Abstract
Human lipoxygenases (hLO) have been implicated in a variety of diseases and cancers and each hLO isozyme appears to have distinct roles in cellular biology. This fact emphasizes the need for discovering selective hLO inhibitors for both understanding the role of specific lipoxygenases in the cell and developing pharmaceutical therapeutics. To this end, we have modified a known lipoxygenase assay for high-throughput (HTP) screening of both the National Cancer Institute (NCI) and the UC Santa Cruz marine extract library (UCSC-MEL) in search of platelet-type 12-hLO (12-hLO) selective inhibitors. The HTP screen led to the characterization of five novel 12-hLO inhibitors from the NCI repository. One is the potent but non-selective michellamine B, a natural product, antiviral agent. The other four compounds were selective inhibitors against 12-hLO, with three being synthetic compounds and one being α-mangostin, a natural product, caspase-3 pathway inhibitor. In addition, a selective inhibitor was isolated from the UCSC-MEL (neodysidenin), which has a unique chemical scaffold for an hLO inhibitor. Due to the unique structure of neodysidenin, steady-state inhibition kinetics were performed and its mode of inhibition against 12-hLO was determined to be competitive (Ki = 17 µM) and selective over reticulocyte 15-hLO-1 (Ki 15-hLO-1/12-hLO > 30).
Keywords: lipoxygenase, high-throughput, NCI, α-mangostin, michellamine B, neodysidenin, dysidenin, kinetics, IC50
Introduction
Human Lipoxygenases (hLOs) are non-heme, iron-containing enzymes that catalyze the dioxygenation of polyunsaturated fatty acids to their hydroperoxy acids1 and have been implicated in several diseases involving inflammation, immune disorders and various types of cancers.2–4 The reticulocyte 15–hLO-1 (15-hLO-1) has been implicated in colorectal5 and prostate6 cancers, while platelet-type 12–LO has been implicated in pancreatic,7 breast8, 9 and prostate cancers.10, 11 In contrast, prostate epithelial 15-hLO-2 (15-hLO-2) appears to be anti-tumorigenic in prostate cancer,12 highlighting the need for specific LO inhibitors.
There are three isozymes of human 12-LO, platelet-type, leukocyte and epidermal, which show distinct biochemical and inhibitory properties.13–15 Platelet-type 12-hLO (12-hLO) is one of the most commonly expressed arachidonic acid metabolizing enzymes in human cancer cells.9, 16–18 Enhanced expression of 12-hLO has been observed in many cancers17 and it is thought to promote angiogenesis,19 cell migration,20 cell adhesion,21 endothelial cell retraction,22 and inhibition of apoptosis,23 all properties essential for tumor growth and metastasis.16 These broad implications in cancer regulation24 underscore the need for small molecule inhibitors against 12-hLO, however, relatively few inhibitors, with selectivity ratios greater than 10 against 15-hLO-1, have been found. Gallocatechin gallate is a 12-hLO selective inhibitor which has an IC50 of 0.14 µM against 12-hLO and an IC50 15-hLO-1/12-hLO ratio of greater than 700.25 Hinokitol, a tropolone,26 is also selective against 12-hLO (IC50 = 0.1 µM and an IC50 15-hLO-1/12-hLO ratio > 1000),27 but it unfortunately undergoes a structural conversion upon exposure to light, making it unsuitable as a drug lead. Baicalein has historically been thought of as a 12-hLO selective inhibitor but this is not the case as seen by recent in vitro data.28 The lack of 12-hLO specific inhibitors in the literature is also reflected in our screening of the UC Santa Cruz marine extract library (UCSC-MEL). Over 20 hLO inhibitors have been characterized from our of marine natural products (MNP) library, but only a few are selective against 12-hLO and none have selectivities greater than 10.29–33 The most selective 12-hLO inhibitors discovered to date in our laboratories are (-)-7-N-methyldibromophakellin33 (IC50 15-hLO-1/12-hLO = 5), jaspic acid29 (IC50 15-hLO-1/12-hLO = 2) and hyrtenone A29 (IC50 15-hLO-1/12-hLO = 2).
In the current investigation, we have modified a known lipoxygenase assay34 to a high-throughput (HTP), 384-well format and screened four different compound libraries for 12-hLO selective inhibitors. These include the National Cancer Institute (NCI) chemical repository, which contains the NCI natural product library (NCI-NPL) (235 structurally diverse compounds), the NCI diversity library (NCI-DL) (1990 known pharmacophores) and the NCI mechanistic library (NCI-ML) (879 known growth inhibitors of tumor cell lines) and a portion of the UCSC-MEL (272 sponge extracts from approximately 90 different sponges). By this screen, we have discovered five novel 12-hLO selective inhibitors and characterized their inhibitory properties.
Results
Expression and Purification of Lipoxygenases
Human platelet-type 12-hLO (12-hLO), human reticulocyte 15-hLO-1 (15-hLO-1) and human prostate epithelial 15-hLO-2 (15-hLO-2) were purified utilizing SF9 insect cells with approximate yields of 50 mg/L. The ICP-MS data indicated that 12-hLO had 12 ± 1% iron content, 15-hLO-1 had 24 ± 2% iron content and 15-hLO-2 had 35 ± 5% iron content. All kinetics data were adjusted for iron content.
High-Throughput Screening Assay Analysis
Initial HTP screening of the 3104 NCI compounds and the 272 sponge extracts was performed against both 12-hLO and 15-hLO-1. Utilizing the Fe3+/xylenol orange screening method,34,35 we miniaturized it to an HTP 384-well format and discovered six 12-hLO selective inhibitors from the NCI library; NSC30552, NSC125034, NSC172033, NSC292213, NSC617570, NSC661755 (Table 1). Four organo-mercurial compounds (NSC20410, NSC268879, NSC321237 and NSC321239) were also found to be selective, but not pursued further due to their potentially toxic side effects.36 Screening of the UCSC-MEL only revealed one clear 12-hLO selective crude extract, the dichloromethane fraction (02155FD) from the sponge Dysiden herbacea (Table 1). All other sponge extracts or semi-pure fractions showed little or no selective inhibition against 12-hLO.
Table 1.
