a basis, we use the average and standard deviation of the values of a property for the whole genome. For bigger genomes (human and mouse), this deviation should be larger than for other species. Empirically, we determined that the optimal threshold is the average plus one standard deviation for all species, except for human and mouse where this is the average plus three standard deviations. Future research with more available genomes, especially ones that fill in the gap between the small genomes (≤300 Mbp) and the large ones (≥2.5 Gbp) may allow us to define a function that can calculate the optimal multiplier for the standard deviation from the genome size. When the window size (400 bp) and the threshold (depends on genome size) are known, the prediction algorithm is straightforward. The program calculates the profile, and each time a value in the profile is above the threshold for this genome, the location is predicted to be a putative promoter region.
Evaluating predictions
A measure for the performance of a PPP is the harmonic mean of the recall (sensitivity) and the precision (specificity), known as the F-measure (van Rijsbergen 1979). The higher this value, the better the program is able to correctly predict promoters. The recall or sensitivity is the number of predicted promoters (TP) divided by the total number of promoters (TP+FN). The precision or specificity is the number of correct predictions (TP) divided by the total number of predictions (TP+FP). To assess the number of TPs, FPs, and FNs, we need to define the maximum allowed mismatch between the prediction and the true TSS. In previous studies, this value was set to 2000 bp (Bajic et al. 2004; Sonnenburg et al. 2006; Xie et al. 2006), although more recently, for the ENCODE project and for two PPPs, a maximum mismatch of 1000 bp also was evaluated (Bajic et al. 2006a). In Tables 1–3, we display the performance of the programs for three different maximum mismatch values: 2000 bp, 1000 bp, and 500 bp. The smaller the mismatch allowed, the stricter the evaluation. We ranked the performance of the different programs according to the smallest window size (500 bp). In the classic way to evaluate a PPP (Bajic et al. 2004), one will start from gene annotations, e.g., as compiled at Ensembl. A TP is then defined as a TSS that has a prediction within the maximum allowed distance from the true TSS as annotated in the database. A FP is a prediction that lies inside the gene but not within the maximum allowed distance from the TSS. A FN is a true TSS from the database that has no prediction within the maximum allowed mismatch. All predictions that fall within an intergenic region and further from a TSS than the maximum allowed mismatch are ignored. Counting a prediction inside a gene as a FP will be incorrect in some cases, as Carninci et al. (2006) showed that transcription can start in the 5′ untranslated region (UTR) of a gene, as well as in an exon or in the 3′ UTR. Therefore, not taking into account any of the intergenic predictions or only considering the TSSs located at the 5′ UTR gives a biased view of a promoter predictor. Using a gene annotation and ignoring the intergenic predictions will not give a real estimate of the performance of the prediction program for two reasons: (1) predictions inside a gene may in fact be true TSSs; TPs might be underestimated, and consequently FPs are then overestimated; (2) intergenic predictions outside the TSS region are ignored because it is impossible to know whether they are real FPs or whether they point to genes missed by the annotation process. This may in turn lead to underestimation or overestimation of FPs. When using gene annotation with all its uncertainties, it is impossible to know the actual performance of the program in a genomic setting, especially in light of the evidence that much more of the genome is transcribed than just the protein-coding regions (Kapranov et al. 2007; Peters et al. 2007; Ponjavic et al. 2007).
To address some of these caveats, we developed an alternative way to assess the performance of a promoter prediction program that does not exhibit these shortcomings. Recently available whole-genome TSS data allow for a new way to count and evaluate. We used a dataset of TSSs for the whole human genome that were characterized experimentally using high-throughput cap-analysis of gene expression (CAGE) (for review, see Carninci 2006). In contrast to previous studies (Bajic et al. 2004, 2006a; Sonnenburg et al. 2006), this dataset allows counting the number of TPs and FPs in an objective way. The dataset contains locations where transcription starts. A TP is a prediction that is within the maximum allowed mismatch from a true TSS, a FN is a true TSS that has no prediction, and a FP is a prediction that is not near a true TSS from the dataset.
Availability
A user-friendly online implementation of our promoter prediction software EP3 is available at http://bioinformatics.psb.ugent.be/. The program allows the user to process FASTA-formatted sequence files. The online implementation is also available as a standalone application.
