Abstract
Mitogen-activated protein (MAP)§ kinase cascades are signal transduction pathways that play pivotal regulatory roles in the biosynthesis of proinflammatory cytokines. MAP kinase phosphatase (MKP)-1, an archetypal member of the MKP family, is essential for the dephosphorylation/deactivation of MAP kinases p38 and JNK. Earlier studies conducted using cultured immortalized macrophages provided compelling evidence indicating that MKP-1 deactivates p38 and JNK, thereby limiting pro-inflammatory cytokine biosynthesis in innate immune cells exposed to microbial components. Recent studies employing MKP-1 knockout mice have confirmed the central function of MKP-1 in the feedback control of p38 and JNK activity as well as the crucial physiological function of MKP-1 as a negative regulator of the synthesis of pro-inflammatory cytokines in vivo. MKP-1 was shown to be a major feedback regulator of the innate immune response and to play a critical role in preventing septic shock and multi-organ dysfunction during pathogenic infection. In this review, we will update the studies on the biochemical properties and the regulation of MKP-1, and summarize our understanding on the physiological function of this key phosphatase in the innate immune response.
Keywords: negative regulator, feedback control, dephosphorylation, innate immunity, inflammation, cytokine, infection, MAP kinase
1. Introduction
The mammalian immune system is a highly efficient cellular and molecular network that protects the organism against pathogenic infections. It is broadly divided into two major components: the innate immunity and the adaptive immunity [1]. The innate immune system detects a wide variety of pathogens through a limited set of germ line-encoded receptors, and serves as a first line of defense against pathogenic organisms [2]. The innate immune system is comprised of a number of cell types, including dendritic cells, macrophages and monocytes, polymorphonuclear cells, natural killer cells, γδ T cells, and natural killer T cells. As these cells recognize a limited array of conserved molecules which are essential for the survival of pathogenic organisms, the innate immune system can provide immediate generic defense and prevent rapidly dividing pathogens from overwhelming the host. On the other hand, the adaptive immunity is a pathogen-specific host defense based on clonally expanded B- and T-lymphocytes generated first through random somatic recombination of immunoglobulin genes and then through positive and negative selections. These pathogen-specific B- and T-lymphocytes not only serve as effector cells for pathogen eradication, but they also function as memory cells for rapid expansion upon re-encountering these pathogens. While the adaptive immunity is highly specific and effective against evolving pathogens, the pathogen-specific immune response takes days or weeks to develop [1]. The innate and the adaptive immunity are thus very different, but they interact closely and synergistically in the host defense. The innate immune system plays a central role in the initiation of inflammatory response that is essential for the mobilization and function of the various immune effector cells. Moreover, by serving as antigen-presenting cells, the innate immune system functions as a gatekeeper for mounting an efficient adaptive immune response [3]. Reciprocally, B- and T-lymphocytes can regulate the innate immune cell functions by secreting a variety of cytokines and pathogen-specific antibodies to facilitate the anti-microbial function of innate immune cells [4].
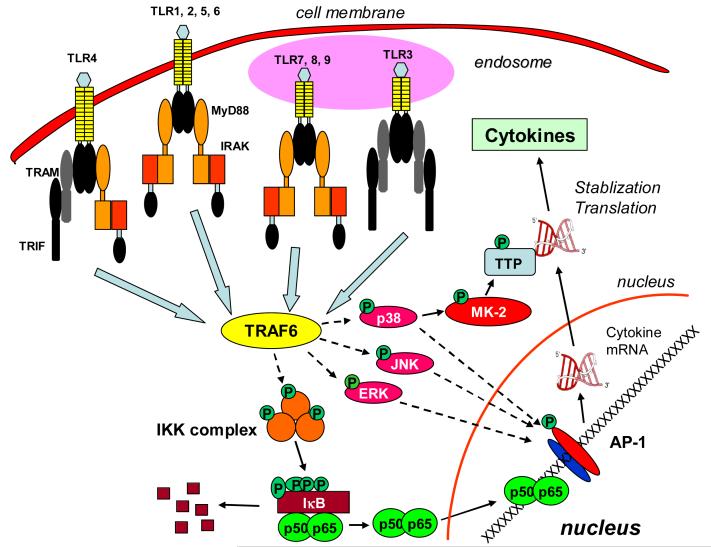
A major function of the innate immune cells during microbial infection is the production of inflammatory mediators such as cytokines, chemokines, nitric oxide, and reactive oxygen species [1]. Cytokines and chemokines are essential for the mobilization of the leukocytes to the site of infection, the initiation of the adaptive immune response, and the initiation of acute phase response; the reactive nitrogen and oxygen species are crucial for the killing of the invading pathogens. Upon microbial infection, innate immune cells such as macrophages and dendritic cells localized in the affected tissues detect the pathogen-associated molecular patterns through their specialized receptors including Toll-like receptors (TLR) and nucleotide-binding oligomerization domain-containing (NOD) proteins [5, 6]. The identification of pathogen components by the pattern recognition receptors triggers a cascade of evolutionarily conserved signal transduction events mediated by adaptor proteins, including myeloid differentiation factor 88 (MyD88) and Toll/IL-1 receptor domain-containing adapter inducing interferon-β (TRIF), which in turn activate the NF-κB and mitogen-activated protein (MAP) kinase pathways [7]. NF-κB can bind to the promoter regions of many pro-inflammatory cytokine and chemokine genes and activate their transcription. MAP kinases, including the extracellular signal-regulated kinases (ERK), c-Jun N-terminal kinases (JNK), and p38, also play a crucial role in the production of pro-inflammatory cytokines and chemokines (Fig. 1). MAP kinases can induce cytokine production by enhancing gene transcription via factors such as AP-1 and by increasing the translation and stability of cytokine and chemokine mRNAs [8-10].