Percent inhibition from the high through-put (HTP) and Manual-Initial (M-I) screens for 12-hLO and 15-hLO-1. Concentrations of 40 µM and 10 µM were used for the NSC compounds and 40 µg/ml and 25 µg/ml for the crude sponge extract for the HTP assay and the M-I, respectively.
| Sample | NSC30552 | NSC125034 | NSC172033 | NSC292213 | NSC617570 | NSC661755 | 02155FD | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin | Nat. Prod. | Syn. Org. | Syn. Org. | Syn. Org. | Syn. Inorg. | Nat. Prod. | Extract | |||||||
| Screen | HTP | M-I | HTP | M-I | HTP | M-I | HTP | M-I | HTP | M-I | HTP | M-I | HTP | M-I |
| 12-hLO | 47 | 100 | 71 | 0 | 70 | 100 | 46 | 88 | 46 | 81 | 52 | 75 | 50 | 62 |
| 15-hLO-1 | 12 | 36 | 39 | 0 | 37 | 36 | 0 | 0 | 0 | 0 | 0 | 69 | 0 | 2 |
Verification of Hits from Initial High-Throughput Screening Assay
Manual, continuous assays, with one inhibitor concentration, were performed on the six NCI compounds and the one marine sponge extract from the HTP screen to verify their selectivity against 12-hLO (Table 1). We determined that NSC30552, NSC172033, NSC292213 and NSC617570 were selective inhibitors against 12-hLO. NSC661755 was not selective but was a potent inhibitor against both 12-hLO and 15-hLO-1. NSC125034, however, did not inhibit either LO isozyme, even though it did show potency in the HTP screen (Table 1). It is unclear why the HTP displayed this false positive, but one possibility is the differences in reaction conditions and inhibitor concentrations between the HTP and manual assays. The 02155 FD fraction from the UCSC-MEL was selective against 12-hLO (Table 1).
IC50 Analysis
The IC50 analysis of 12-hLO, 15-hLO-1 and 15-hLO-2 was performed on the NCI and UCSC compounds as previously described31 and were consistent with the one point, manual assay. Four compounds, NSC30552, NSC172033, NSC292213 and NSC617570, had selectivity ratios greater than 5 (Table 2). Compound NSC661755 was determined to be non-selective with an IC50 15-hLO-1/12-hLO ratio of 2. None of these compounds inhibited 15-hLO-2, with all exhibiting IC50 values greater than 50 µM (Table 2). These compounds were analyzed by LC-MS and shown to be greater than 95% pure as received from NCI. For the UCSC-MNP, neodysidenin had an IC50 greater than 100 µM against 12-hLO and had no effect against 15-hLO-1 (Figure 2, Table 2).
Table 2.
Compound IC50 values (µM) for 12-hLO, 15-hLO-1 and 15-hLO-2.
| Sample | Structure | 12-hLO | 15-hLO-1 | 15-hLO-2 | 15-hLO-1 /12-hLO |
|---|---|---|---|---|---|
| NSC30552 (α-mangostin) | 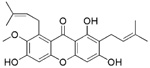 |
0.58 ± 0.09 | 3.1 ± 0.69 | > 50 | 5 |
| NSC172033 | 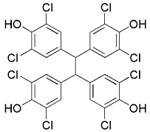 |
0.21 ± 0.05 | 9.4 ± 2.0 | >50 | 45 |
| NSC292213 | 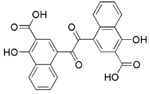 |
0.15 ± 0.02 | >25 | >50 | >167 |
| NSC617570 | 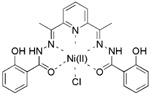 |
0.19 ± 0.08 | > 500 | > 50 | >2632 |
| NSC661755 (michellamine B) | 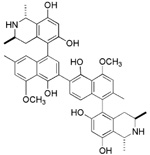 |
4.9 ± 0.8 | 7.6 ± 2.5 | >50 | 2 |
| neodysidenin | 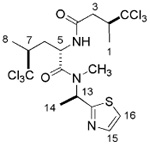 |
>100 | >500 | n.d.a | >5 |
n.d. = not determined
Figure 2.
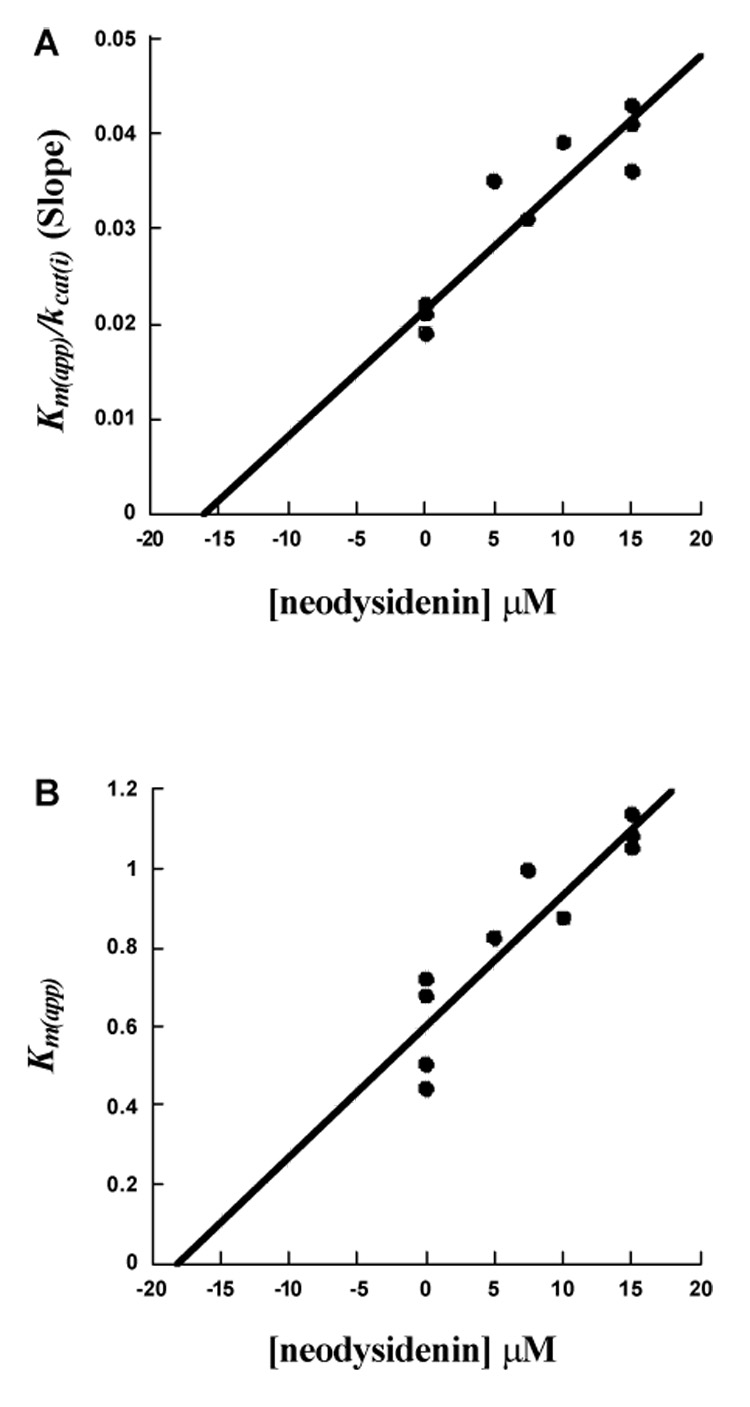
Competitive inhibition steady-state kinetic data for Ki determination for 12-hLO with neodysidenin. (A), Km(app) / kcat(i) (slope) vs. [neodysidenin] µM (B), Km(app) vs. [neodysidenin] µM.