Acknowledgments
We thank Pierre Hilson for fruitful discussions and three anonymous referees for their constructive comments that greatly helped improve the original version of the manuscript. This work was supported by a grant from the Research Foundation–Flanders (3G031805). T.A. thanks the Institute for the Promotion of Innovation by Science and Technology in Flanders for a predoctoral fellowship. Y.S. is a Postdoctoral Fellow of the Research Foundation–Flanders.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6991408
References
- Aerts S., Thijs G., Dabrowski M., Moreau Y., De Moor B., Thijs G., Dabrowski M., Moreau Y., De Moor B., Dabrowski M., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics. 2004;5:34. doi: 10.1186/1471-2164-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaldi F., Bozzoni I., Beccari E., Pierandrei-Amaldi P., Bozzoni I., Beccari E., Pierandrei-Amaldi P., Beccari E., Pierandrei-Amaldi P., Pierandrei-Amaldi P. Expression of ribosomal protein genes and regulation of ribosome biosynthesis in Xenopus development. Trends Biochem. Sci. 1989;14:175–178. doi: 10.1016/0968-0004(89)90269-7. [DOI] [PubMed] [Google Scholar]
- Baek D., Davis C., Ewing B., Gordon D., Green P., Davis C., Ewing B., Gordon D., Green P., Ewing B., Gordon D., Green P., Gordon D., Green P., Green P. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007;17:145–155. doi: 10.1101/gr.5872707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic V.B., Seah S.H., Chong A., Zhang G., Koh J.L.Y., Brusic V., Seah S.H., Chong A., Zhang G., Koh J.L.Y., Brusic V., Chong A., Zhang G., Koh J.L.Y., Brusic V., Zhang G., Koh J.L.Y., Brusic V., Koh J.L.Y., Brusic V., Brusic V. Dragon Promoter Finder: Recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- Bajic V.B., Seah S.H., Chong A., Krishnan S.P.T., Koh J.L.Y., Brusic V., Seah S.H., Chong A., Krishnan S.P.T., Koh J.L.Y., Brusic V., Chong A., Krishnan S.P.T., Koh J.L.Y., Brusic V., Krishnan S.P.T., Koh J.L.Y., Brusic V., Koh J.L.Y., Brusic V., Brusic V. Computer model for recognition of functional transcription start sites in RNA polymerase II promoters of vertebrates. J. Mol. Graph. Model. 2003;21:323–332. doi: 10.1016/s1093-3263(02)00179-1. [DOI] [PubMed] [Google Scholar]
- Bajic V.B., Tan S.L., Suzuki Y., Sugano S., Tan S.L., Suzuki Y., Sugano S., Suzuki Y., Sugano S., Sugano S. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 2004;22:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]
- Bajic V.B., Brent M.R., Brown R.H., Frankish A., Harrow J., Ohler U., Solovyev V.V., Tan S.L., Brent M.R., Brown R.H., Frankish A., Harrow J., Ohler U., Solovyev V.V., Tan S.L., Brown R.H., Frankish A., Harrow J., Ohler U., Solovyev V.V., Tan S.L., Frankish A., Harrow J., Ohler U., Solovyev V.V., Tan S.L., Harrow J., Ohler U., Solovyev V.V., Tan S.L., Ohler U., Solovyev V.V., Tan S.L., Solovyev V.V., Tan S.L., Tan S.L. Performance assessment of promoter predictions on ENCODE regions in the EGASP experiment. Genome Biol. 2006a;7:S3.1–3.13. doi: 10.1186/gb-2006-7-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic V.B., Tan S.L., Christoffels A., Schonbach C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Tan S.L., Christoffels A., Schonbach C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Christoffels A., Schonbach C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Schonbach C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Yang L., Hofmann O., Kruger A., Hide W., Kai C., Hofmann O., Kruger A., Hide W., Kai C., Kruger A., Hide W., Kai C., Hide W., Kai C., Kai C., et al. Mice and men: Their promoter properties. PLoS Genet. 2006b;2:e54. doi: 10.1371/journal.pgen.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Chauvin Y., Brunak S., Gorodkin J., Pedersen A.G., Chauvin Y., Brunak S., Gorodkin J., Pedersen A.G., Brunak S., Gorodkin J., Pedersen A.G., Gorodkin J., Pedersen A.G., Pedersen A.G. Computational applications of DNA structural scales. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:35–42. [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blake R.D., Delcourt S.G., Delcourt S.G. Thermal stability of DNA. Nucleic Acids Res. 1998;26:3323–3332. doi: 10.1093/nar/26.14.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R.D., Bizzaro J.W., Blake J.D., Day G.R., Delcourt S.G., Knowles J., Marx K.A., SantaLucia J., Bizzaro J.W., Blake J.D., Day G.R., Delcourt S.G., Knowles J., Marx K.A., SantaLucia J., Blake J.D., Day G.R., Delcourt S.G., Knowles J., Marx K.A., SantaLucia J., Day G.R., Delcourt S.G., Knowles J., Marx K.A., SantaLucia J., Delcourt S.G., Knowles J., Marx K.A., SantaLucia J., Knowles J., Marx K.A., SantaLucia J., Marx K.A., SantaLucia J., SantaLucia J. Statistical mechanical simulation of polymeric DNA melting with MELTSIM. Bioinformatics. 1999;15:370–375. doi: 10.1093/bioinformatics/15.5.370. [DOI] [PubMed] [Google Scholar]