Figure 1. Diagram of the signal transduction pathways initiated at TLRs by microbial components.
Binding of microbial components (ligands) to TLRs triggers conformational changes that lead to the recruitment of IRAK and TRAF-6 mediated by adaptor proteins MyD88 and TRIF. TRAF-6 can activate both the NFκB and MAP kinase pathways. NF-κB is critical for the transcription of inflammatory response genes, including genes of various cytokines and chemokines. MAP kinases, including ERK, JNK, and p38, also regulate the expression of many inflammatory genes. MAP kinases can activate AP-1 transcription factor, thus enhancing gene transcription. MAP kinases, p38 in particular, also enhance cytokine production through post-transcriptional mechanisms. p38 phosphorylates/activates MK-2, which in turn phosphorylates TTP, leading to both enhanced cytokine mRNA stability and accelerated cytokine mRNA translation.
While activation of the signal transduction cascades is critical for mounting an aggressive immune response to eliminate invading pathogens, deactivation of the signaling pathways restrains the potentially devastating actions of the immunological system on the host, thus preventing self destruction. A variety of negative regulators operate at multiple steps along the signal transduction pathways downstream of TLRs. These negative regulators modulate the strength and duration of the transduced signals and control the production of inflammatory cytokines [11]; for example, TLR-4 was shown to be transiently suppressed in response to LPS [12]. In addition to the modulation at the receptor level, a number anti-inflammatory proteins are also induced, including the IL-1 receptor-associated kinase (IRAK)-M [13], the suppressor of cytokine-signaling (SOCS)-1 [14], the NF-κB inhibitor (IκB), and anti-inflammatory cytokines such as IL-10 [11]. Through these inhibitory proteins, cells not only terminate the signaling cascade at the cell surface, but also switch off downstream mediators, thereby silencing the signaling pathways and stopping the production of pro-inflammatory cytokines. In mammalian cells, MAP kinases are primarily inactivated by a group of dual-specificity protein phosphatases through dephosphorylation of the critical tyrosine and threonine residues of activated MAP kinases [15]. In this capacity, this group of protein phosphatases may serve as pivotal feedback control regulators in the innate immune response during microbial infection and thus, play a significant role in the resolution of sepsis pathobiology. Our laboratory has explored the role of MAP kinase phosphatase (MKP)-1 in the negative control of cytokine expression in innate immune cells. Our studies indicated that MKP-1 functions as a feedback control mechanism that governs the production of pro-inflammatory cytokines by limiting the activation of MAP kinases [16-18]. More recently, several laboratories including ours have employed MKP-1 knockout mice to obtain unequivocal support for a central role of MKP-1 in the restraint of innate immune response and the prevention of septic shock syndrome during pathogenic microbial infection [19-22]. A number of excellent review articles have been published on the function of various MKP family members in the immune response [23-26]. Here, we will briefly summarize our understanding on the biochemical properties and the regulation of MKP-1, and focus on the role of MKP-1 in controlling cytokine expression during microbial infection. We will discuss the influence of MKP-1 in the mode of actions of key immunomodulatory agents.
2. MAP Kinases and their Functions in Regulating Cytokine Production
MAP kinases are a group of highly conserved serine/threonine protein kinases in eukaryotes [27]. They play pivotal roles in a variety of cellular processes including proliferation, differentiation, stress response, apoptosis, and host immune defense. In innate immune cells, MAP kinases are critical for the syntheses of numerous cytokines, chemokines, and other inflammatory mediators that mobilize the immune system to combat pathogenic infections [28]. In adaptive immune cells, MAP kinases also serve as critical regulators in the clonal expansion of effector T- and B-lymphocytes through modulation of cytokine production, cell proliferation, and survival [28].