Neodysidenin Steady-State Inhibition Studies of 12-hLO and 15-hLO-1
For 12-hLO, neodysidenin showed competitive inhibition. The plots Km(app) / kcat(i) (slope) and Km(app) versus neodysidenin concentration for 12-hLO are shown in Figure 2A and 2B, respectively. Both plots showed linear graphs with similar inhibition constants, where the Km/kcat plot yields a Ki of 16 ± 1 µM (Figure 2A) and the Km(app) plot yields a Ki of 18 ± 1 µM (Figure 2B), indicating competitive inhibition.37 The average of the Ki values is 17 ± 1 µM (Table 3). It is interesting to note that these inhibition constants are lower than those seen from the IC50 data. It is unclear why there is this discrepancy, except for the fact that the steady-state data is much more accurate than the IC50 data. Neodysidenin had no effect on 15-hLO-1 at concentrations up to 500 µM, indicating no appreciable inhibition (Table 3). The specific activities of both 12-hLO and 15-hLO-1 were comparable to our previously published values.38
Table 3.
UCSC library compound steady-state inhibition data for 12-hLO and 15-hLO-1.
| Neodysidenin | Dysidenin | |
|---|---|---|
| 12-hLO | Ki = 17 ± 1 µM | Ki = 9 ± 2 µM |
| Ki’ = 55 ± 24 µM | ||
| 15-hLO-1 | Ki > 500 µM | Ki = 8 ± 3 µM |
| Ki’ = 54 ± 27 µM | ||
| 15-hLO-1/12hLO | > 30 | 1 |
Dysidenin Steady-State Inhibition Studies of 12-hLO and 15-hLO-1
Dysidenin was obtained from the UC Santa Cruz marine compound library for SAR comparison to neodysidenin. For 12-hLO, plots of Km(app) / kcat(i) (slope) and 1 / kcat(i) (y-intercept) versus dysidenin are shown in Figure 3A and 3B.37 The plots are linear and give two different inhibitor constants, Ki and Ki’, which are defined as the equilibrium constant of the dissociation of inhibitor from the catalytic site and a secondary site, possibly an allosteric binding site,39 respectively. The Km(app) / kcat(i) versus [I] plot yields a Ki of 9 ± 2 µM, while 1 / kcat(i) versus [I] plot yields a Ki’ of 55 ± 24 µM (Table 3), representing a linear mixed-type inhibition.37 For 15-hLO-1, dysidenin also showed linear mixed-type inhibition with a Ki of 8 ± 3 µM and a Ki’ of 54 ± 27 µM (Table 3).
Figure 3.
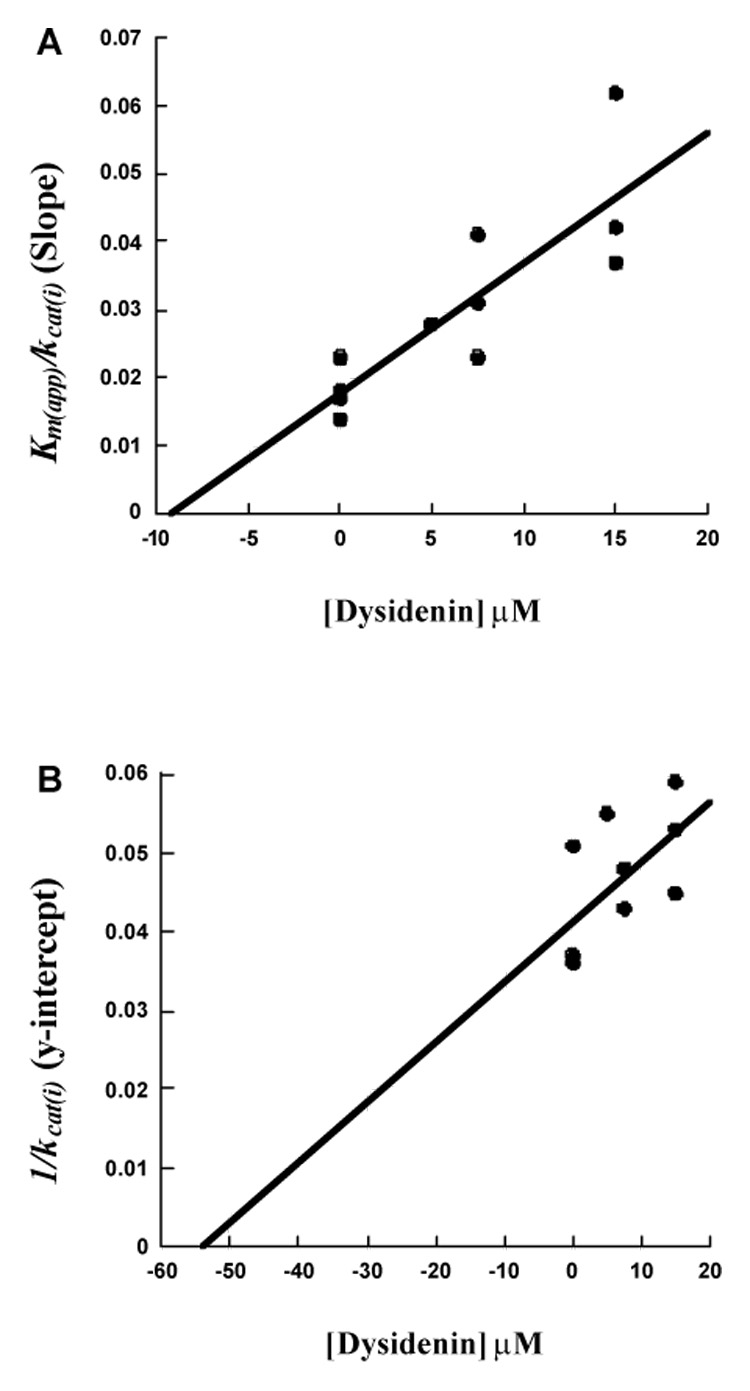
Linear mixed-type inhibition steady-state kinetic data for determination of Ki and Ki’ for 12-hLO with dysidenin. (A), Km(app) / kcat(i) (slope) vs. [dysidenin] µM (B), 1/kcat(i) (y-intercept) vs. [dysidenin] µM.