- Bode J., Winkelmann S., Gotze S., Spiker S., Tsutsui K., Bi C., Prashanth A.K., Benham C., Winkelmann S., Gotze S., Spiker S., Tsutsui K., Bi C., Prashanth A.K., Benham C., Gotze S., Spiker S., Tsutsui K., Bi C., Prashanth A.K., Benham C., Spiker S., Tsutsui K., Bi C., Prashanth A.K., Benham C., Tsutsui K., Bi C., Prashanth A.K., Benham C., Bi C., Prashanth A.K., Benham C., Prashanth A.K., Benham C., Benham C. Correlations between scaffold/matrix attachment region (S/MAR) binding activity and DNA duplex destabilization energy. J. Mol. Biol. 2006;358:597–613. doi: 10.1016/j.jmb.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Breslauer K.J., Frank R., Blocker H., Marky L.A., Frank R., Blocker H., Marky L.A., Blocker H., Marky L.A., Marky L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I., Sanchez R., Suck D., Pongor S., Sanchez R., Suck D., Pongor S., Suck D., Pongor S., Pongor S. Trinucleotide models for DNA bending propensity: Comparison of models based on DNaseI digestion and nucleosome packaging data. J. Biomol. Struct. Dyn. 1995;13:309–317. doi: 10.1080/07391102.1995.10508842. [DOI] [PubMed] [Google Scholar]
- Burden S., Lin Y.X., Zhang R., Lin Y.X., Zhang R., Zhang R. Improving promoter prediction for the NNPP2.2 algorithm: A case study using Escherichia coli DNA sequences. Bioinformatics. 2004;21:601–607. doi: 10.1093/bioinformatics/bti047. [DOI] [PubMed] [Google Scholar]
- Butler J.E.F., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;22:501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., Taylor M.S., Engström P.G., Frith M.C., Engström P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Chen Q.K., Hertz G.Z., Stormo G.D., Hertz G.Z., Stormo G.D., Stormo G.D. PromFD 1.0: A computer program that predicts eukaryotic pol II promoters using strings and IMD matrices. Comput. Appl. Biosci. 1997;13:29–35. doi: 10.1093/bioinformatics/13.1.29. [DOI] [PubMed] [Google Scholar]
- Choi C.H., Kalosakas G., Rasmussen K.O., Hiromura M., Bishop A.R., Usheva A., Kalosakas G., Rasmussen K.O., Hiromura M., Bishop A.R., Usheva A., Rasmussen K.O., Hiromura M., Bishop A.R., Usheva A., Hiromura M., Bishop A.R., Usheva A., Bishop A.R., Usheva A., Usheva A. DNA dynamically directs its own transcription initiation. Nucleic Acids Res. 2004;32:1584–1590. doi: 10.1093/nar/gkh335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J.M., Poirot O., Lopez F., Poirot O., Lopez F., Lopez F. The difficulty of identifying genes in anonymous vertebrate sequences. Comput. Chem. 1997;21:203–214. doi: 10.1016/s0097-8485(96)00039-3. [DOI] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R., Tanese N., Tjian R., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Cooper S.J., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Anton E.D., Nguyen L., Myers R.M., Nguyen L., Myers R.M., Myers R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P., Struhl K., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Davuluri R.V., Grosse I., Zhang M.Q., Grosse I., Zhang M.Q., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Delgado S., Gómez M., Bird A., Antequera F., Gómez M., Bird A., Antequera F., Bird A., Antequera F., Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Roberts S.G., Roberts S.G. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes & Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Roberts S.G., Roberts S.G. Core promoter elements recognized by transcription factor IIB. Biochem. Soc. Trans. 2006;34:1051–1053. doi: 10.1042/BST0341051. [DOI] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J.P., Hubbard T.J.P. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- El Hassan M.A., Calladine C.R., Calladine C.R. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J. Mol. Biol. 1996;259:95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- Faiger H., Ivanchenko M., Cohen I., Haran T.E., Ivanchenko M., Cohen I., Haran T.E., Cohen I., Haran T.E., Haran T.E. TBP flanking sequences: Asymmetry of binding, long-range effects and consensus sequences. Nucleic Acids Res. 2006;34:104–119. doi: 10.1093/nar/gkj414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J.W., Hatzigeorgiou A.G., Hatzigeorgiou A.G. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- Fitzgerald P., Sturgill D., Shyakhtenko A., Oliver B., Vinson C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C., Shyakhtenko A., Oliver B., Vinson C., Oliver B., Vinson C., Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florquin K., Saeys Y., Degroeve S., Rouzé P., de Van Peer Y., Saeys Y., Degroeve S., Rouzé P., de Van Peer Y., Degroeve S., Rouzé P., de Van Peer Y., Rouzé P., de Van Peer Y., de Van Peer Y. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res. 2005;33:4255–4264. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukue Y., Sumida N., Nishikawa J.-i., Ohyama T., Sumida N., Nishikawa J.-i., Ohyama T., Nishikawa J.-i., Ohyama T., Ohyama T. Core promoter elements of eukaryotic genes have a highly distinctive mechanical property. Nucleic Acids Res. 2004;32:5834–5840. doi: 10.1093/nar/gkh905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukue Y., Sumida N., Tanase J.-i., Ohyama T., Sumida N., Tanase J.-i., Ohyama T., Tanase J.-i., Ohyama T., Ohyama T. A highly distinctive mechanical property found in the majority of human promoters and its transcriptional relevance. Nucleic Acids Res. 2005;33:3821–3827. doi: 10.1093/nar/gki700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin A.A., Zhurkin V.B., Olson W.K., Zhurkin V.B., Olson W.K., Olson W.K. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- Greenbaum J.A., Pang B., Tullius T.D., Pang B., Tullius T.D., Tullius T.D. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007;17:947–953. doi: 10.1101/gr.6073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.S., Brent M.R., Brent M.R. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Hannenhalli S., Levy S., Levy S. Promoter prediction in the human genome. Bioinformatics. 2001;17:S90–S96. doi: 10.1093/bioinformatics/17.suppl_1.s90. [DOI] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.S., Zhou G.W., Clark L.B., Zhou G.W., Clark L.B., Clark L.B. Polarized electronic spectra of Z-DNA single crystals. Biopolymers. 1990;30:151–163. doi: 10.1002/bip.360300115. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I.P., Zhang M.Q., Zhang M.Q. Large-scale human promoter mapping using CpG islands. Nat. Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- Ivanov V.I., Minchenkova L.E., Minchenkova L.E. The A-form of DNA: In search of the biological role. Mol. Biol. (Mosk.) 1994;28:1258–1271. [PubMed] [Google Scholar]
- Jacob S.T. Regulation of ribosomal gene transcription. Biochem. J. 1995;306:617–626. doi: 10.1042/bj3060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V.X., Singer G.A.C., Agosto-Pérez F.J., Liyanarachchi S., Davuluri R.V., Singer G.A.C., Agosto-Pérez F.J., Liyanarachchi S., Davuluri R.V., Agosto-Pérez F.J., Liyanarachchi S., Davuluri R.V., Liyanarachchi S., Davuluri R.V., Davuluri R.V. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A., Bansal M., Bansal M. Structural properties of promoters: Similarities and differences between prokaryotes and eukaryotes. Nucleic Acids Res. 2005;33:3165–3175. doi: 10.1093/nar/gki627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R., Willingham A.T., Gingeras T.R., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kawaji H., Frith M.C., Katayama S., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Frith M.C., Katayama S., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Katayama S., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kawai J., Carninci P., Hayashizaki Y., Carninci P., Hayashizaki Y., Hayashizaki Y. Dynamic usage of transcription start sites within core promoters. Genome Biol. 2006;7:R118. doi: 10.1186/gb-2006-7-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Stanke M., Smith K.E., Siepel A., Smith K.E., Siepel A., Siepel A., et al. The UCSC genome browser database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H., Tang H., Reinberg D., Ebright R.H., Reinberg D., Ebright R.H., Ebright R.H. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G-c., Rehm E.J., Rubin G.M., Rehm E.J., Rubin G.M., Rubin G.M. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K., Tavernarakis N., Hugenholtz P., Kyrpides N.C., Tavernarakis N., Hugenholtz P., Kyrpides N.C., Hugenholtz P., Kyrpides N.C., Kyrpides N.C. The Genomes On Line Database (GOLD) v.2: A monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., States D.J., States D.J. Consensus promoter identification in the human genome utilizing expressed gene markers and gene modeling. Genome Res. 2002;12:462–469. doi: 10.1101/gr.198002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu X.S., Wei L., Altman R.B., Batzoglou S., Liu X.S., Wei L., Altman R.B., Batzoglou S., Wei L., Altman R.B., Batzoglou S., Altman R.B., Batzoglou S., Batzoglou S. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res. 2004;14:451–458. doi: 10.1101/gr.1327604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Tostesen E., Sundet J.K., Jenssen T.K., Bock C., Jerstad G.I., Thilly W.G., Hovig E., Tostesen E., Sundet J.K., Jenssen T.K., Bock C., Jerstad G.I., Thilly W.G., Hovig E., Sundet J.K., Jenssen T.K., Bock C., Jerstad G.I., Thilly W.G., Hovig E., Jenssen T.K., Bock C., Jerstad G.I., Thilly W.G., Hovig E., Bock C., Jerstad G.I., Thilly W.G., Hovig E., Jerstad G.I., Thilly W.G., Hovig E., Thilly W.G., Hovig E., Hovig E. The human genomic melting map. PLoS Comput. Biol. 2007;3:e93. doi: 10.1371/journal.pcbi.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K., Smale S.T., Smale S.T. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathé C., Sagot M.-F., Schiex T., Rouzé P., Sagot M.-F., Schiex T., Rouzé P., Schiex T., Rouzé P., Rouzé P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merling A., Sagaydakova N., Haran T.E., Sagaydakova N., Haran T.E., Haran T.E. A-tract polarity dominate the curvature in flanking sequences. Biochemistry. 2003;42:4978–4984. doi: 10.1021/bi020662w. [DOI] [PubMed] [Google Scholar]
- Munch K., Krogh A., Krogh A. Automatic generation of gene finders for eukaryotic species. BMC Bioinformatics. 2006;7:263. doi: 10.1186/1471-2105-7-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov D.B., Chen H., Halay E.D., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K., Chen H., Halay E.D., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K., Halay E.D., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K., Hisatake K., Lee D.K., Roeder R.G., Burley S.K., Lee D.K., Roeder R.G., Burley S.K., Roeder R.G., Burley S.K., Burley S.K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Ohler U. Promoter prediction on a genomic scale—The Adh experience. Genome Res. 2000;10:539–542. doi: 10.1101/gr.10.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U. Computational promoter recognition in eukaryotic genomic DNA. Technische Fakultät der Universität Erlangen; Nürnberg: 2001. [Google Scholar]
- Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Niemann H., Niemann H. Identification and analysis of eukaryotic promoters: Recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- Ohler U., Stemmer G., Harbeck S., Niemann H., Stemmer G., Harbeck S., Niemann H., Harbeck S., Niemann H., Niemann H. Stochastic segment models of eukaryotic promoter regions. Pac. Symp. Biocomput. 2000;5:377–388. doi: 10.1142/9789814447331_0036. [DOI] [PubMed] [Google Scholar]
- Ohler U., Niemann H., Liao G.-c., Rubin G.M., Niemann H., Liao G.-c., Rubin G.M., Liao G.-c., Rubin G.M., Rubin G.M. Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics. 2001;17:S199–S206. doi: 10.1093/bioinformatics/17.suppl_1.s199. [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G.-c., Niemann H., Rubin G.M., Liao G.-c., Niemann H., Rubin G.M., Niemann H., Rubin G.M., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W.K., Gorin A.A., Lu X.-J., Hock L.M., Zhurkin V.B., Gorin A.A., Lu X.-J., Hock L.M., Zhurkin V.B., Lu X.-J., Hock L.M., Zhurkin V.B., Hock L.M., Zhurkin V.B., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl. Acad. Sci. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornstein R.L., Rein R., Breen D.L., MacElroy R.D., Rein R., Breen D.L., MacElroy R.D., Breen D.L., MacElroy R.D., MacElroy R.D. Optimized potential function for calculation of nucleic-acid interaction energies. 1. Base stacking. Biopolymers. 1978;17:2341–2360. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]
- Pedersen A.G., Baldi P., Chauvin Y., Brunak S.O., Baldi P., Chauvin Y., Brunak S.O., Chauvin Y., Brunak S.O., Brunak S.O. DNA structure in human RNA polymerase II promoters. J. Mol. Biol. 1998;281:663–673. doi: 10.1006/jmbi.1998.1972. [DOI] [PubMed] [Google Scholar]