There are three well-defined MAP kinase subfamilies: ERK, JNK, and p38 [29]. The MAP kinase pathway is activated through a cascade of sequential phosphorylation events, beginning with the activation of MAP kinase kinase kinase. MAP kinase kinase kinase activates MAP kinase kinase by phosphorylating two serine residues. MAP kinase kinase, in turn, interacts with and phosphorylates MAP kinase at the adjacent threonine and tyrosine residues in the conserved TXY motif located in a regulatory loop between the kinase subdomains VII and VIII. Activated MAP kinases can phosphorylate a wide array of downstream targets, including protein kinases and other enzymes. In addition, activated MAP kinases can translocate to the nucleus and phosphorylate proteins that control chromatin structure as well as numerous transcription factors such as AP-1, thereby influencing the transcription of MAP kinase-regulated genes [29]. In addition to regulating gene transcription, MAP kinases can also modulate protein expression by altering the stability, transport, and translation of mRNA species that contain AU-rich elements (Ares) [28]. It was demonstrated that tristetraprolin (TTP) bound to the AREs present in the 3′-untranslated regions of several cytokine mRNAs and promoted the deadenylation and destabilization of the mRNA [30]. By phosphorylating the MAP kinase-activated protein kinase (MK)-2, p38 MAP kinase can enhance the activity of MK-2 [31], which in turn phosphorylates and thereby inhibits the TTP-mediated degradation of ARE-containing mRNAs (Fig. 1). Many pro-inflammatory cytokine transcripts, including TNF-α, IL-1β, IL-6, granulocyte macrophage colony stimulating factor (GM-CSF), and IL-2, contain AREs in their 3′-untranslated regions and have been shown to be targets of TTP-mediated mRNA decay [32].
3. Structure, Function, and Regulation of MAP Kinase Phosphatase (MKP)-1
3.1. Overview of MAP Kinase Phosphatase Family
Since MAP kinase pathways are activated through phosphorylation, dephosphorylation of MAP kinases mediated by phosphatases represents a highly efficient mode of kinase deactivation. A number of protein phosphatases are known to deactivate MAP kinases, including tyrosine, serine/threonine, and dual-specificity phosphatases [15]. In mammalian cells, the dual-specificity protein phosphatases are the primary phosphatases responsible for dephosphorylation/deactivation of MAP kinases [15]. Therefore, these phosphatases are often referred to as MAP kinase phosphatases (MKPs). To date, at least 10 MKPs have been identified in mammalian cells, with MKP-1 being the archetype [15]. These MKPs exhibit distinct biochemical properties with regard to their substrate specificity, subcellular localization, and regulation (see review [15]). They are presumed to act cooperatively to modulate the MAP kinase pathways and to orchestrate appropriate cellular responses. To determine the importance of MKPs in modulating MAP kinase pathways, Bhalla et al. have studied the dynamic response of MAP kinases using a combination of computational and experimental approaches. Their findings demonstrated that MKPs vitally regulate the dynamic properties of MAP kinase activation. Their studies also indicate that at low concentrations of MKP, MAP kinase exhibits a switch-like (or bistable) behavior, such that a brief mitogenic stimulus results in sustained MAP kinase activation. At high concentrations of MKP, MAP kinase responds proportionately over a range of stimulus strengths (or monostable state) [33].
3.2. Biochemical Properties of MKP-1
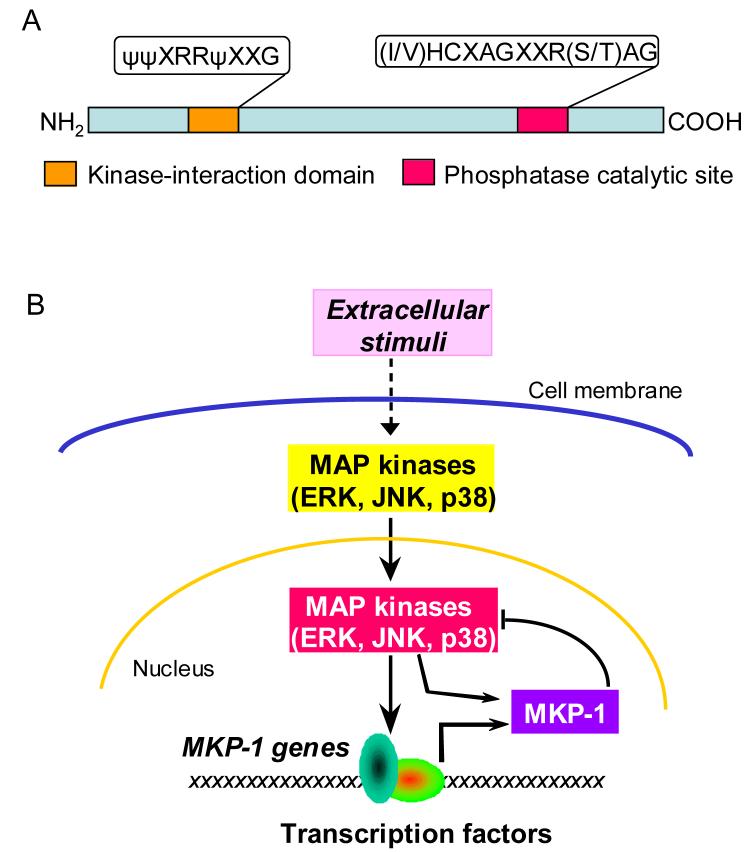