Discussion
For many years, our laboratories have been interested in discovering hLO selective inhibitors, however, none have had a greater IC50 15-hLO-1/12-hLO ratio than 5.29, 30 To accelerate our ability to screen for 12-hLO selective inhibitors, we modified the known xylenol orange lipoxygenase assay34, 35, 40–42 into an HTP 384-well format and screened the 3104 compounds of the NCI mechanistic, diversity and natural product library (Scheme 1). Sixteen potent 12-hLO inhibitors were found, 10 being relatively selective, which represents a 0.3% proportion of selective 12-hLO inhibitors. By comparison, the HTP screen of the NCI repository found 43 potent inhibitors against 15-hLO-1, with 33 being selective, representing a 1.4% proportion of selective inhibitors. The lower percentage of 12-hLO selective inhibitors versus 15-hLO-1 selective inhibitors is consistent with our previous work with UCSC-MNPs and illustrates the difficulty in targeting 12-hLO. Of the 10 selective 12-hLO inhibitors found by the HTP screen, 4 were organo-mercurials, which were discarded due to potential toxicity.36 The remaining 6 were subjected to secondary manual screening, with NSC125034 losing all inhibitory activity and NSC661755 (michellamine B)43 losing its inhibitory selectivity, however, not its potency (IC50 for 12-hLO is 4.9 µM and 7.6 µM for 15-hLO-1). Michellamine B is a natural product, anti-HIV agent, which inhibits protein kinase C (PKC)44 and has been implicated as a potential anti-cancer treatment. Our discovery that michellamine B inhibits both 12-hLO and 15-hLO-1 but does not inhibit 15-hLO-2, may stimulate further interest in its study as an anti-cancer agent.
Scheme 1.
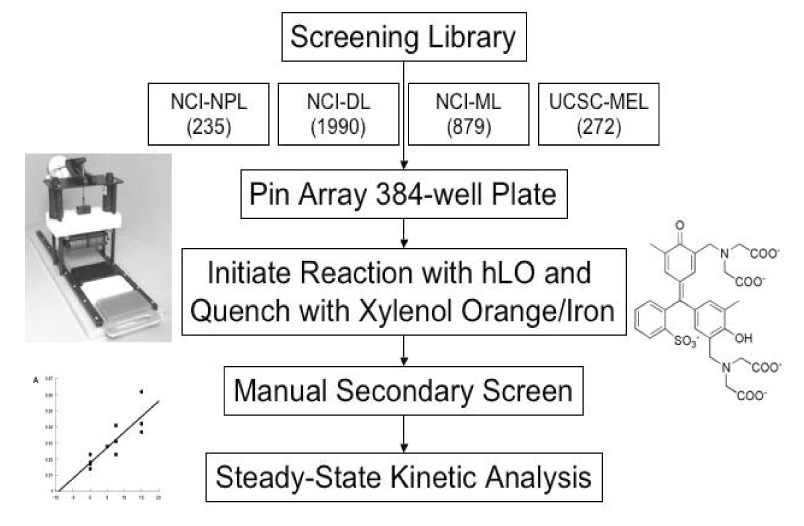
The remaining 4 compounds (NSC30552, NSC172033, NSC292213 and NSC617570) are all selective against 12-hLO but have little structural similarity. The only common structural feature is that they all contain two or more aromatic hydroxyl groups, which typically reduce the active site ferric iron of LO.45 The most unusual inhibitor is NSC617570, a nickel complex, which is not only exceptionally potent (IC50 = 190 nM), but also has a high IC50 15-hLO-1/12-hLO ratio of 2631. Its structure is not amenable to drug discovery but it could be useful as a molecular probe against LO activity in the cell. Of the remaining three compounds, Lipinski’s “rule of five” (LRoF) was used to assess the extent that their physical and structural features were drug-like46 and only NSC292213, a known 5-aminoimidazole-4-carboxamide ribonucleotide transformylase inhibitor,47 adhered to LRoF (Table 4). NSC172033, a known human mitotic kinesin inhibitor,48, 49 has a large MW (674.018 Da), while NSC30552 has a cLogP greater than 5, both values violate LRoF. Despite its high cLogP, NSC30552 (α-mangostin) is used as a traditional medicine for the treatment of skin infections, wounds and diarrhea50 and has been shown to inhibit sphingomyelinase.51,52 α-Mangostin also inhibits cyclooxygenase53 and the caspase-3 pathway through direct inhibition of Ca(II)-ATPase, inducing apoptosis.54 It is interesting to note that 12-hLO inhibitors also induce apoptosis through the caspase-3 pathway,55 which could imply that α-mangostin has dual activity against cancer proliferation. This hypothesis should be tempered by the fact that its IC50 15-hLO-1/12-hLO ratio is only 5, however further study is still warranted.
Table 4.
Physical and structural features of 12-hLO inhibitors
| Compound | MW (Da) | cLogPa | H-bond Acceptors | H-bond donors |
|---|---|---|---|---|
| NSC30552 (α-mangostin) | 410.47 | 6.3 | 6 | 3 |
| NSC172033 | 674.02 | 9.6 | 4 | 4 |
| NSC292213 | 430.37 | 4.2 | 8 | 4 |
| NSC617570 | 527.61 | 1.8 | 9 | 6 |
| NSC661755 (michellamine B) | 756.90 | 9.9 | 10 | 8 |
| neodysidenin | 543.97 | 4.9 | 5 | 2 |
clogP (i.e. miLogP) determined with Molinspiration Calculation Service, www.molinspiration.com.
The HTP screening of the 272 sponge extracts from the UCSC-MEL yielded only one extract, which was selective against 12-hLO (02155 FD). Fractionation of this crude extract yielded the 12-hLO inhibitor, neodysidenin, an isomer of dysidenin, a known inhibitor of iodide transport in thyroid cells.56 Steady-state inhibition kinetics demonstrated that neodysidenin was a competitive inhibitor against 12-hLO (Ki = 17 ± 1 µM) but it was inactive against 15-hLO-1 (Ki > 500 µM). Even though the potency of neodysidenin is relatively low, its selectivity against 12-hLO is the highest recorded for our MNP hLO inhibitors (Ki 15-hLO-1/12-hLO ratio > 30). In contrast, steady-state inhibition kinetics determined that dysidenin was a linear mixed inhibitor against both 12-hLO (Ki = 9 ± 2 µM, Ki ’ = 55 ± 24 µM) and 15-hLO-1 (Ki = 8 ± 3 µM, Ki’ = 54 ± 27 µM). These data suggest that neodysidenin binds only to the active site of 12-hLO, while dysidenin binds to both the active site and a secondary site of both 12-hLO and 15-hLO-1, possibly the allosteric site.57 This is an intriguing result because neodysidenin and dysidenin58 are parallel in structure and differ only in the regiochemistry of the N-methyl, with epimeric geometries at carbon-13. These minor structural changes modulate hLO selectivity from 1 for dysidenin to >30 for neodysidenin. Nevertheless, this gain in selectivity for neodysidenin has a concomitant loss of potency, with neodysidenin’s Ki values to the active site of 12-hLO being twice that of dysidenin. A similar effect was seen previously in our laboratories, where hyrtenone A, which is selective against 12-hLO, is over 10-fold less potent than its analogue, puupehenone, a selective 15-hLO-1 inhibitor.29 The potency and selectivity of neodysidenin against 12-hLO versus 15-hLO-1 make it a novel class of LO inhibitor that could potentially be optimized by further structural modifications to increase its potency against 12-hLO, while maintaining its high selectivity. In addition, it will be important to determine whether neodysidenin is also selective against platelet-type 12-hLO versus leukocyte and epidermal 12-hLO.