- Pedersen A.G., Baldi P., Chauvin Y., Brunak S.O., Baldi P., Chauvin Y., Brunak S.O., Chauvin Y., Brunak S.O., Brunak S.O. The biology of eukaryotic promoter prediction—A review. Comput. Chem. 1999;23:191–207. doi: 10.1016/s0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- Peters B.A., St Croix B., Sjoblom T., Cummins J.M., Silliman N., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., St Croix B., Sjoblom T., Cummins J.M., Silliman N., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Sjoblom T., Cummins J.M., Silliman N., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Cummins J.M., Silliman N., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Silliman N., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Ptak J., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Saha S., Kinzler K.W., Hatzis C., Velculescu V.E., Kinzler K.W., Hatzis C., Velculescu V.E., Hatzis C., Velculescu V.E., Velculescu V.E. Large-scale identification of novel transcripts in the human genome. Genome Res. 2007;17:287–292. doi: 10.1101/gr.5486607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponger L., Mouchiroud D., Mouchiroud D. CpGProD: Identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics. 2002;18:631–633. doi: 10.1093/bioinformatics/18.4.631. [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C.P., Lunter G., Ponting C.P., Lunter G., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Presutti C., Ciafre S.A., Bozzoni I., Ciafre S.A., Bozzoni I., Bozzoni I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991;10:2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsch G., Sonnenburg S., Schafer C., Sonnenburg S., Schafer C., Schafer C. Learning interpretable SVMs for biological sequence classification. BMC Bioinformatics. 2006;7:S9. doi: 10.1186/1471-2105-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Rombauts S., Florquin K., Lescot M., Marchal K., Rouzé P., de Van Peer Y., Florquin K., Lescot M., Marchal K., Rouzé P., de Van Peer Y., Lescot M., Marchal K., Rouzé P., de Van Peer Y., Marchal K., Rouzé P., de Van Peer Y., Rouzé P., de Van Peer Y., de Van Peer Y. Computational approaches to identify promoters and cis-regulatory elements in plant genomes. Plant Physiol. 2003;132:1162–1176. doi: 10.1104/pp.102.017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J.S., Newburger D., Sayward F., Wu J., Jordan G., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Newburger D., Sayward F., Wu J., Jordan G., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Sayward F., Wu J., Jordan G., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Wu J., Jordan G., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Jordan G., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Korbel J.O., Nagalakshmi U., Yang J., Zheng D., Guigo R., Nagalakshmi U., Yang J., Zheng D., Guigo R., Yang J., Zheng D., Guigo R., Zheng D., Guigo R., Guigo R., et al. The DART classification of unannotated transcription within the ENCODE regions: Associating transcription with known and novel loci. Genome Res. 2007;17:732–745. doi: 10.1101/gr.5696007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S.C., Drew H.R., Travers A.A., Drew H.R., Travers A.A., Travers A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Scherf M., Klingenhoff A., Werner T., Klingenhoff A., Werner T., Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: A novel context analysis approach. J. Mol. Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- Scherf M., Klingenhoff A., Frech K., Quandt K., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Klingenhoff A., Frech K., Quandt K., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Frech K., Quandt K., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Quandt K., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Gailus-Durner V., Seidel A., Brack-Werner R., Seidel A., Brack-Werner R., Brack-Werner R., et al. First pass annotation of promoters on human chromosome 22. Genome Res. 2001;11:333–340. doi: 10.1101/gr.154601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.D., Perier R., Praz V., Bucher P., Perier R., Praz V., Bucher P., Praz V., Bucher P., Bucher P. EPD in its twentieth year: Towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–D85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Watahiki A., Nakamura M., Arakawa T., Nakamura M., Arakawa T., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivolob A.V., Khrapunov S.N., Khrapunov S.N. Translational positioning of nucleosomes on DNA: The role of sequence-dependent isotropic DNA bending stiffness. J. Mol. Biol. 1995;247:918–931. doi: 10.1006/jmbi.1994.0190. [DOI] [PubMed] [Google Scholar]