Conclusion
In summary, our data provides a number of important insights regarding rapid inhibitor discovery against platelet-type 12-hLO. First, we demonstrated that the xylenol orange assay can be miniaturized to an HTP 384-well format, which greatly accelerates our discovery of platelet-type 12-hLO selective inhibitors versus 15-hLO-1. Second, the NCI repository contains 4 potent and selective 12-hLO inhibitors, which can be used as investigative tools for the role of hLO in the cell. Third, neodysidenin represents a novel structural class of LO inhibitors that is selective against platelet-type 12-hLO and whose potency could be optimized with structural modifications.
Experimental
Materials
Arachidonic acid (AA), linoleic acid (LA), xylenol orange and ferrous ammonium sulfate were purchased from Sigma-Aldrich Chemical Company. Mechanistic, diversity and natural products 384-well library plates were obtained from the National Cancer Institute (NCI). Larger quantities of selected compounds required for further inhibition investigations were also obtained from the NCI and shown to be pure with a Thermo LCQ LC-MS. The marine sponge crude extracts and dysidenin were obtained from the marine natural products repository at UCSC. All other chemicals were reagent grade or better and were used without further purification.
Reverse phase-HPLC purification of AA and LA
AA and LA were purified as published,38 using a Higgins Preparative Haisil (250 × 10mm) C-18 5 µM column. An isocratic elution of 85% A and 15% B (Solvent A: 99.9% MeOH 0.1% acetic acid, Solvent B: 99.9% H2O and 0.1% acetic acid) was used to purify the fatty acids and both were stored in 95% EtOH at -20°C.
Expression and Purification of Lipoxygenases
Human platelet-type 12-lipoxygenase (12-hLO) and reticulocyte 15-lipoxygenase (15-hLO-1) are N-terminus, His6-tagged proteins and were expressed and purified as described previously.29 The SF9 expression vector for human prostate epithelial 15-lipoxygenase-2 (15-hLO-2) was constructed as follows. The previously published plasmid, pCRII-TOPO- 15-hLO-2,12 was cut with EcoRI to liberate the 15-hLO-2 fragment. This 15-hLO-2 fragment was then ligated into EcoRI cut, pFastBac1 (GibcoBRL) to generate the complete plasmid, pFastBac-15-hLO-2, which was digested to determine the correct orientation. The pFastBac-15-hLO-2 was then transposed into a recombinant FastBac bacmid by DH10Bac cells (GibcoBRL) and then transfected into SF9 cells, as described in the product literature for pFastBac1 (GibcoBRL). The virus was subsequently amplified to 2×1010 plaque forming units (pfu), added to SF9 cells (2×106 cells/ml) at a concentration of 2×107 pfu/ml and allowed to shake for 72 hours. The cells were then harvested and frozen. 15-hLO-2 was purified by douncing the thawed cells and loading the cell extracts onto a 20 mL Macro-prep High Q, strong anion exchange column, from Bio-Rad. The protein was eluted with a 50-300 mM sodium chloride gradient. The collected fractions, containing 95% pure protein and 5% glycerol, were then frozen at −80° C. Subsequent thawing produced no decrease in enzymatic activity. Iron contents of all three lipoxygenase enzymes were determined with a Finnigan inductively coupled plasma mass spectrometer (ICP-MS), using cobalt-EDTA as an internal standard. Enzyme iron concentrations were compared to standardized iron solutions.
Collection and Identification of Dysidea herbacea
The crude extract (02155FD) was obtained in 2002 near Duchess Island, Papua New Guinea (9_57.228 S, 150_51.059 E) using SCUBA at -30 ft (UCSC coll. nos. 02155). The sample was fan shaped, green in color with a purple underside, and had a ridged surface punctuated with digitate projections (3–5 cm). Taxonomy of 02155 was performed by R.W.M. van Soest (Zoological Museum of Amsterdam) and was identified as Lamellodysidea herbacea (order Dictyoceratida; family Dysideidae). The recent re-classification of the family Dysideidae has led to the novel genus Lamellodysidea,59 which is marked by its massive, lamellate to digitate morphology, and thin basal plate. This newly proposed taxonomic nomenclature is synonymous with many previously designated Dysidea herbacea, and has only recently begun to enjoy proper usage. Thus, we have chosen to indicate these sponges as Dysidea (Lamellodysidea) herbacea and note that such samples are similar to our previous disclosures of Dysidea herbacea.60, 61
Extraction and Isolation of Neodysidenin
The D. herbacea collection 02155 was preserved, transported, and stored at 4°C until processed, as previously described.60, 62 The UCSC Kupchan-like extraction method was utilized to produce six crude extracts.33 These six crude extracts were then subjected to LCMS analysis using an HPLC system equipped with a Waters 717plus Autosampler, and Waters 996 photodiode array detector (PDAD), Sedere Sedex 75 evaporative light scattering detector (ELSD), and a Mariner MS (ESI-TOF-MS). Isotopic distribution patterns from MS were then used to determine HPLC peaks corresponding to halogenated metabolites and ELSD data provided approximate relative amounts. It was readily apparent from LCMS that collection 02155 contained a number of polychlorinated species with molecular weights corresponding to known peptides from D. herbacea. The most abundant chlorinated metabolite in the dichloromethane crude extract (FD) of the 02155 bulk collection was pursued and identified as a hexachlorinated species of m/z 544 [M+H]+ by LCMS. Approximately 100 mg of 02155 FD was directly applied to reversed-phase HPLC separation, leading to the isolation of (-)-neodysidenin (5.0 mg). The following properties of (-)- neodysidenin were compared with literature values: Neodysidenin: white powder: [α]26d - 52.7° (c 0.10, MeOH); UV ?max (MeOH) 241 nm; ¹H NMR (500 MHz, CDCL3) d 1.39 (3H, d, J = 6.5 Hz, H-1), 1.50 (H3, d, J = 6.0 Hz, H-8), 1.69 (3H, d, J = 7.5 Hz, H-14), 1.81a (m), 2.32 (H, m, H-3), 2.34 (H, m, H-6′), 2.62 (H, m, H-7), 2.96 (3H, s, N-Me), 3.09 (H, dd, J = 14.5, 2.0 Hz, H-3′), 3.22 (H, m, H-2), 5.09, (H, ddd, J = 11.5, 8.5, 2.5 Hz, H-5), 6.17 (H, q, J = 7.0 Hz, H-13), 6.80 (H, bd, J = 8.0 Hz, NH), 7.35 (H, d, J = 3.5 Hz, (H-17), 7.75 (H, d, J = 3.5 Hz, H-16); and 13C NMR and DEPT (125.5 MHz, MeOD) d 15.2 CH3, 15.3 CH3, 15.7 CH3, 29.2 CH3, 34.9 CH2, 38.9 CH2, 49.0 CH, 50.7 CH, 51.5 CH, 51.6 CH, 104.7 C, 105.3 C, 120.0 CH, 141.9 CH, 170.5 C, 171.5 C, 171.6 C. HRESITOFMS [M+H]+ m/z 543.9712 (calcd for C17H24N3O2SCl6 543.9715). a Obscured by overlap.
Dysidenin was obtained from the UCSC-MNP pure compound repository. The following properties of (-)-dysidenin were compared with literature values: Dysidenin: white powder; [α]23d - 68° (c 0.034, CHCl3); UV ?max (CHCl3) 248 nm; ¹H NMR (250 MHz, CDCL3) d 1.36 (3H, d, J = 6.5 Hz, H-8), 1.39 (H3, d, J = 6.9 Hz, H-1), 1.59 (H3, d, J = 6.8 Hz, H-14), 1.92 (H, ddd, J =14.8, 10.5, 2.5, 2.22 Hz, H-6′), 2.22 (H, ddq, J =10.5, 6.5, 2.5 Hz, H-7), 2.49 (H, dd, J = 16.3, 10.0 Hz, H-3′), 2.62 (H, dd, J = 14.8 10.5 Hz, H-6), 3.04 (H, s, H-10), 3.13 (H, dd, J = 16.3, 2.5 Hz, H-3), 3.38 (H, ddq, J = 10.0, 6.5, 2.5 Hz, H-2), 5.36 (H, q, J = 6.8 Hz, H-13), 5.37 (H, 13 dd, J =10.5, 2.5 Hz, H-5), 6.84 (H, bd, J = 7.0 Hz, N-H), 7.28 (H, d, J = 3.3 Hz, H-17), 7.71, (H, d, J = 3.3 Hz, H-16); 13C NMR (75.5 MHz, CDCl3) d 16.3 (C-8), 17.4 (C-1), 21.9 (C-14), 30.9 (C-10), 31.0 (C-6), 37.5 (C-3), 47.3 (C-13), 51.5 (C-2), 51.9 (C-7), 54.1 (C-5), 105.2 (C-11), 105.6 (C-9), 119.2 (C-17), 142.5 (C-16), 168.9 (C-12), 172.0 (C-15), 172.1 (C-4); LRFABMS (m/z 544 [M+H]+).
High-Throughput Screening Assay
The high-throughput (HTP) screening assay was modified from a previously published procedure to utilize a 384-well assay plate.34,35 Briefly, the lipid peroxide product (HPETE) formed by the LO reaction converts ferrous iron (Fe2+) into ferric iron (Fe3+), which forms a Fe3+/xylenol orange complex that absorbs at 560 nm. The HTP screen was performed as published earlier with the following modifications.34,35 Reactions with 12-hLO were carried out in 25 mM Hepes buffer (pH 8.0) in the presence of 0.1% Triton X-100. Reactions with 15-hLO-1 and 15-hLO-2 were carried out in 25 mM HEPES (pH 7.5) in the presence of 0.1% Triton X-100. A 30 µM (45 µl) substrate buffer solution (AA for 12-hLO and 15-hLO-2 and LA for 15-hLO-1) was added to each well using a Thermo Labsystems 384-well Multidrop. The compounds from the 384 assay plate libraries, dissolved in DMSO, were added using a specially designed automated liquid pin transfer device from V & P Scientific, which delivers 0.2 µL of sample. The NCI plates are at 10 mM concentrations and the UC Santa Cruz marine extracts are at 10 mg/ml, which yields final concentrations of inhibitors to be 40 µM and 40 µg/ml for the NCI pure samples and MNP crude extracts, respectively. Using the Multidrop, the reactions were initiated by adding 5 µL of an enzyme concentration that produces a linear rate for the duration of the experiment with approximately 40 nM for 12-hLO and 15-hLO-1 (final volume of 50 µL). The reactions were then stopped at approximately 35% completion, by the addition of the appropriate amounts of regents to give a final concentration of 100 µM of xylenol orange and 150 µM ferrous ammoniums sulfate in 25 mM H2SO4, with a final volume of 100 µl in each well. The plates were allowed to shake for 30 minutes in the dark on an orbital shaker to allow the Fe3+/xylenol orange complex to form. After 30 minutes the plates were read by measuring the absorption at 560 nm using a Victor2 Perkin-Elmer plate reader. The negative controls for the assay contained no enzyme and 0.2 µl of DMSO (with no inhibitor) in the reaction buffer. The positive controls for the assay contained enzyme and 0.2 µl of DMSO in the reaction buffer, as mentioned above. All assays were done in triplicate. The percent inhibitions were calculated by subtracting the absorbance of the negative controls from the absorbance of the positive controls. The values were then subtracted from 1 and multiplied by 100 to get the percent inhibition.
Manual-Initial Screens
The one-point inhibition percentages were determined by following the formation of the conjugated diene product at 234 nm (ε = 25,000 M¹cm−1) with a Perkin-Elmer Lambda 40 UV/Vis spectrophotometer at one inhibitor concentration. All reactions were 2 mL in volume and constantly stirred using a magnetic stir bar at room temperature (23° C) with approximately 40 nM for 12-hLO and 15-hLO and 1 µM for 15-hLO-2. Reactions with 12-hLO were carried out in 25 mM Hepes buffer (pH 8) in the presence of 0.01% Triton-X-100. Reactions with 15-hLO-1 and 15-hLO-2 were carried out in 25 mM Hepes buffer (pH 7.5) in the presence of 0.01% Triton-X-100. The concentration of AA (for 12-hLO and 15-hLO-2) and LA (for 15-hLO-1) were quantitatively determined by allowing the enzymatic reaction to go to completion. For the NSC compounds, 10 µM inhibitor was used and for the crude sponge extract, 25 µg/ml was used.
IC50 Assay
IC50 values were determined using the same method as previously described in the one point, manual screen section above. IC50 values were obtained by determining the enzymatic rate at various inhibitor concentrations then plotting them against inhibitor concentration. The data was fit to a saturation curve and the inhibitor concentration at 50% activity was determined (IC50). Inhibitors were stored at −20°C in MeOH or DMSO depending on their solubility.
Steady-State Inhibition Kinetics Studies
Lipoxygenase rates were determined using the same method as previously described in the IC50 section. Michaelis-Menten kinetics were determined for 12-hLO and 15-hLO-1 with their respective substrates and at varying inhibitor concentrations, from 0.38 to 80 µM. Enzymatic reactions were initiated by the addition of 5 nM 12-hLO and 9 nM 15-hLO-1. Kinetic data were obtained by recording initial enzymatic rates at each substrate concentration and then fitting them to the Michaelis-Menten equation using the KaleidaGraph (Synergy) program. All inhibitors were studied in separate experiments against each enzyme at least three times to determine their inhibitor binding constants (Ki and Ki’).37 The Km(app) and kcat(i) values were obtained from hyperbolic fits at various inhibitor concentrations. The percent error for both Ki and Ki’ were determined from the linear Km(app) / kcat(i) (slope) and 1 / kcat(i) (y-intercept) plots, utilizing the LINEST function of Excel (Microsoft).