- Smale S.T. Core promoters: Active contributors to combinatorial gene regulation. Genes & Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Solovyev V.V., Shahmuradov I.A., Shahmuradov I.A. PromH: Promoters identification using orthologous genomic sequences. Nucleic Acids Res. 2003;31:3540–3545. doi: 10.1093/nar/gkg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V., Kosarev P., Seledsov I., Vorobyev D., Kosarev P., Seledsov I., Vorobyev D., Seledsov I., Vorobyev D., Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7:11–12. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg S.O., Zien A., Rätsch G., Zien A., Rätsch G., Rätsch G. ARTS: Accurate recognition of transcription starts in human. Bioinformatics. 2006;22:472–480. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu. Rev. Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Nakano S., Yoneyama M., Honda K., Nakano S., Yoneyama M., Honda K., Yoneyama M., Honda K., Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Tsunoda T., Sese J., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Tsunoda T., Sese J., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Sese J., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Ota T., Isogai T., Tanaka T., Nakamura Y., Isogai T., Tanaka T., Nakamura Y., Tanaka T., Nakamura Y., Nakamura Y., et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y., Brosh R., Buganim Y., Reiner A., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Brosh R., Buganim Y., Reiner A., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Buganim Y., Reiner A., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Reiner A., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Yitzhaky A., Koudritsky M., Rotter V., Domany E., Koudritsky M., Rotter V., Domany E., Rotter V., Domany E., Domany E. Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site. PLoS ONE. 2007;2:e807. doi: 10.1371/journal.pone.0000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Schroeder D.I., Otillar R.P., Myers R.M., Otillar R.P., Myers R.M., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren P., Cameron-Jones M., Sale A., Cameron-Jones M., Sale A., Sale A. Promoter prediction using physic-chemical properties of DNA. Lect. Notes Comput. Sci. 2006;4216:21–31. [Google Scholar]
- van Rijsbergen C.J. Information retrieval. Butterworths; London: 1979. [Google Scholar]
- Wang H., Benham C.J., Benham C.J. Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics. 2006;7:248. doi: 10.1186/1471-2105-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang W., Zhang W. A steganalysis-based approach to comprehensive identification and characterization of functional regulatory elements. Genome Biol. 2006;7:R49. doi: 10.1186/gb-2006-7-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen Y., Li Y., Chen Y., Li Y., Li Y. A brief review of computational gene prediction methods. Genom. Proteom. Bioinformatics. 2004;2:216–221. doi: 10.1016/S1672-0229(04)02028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Thompson W., Fickett J.W., Lawrence C.E., Fickett J.W., Lawrence C.E., Lawrence C.E. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Werner T. The state of the art in mammalian promoter recognition. Brief. Bioinform. 2003;4:22–30. doi: 10.1093/bib/4.1.22. [DOI] [PubMed] [Google Scholar]
- White R.J., Jackson S.P., Jackson S.P. The TATA-binding protein: A central role in transcription by RNA polymerases I, II and III. Trends Genet. 1992;8:284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- Xie X., Wu S., Lam K.M., Yan H., Wu S., Lam K.M., Yan H., Lam K.M., Yan H., Yan H. PromoterExplorer: An effective promoter identification method based on the AdaBoost algorithm. Bioinformatics. 2006;22:2722–2728. doi: 10.1093/bioinformatics/btl482. [DOI] [PubMed] [Google Scholar]
- Zhang M.Q. Computational prediction of eukaryotic protein-coding genes. Nat. Rev. Genet. 2002;3:698–709. doi: 10.1038/nrg890. [DOI] [PubMed] [Google Scholar]
- Zhou X., Ruan J., Wang G., Zhang W., Ruan J., Wang G., Zhang W., Wang G., Zhang W., Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput. Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]