Figure 1.
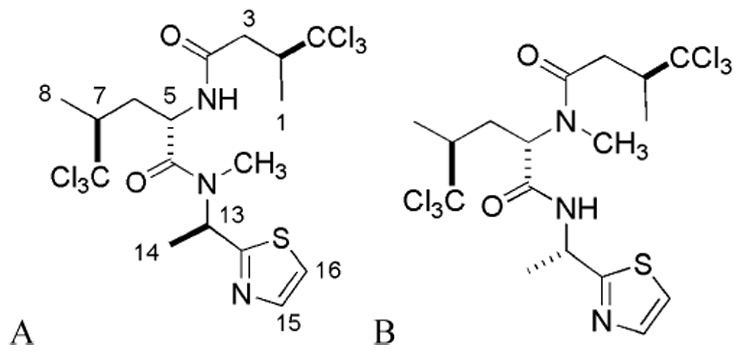
Structures of the UCSC library natural products, neodysidenin (A) and dysidenin (B)
Acknowledgment
The research was supported by grants NIH-GM56062-6 (TRH), American Cancer Society-RPG-00-219-01-CDD (TRH), NIH-CA47135 (PC) and NIH/NIGMS-R25GM51765 (PC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Solomon EI, Zhou J, Neese F, Pavel EG. Chem. Biol. 1997;4:795. doi: 10.1016/s1074-5521(97)90113-7. [DOI] [PubMed] [Google Scholar]
- 2.Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Science. 1987;237:1171. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 3.Ford-Hutchinson AW, Gresser M, Young RN. Annu. Rev. Biochem. 1994;63:383. doi: 10.1146/annurev.bi.63.070194.002123. [DOI] [PubMed] [Google Scholar]
- 4.Hussain H, Shornick LP, Shannon VR, Wilson JD, Funk CD, Pentland AP, Holtzman MJ. Am. J. Physiol. Cell Physiol. 1994;266:C243. doi: 10.1152/ajpcell.1994.266.1.C243. [DOI] [PubMed] [Google Scholar]
- 5.Kamitani H, Geller M, Eling TJ. Biol. Chem. 1998;273:21569. doi: 10.1074/jbc.273.34.21569. [DOI] [PubMed] [Google Scholar]
- 6.Kelavkar UP, Cohen C, Kamitani H, Eling TE, Badr KF. Carcinogenesis. 2000;21:1777. doi: 10.1093/carcin/21.10.1777. [DOI] [PubMed] [Google Scholar]
- 7.Ding XZ, Iversen P, Cluck MW, Knezetic JA, Adrian TE. Biochem. Biophys. Res. Commun. 1999;261:218. doi: 10.1006/bbrc.1999.1012. [DOI] [PubMed] [Google Scholar]
- 8.Connolly JM, Rose DP. Cancer Lett. 1998;132:107. doi: 10.1016/s0304-3835(98)00171-2. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan R, Nadler J. Front. Biosci. 1998;3:E81. doi: 10.2741/a369. [DOI] [PubMed] [Google Scholar]
- 10.Nie D, Hillman GG, Geddes T, Tang K, Pierson C, Grignon DJ, Honn KV. Cancer Res. 1998;58:4047. [PubMed] [Google Scholar]
- 11.Shappell SB, Manning S, Boeglin WE, Guan YF, Roberts RL, Davis L, Olson SJ, Jack GS, Coffey CS, Wheeler TM, Breyer MD, Brash AR. Neoplasia. 2001;3:287. doi: 10.1038/sj.neo.7900166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang S, Bhatia B, Maldonado CJ, Yang P, Newman RA, Liu J, Chandra D, Traag J, Klein RD, Fischer SM, Chopra D, Shen J, Zhau HE, Chung LW, Tang DG. J. Biol. Chem. 2002;277:16189. doi: 10.1074/jbc.M111936200. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto S. Biochim. Biophys. Acta. 1992;1128:117. doi: 10.1016/0005-2760(92)90297-9. [DOI] [PubMed] [Google Scholar]
- 14.Funk CD, Keeney DS, Oliw EH, Boeglin WE, Brash AR. J. Biol. Chem. 1996;271:23338. doi: 10.1074/jbc.271.38.23338. [DOI] [PubMed] [Google Scholar]
- 15.Kinzig A, Furstenberger G, Burger F, Vogel S, Muller-Decker K, Mincheva A, Lichter P, Marks F, Krieg P. FEBS Lett. 1997;402:162. doi: 10.1016/s0014-5793(96)01517-7. [DOI] [PubMed] [Google Scholar]
- 16.Tang K, Honn KV. Adv. Exp. Med. Biol. 1999;447:181. doi: 10.1007/978-1-4615-4861-4_17. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto S, Suzuki H, Ueda N. Prog. Lipid Res. 1997;36:23. doi: 10.1016/s0163-7827(97)00002-7. [DOI] [PubMed] [Google Scholar]
- 18.Honn KV, Tang DG, Gao X, Butovich IA, Liu B, Timar J, Hagmann W. Cancer Metastasis Rev. 1994;13:365. doi: 10.1007/BF00666105. [DOI] [PubMed] [Google Scholar]
- 19.Nie D, Tang K, Diglio C, Honn KV. Blood. 2000;95:2304. [PubMed] [Google Scholar]
- 20.Timar J, Silletti S, Bazaz R, Raz A, Honn KV. Int. J. Cancer. 1993;55:1003. doi: 10.1002/ijc.2910550621. [DOI] [PubMed] [Google Scholar]
- 21.Chen YQ, Duniec ZM, Liu B, Hagmann W, Gao X, Shimoji K, Marnett LJ, Johnson CR, Honn KV. Cancer Res. 1994;54:1574. [PubMed] [Google Scholar]
- 22.Gao X, Grignon DJ, Chbihi T, Zacharek A, Chen YQ, Sakr W, Porter AT, Crissman JD, Pontes JE, Powell IJ, et al. Urology. 1995;46:227. doi: 10.1016/s0090-4295(99)80198-8. [DOI] [PubMed] [Google Scholar]
- 23.Tang DG, Chen YQ, Honn KV. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5241. doi: 10.1073/pnas.93.11.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steele VE, Holmes CA, Hawk ET, Kopelovich L, Lubet RA, Crowell JA, Sigman CC, Kelloff GJ. Cancer Epidemiol. Biomark. Prev. 1999;8:467. [PubMed] [Google Scholar]
- 25.Yamamoto S, Katsukawa M, Nakano A, Hiraki E, Nishimura K, Jisaka M, Yokota K, Ueda N. Biochem. Biophys. Res. Commun. 2005;338:122. doi: 10.1016/j.bbrc.2005.08.214. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura S, Iida T, Shirahata K, Kase H. J. Antibiot. 1986;39:589. doi: 10.7164/antibiotics.39.589. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki H, Ueda T, Juranek I, Yamamoto S, Katoh T, Node M, Suzuki T. Biochem. Biophys. Res. Commun. 2000;275:885. doi: 10.1006/bbrc.2000.3390. [DOI] [PubMed] [Google Scholar]
- 28.Deschamps JD, Kenyon VA, Holman TR. Bioorg. Med. Chem. 2006;14:4295. doi: 10.1016/j.bmc.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 29.Amagata T, Whitman S, Johnson TA, Stessman CC, Loo CP, Lobkovsky E, Clardy J, Crews P, Holman TR. J. Nat. Prod. 2003;66:230. doi: 10.1021/np020462l. [DOI] [PubMed] [Google Scholar]
- 30.Cichewicz RH, Kenyon VA, Whitman S, Morales NM, Arguello JF, Holman TR, Crews PJ. Am. Chem. Soc. 2004;126:14910. doi: 10.1021/ja046082z. [DOI] [PubMed] [Google Scholar]
- 31.Carroll J, Jonsson EN, Ebel R, Hartman MS, Holman TR, Crews P. J Org. Chem. 2001;66:6847. doi: 10.1021/jo015784t. [DOI] [PubMed] [Google Scholar]
- 32.Cichewicz RH, Clifford LJ, Lassen PR, Cao X, Freedman TB, Nafie LA, Deschamps JD, Kenyon VA, Flanary JR, Holman TR, Crews P. Bioorg. Med. Chem. 2005;13:5600. doi: 10.1016/j.bmc.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 33.Gautschi JT, Whitman S, Holman TR, Crews P. J. Nat. Prod. 2004;67:1256. doi: 10.1021/np0340495. [DOI] [PubMed] [Google Scholar]
- 34.Waslidge NB, Hayes DJ. Anal. Biochem. 1995;231:354. doi: 10.1006/abio.1995.0063. [DOI] [PubMed] [Google Scholar]
- 35.Gay C, Collins J, Gebicki JM. Anal. Biochem. 1999;273:149. doi: 10.1006/abio.1999.4208. [DOI] [PubMed] [Google Scholar]
- 36.Magos L, Clarkson TW. Ann. Clin. Biochem. 2006;43:257. doi: 10.1258/000456306777695654. [DOI] [PubMed] [Google Scholar]
- 37.Segal IH. Enzyme Kinetics: Behavior and Analysis of Equilibrium and Steady-State Enzyme Systems. New York, N. Y.: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 38.Segraves EN, Holman TR. Biochemistry. 2003;42:5236. doi: 10.1021/bi0273462. [DOI] [PubMed] [Google Scholar]
- 39.Mogul R, Johansen E, Holman TR. Biochemistry. 2000;39:4801. doi: 10.1021/bi992805t. [DOI] [PubMed] [Google Scholar]
- 40.Gupta BL, Gomathy KR. Int. J. Appl. Radiat. Isot. 1974;25:509. doi: 10.1016/0020-708x(74)90077-5. [DOI] [PubMed] [Google Scholar]
- 41.Jiang ZY, Hunt JV, Wolff SP. Anal. Biochem. 1992;202:384. doi: 10.1016/0003-2697(92)90122-n. [DOI] [PubMed] [Google Scholar]
- 42.Gay C, Gebicki JM. Anal. Biochem. 2000;284:217. doi: 10.1006/abio.2000.4696. [DOI] [PubMed] [Google Scholar]
- 43.Bringmann G, Zagst R, Schaffer M, Hallock Y, Cardellina J, Boyd M. Angew. Chem. Int. Ed. Engl. 1993;32:1190. [Google Scholar]
- 44.White EL, Chao WR, Ross LJ, Borhani DW, Hobbs PD, Upender V, Dawson MI. Arch. Biochem. Biophys. 1999;365:25. doi: 10.1006/abbi.1999.1145. [DOI] [PubMed] [Google Scholar]
- 45.Nelson MJ, Brennan BA, Chase DB, Cowling RA, Grove GN, Scarrow RC. Biochemistry. 1995;34:15219. doi: 10.1021/bi00046a030. [DOI] [PubMed] [Google Scholar]
- 46.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Deliv. Rev. 2001;46:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 47.Li C, Xu L, Wolan DW, Wilson IA, Olson AJ. J. Med. Chem. 2004;47:6681. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- 48.DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. Mol. Cancer Ther. 2004;3:1079. [PubMed] [Google Scholar]
- 49.Liu F, You QD, Chen YD. Bioorg. Med. Chem. Lett. 2007;17:722. doi: 10.1016/j.bmcl.2006.10.083. [DOI] [PubMed] [Google Scholar]
- 50.Mahabusarakam W, Wiriyachitra P, Taylor WC. J. Nat. Prod. 1987;50:474. [Google Scholar]
- 51.Iikubo K, Ishikawa Y, Ando N, Umezawa K, Nishiyama S. Tet. Let. 2002;43:291. [Google Scholar]
- 52.Okudaira C, Ikeda Y, Kondo S, Furuya S, Hirabayashi Y, Koyano T, Saito Y, Umezawa K. J. Enzyme Inhib. 2000;15:129. doi: 10.1080/14756360009030346. [DOI] [PubMed] [Google Scholar]
- 53.Nakatani K, Nakahata N, Arakawa T, Yasuda H, Ohizumi Y. Biochem. Pharmacol. 2002;63:73. doi: 10.1016/s0006-2952(01)00810-3. [DOI] [PubMed] [Google Scholar]
- 54.Sato A, Fujiwara H, Oku H, Ishiguro K, Ohizumi Y. J Pharmacol. Sci. 2004;95:33. doi: 10.1254/jphs.95.33. [DOI] [PubMed] [Google Scholar]
- 55.Leung HW, Yang WH, Lai MY, Lin CJ, Lee HZ. Food Chem. Toxicol. 2007;45:403. doi: 10.1016/j.fct.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Van Sande J, Deneubourg F, Beauwens R, Braekman JC, Daloze D, Dumont JE. Mol. Pharmacol. 1990;37:583. [PubMed] [Google Scholar]
- 57.Mogul R, Johansen E, Holman TR. Biochemistry. 2000;39:4801. doi: 10.1021/bi992805t. [DOI] [PubMed] [Google Scholar]
- 58.MacMillan JB, Trousdale EK, Molinski TF. Org. Lett. 2000;2:2721. doi: 10.1021/ol006326u. [DOI] [PubMed] [Google Scholar]
- 59.van Soest RWM, Hooper JNA. In: van Soest RWM, Hooper JNA, editors. Vol. 1. New York: Kluwer Academic/Plenum Publishers; 2002. p. 1061. [Google Scholar]
- 60.Clark WD, Crews P. Tetrahedron Lett. 1995;36:1185. [Google Scholar]
- 61.Horton P, Inmsn WD, Crews P. J. Nat. Prod. 1990;53:143. [Google Scholar]
- 62.Jimenez C, Crews P. Tetrahedron. 1991;47:2097. [Google Scholar]