The mouse MKP-1 cDNA was initially cloned through differential hybridization screening of a BALB/c 3T3 cDNA library [34]. The cDNA clone, initially referred to as 3CH134, was identified as corresponding to an immediate-early transcript induced by growth factors [35]. The human homolog of 3CH134 (CL100) was identified as a tyrosine phosphatase gene induced by oxidative stress shortly after [36]. Both 3CH134 and CL100 encode a predominantly nuclear protein with a molecular mass of 40 kDa, which contains a (I/V)HCXAGXXR(S/T)AG signature motif characteristic of the catalytic domain of tyrosine phosphatases (Fig. 2). They also share considerable homology with the dual-specificity phosphatase of vaccinia virus, VH1, particularly within the catalytic site. 3CH134 and CL100 exhibited highly specific phosphatase activity towards the ERK MAP kinase, both in vitro and in cultured cells [37-40]. As this was the first MAP kinase-selective protein phosphatase discovered, capable of dephosphorylating both their phosphotyrosine and phosphothreonine residues, it was designated as MAP kinase phosphatase (MKP)-1 [40]. Since MKP-1 deactivates MAP kinases and is robustly induced by mitogenic stimulation that also activates MAP kinases, MKP-1 is regarded as an important feedback control mechanism that regulates the MAP kinases (Fig. 2). While MKP-1 was initially thought to be a phosphatase specific for the ERK MAP kinases, subsequent studies demonstrated that MKP-1 also efficiently inactivated JNK and p38, MAP kinase subfamilies preferentially activated by stress [41, 42]. The fact that MKP-1 was also robustly induced by genotoxic stress such as UV irradiation and methyl methanesulfonate that potently activated JNK and p38 but had little effect on ERK, suggested that MKP-1 might likewise play an important role in the feedback control of MAP kinase signaling following exposure to stress stimuli [41]. To address the substrate preference of MKP-1, Franklin and Kraft established a U937 cell line that conditionally expressed MKP-1. By titrating the levels of MKP-1 expression, Franklin and Kraft found that p38 and JNK were much more sensitive to inhibition by MKP-1 than ERK [43]. Several recent studies conducted using MKP-1 knockout cells further support the conclusion that p38 and JNK, but not ERK, are the preferred substrates of MKP-1 [19, 20, 22]. However, these studies do not exclude the possibility that MKP-1 may also participate in the inactivation of ERK in response to certain stimuli, particularly when MKP-1 is expressed in high levels.
Figure 2. Diagram of the structure and function of MKP-1.
(A) Primary structure of MKP-1. MKP-1 has an amino terminal domain responsible for interaction with MAP kinases. The catalytic domain is located at the carboxyl terminus. (B) Feedback control of MAP kinases by MKP-1. Extracellular stimulation triggers the activation of MAP kinases. Upon activation, MAP kinases translocate to nucleus where they phosphorylate and activate transcription factors, leading to altered gene transcription. Among the genes activated by MAP kinases is MKP-1. MKP-1 protein can dephosphorylate MAP kinases, thus terminating MAP kinase-regulated gene transcription. By phosphorylating MKP-1 protein, MAP kinases can regulate the stability of MKP-1 protein.
3.3. Regulation of MKP-1
The activity of MKP-1 can be regulated at multiple levels. First, MKP-1 expression can be robustly induced by growth factors and stress [15], independently of de novo protein synthesis [34]. In response to extracellular stimulation, MKP-1 mRNA levels are usually increased by 10-100 fold within 15-60 minutes. Nuclear run-on studies have clearly demonstrated that transcriptional induction plays an important role in the induction of MKP-1 in response to extracellular stimuli. Although we have known for over two decades that MKP-1 is highly induced by a variety of extracellular stimuli [34], the mechanisms underlying its transcriptional activation remain poorly understood. This lack of progress in delineating the transcriptional regulation of MKP-1 gene is largely due to the fact that MKP-1 promoter is constitutively active in transient transfection reporter assays [44, 45]. It appears that transcriptional induction of MKP-1 also involves chromatin remodeling at the MKP-1 gene which are not properly recapitulated in the transcriptional reporter analyses [45].
In addition to the transcriptional mechanism, MKP-1 expression may also subject to posttranscriptional regulation. Like many other immediate-early genes known to be regulated post-transcriptionally, such as c-jun and c-fos [46, 47], the mRNA of MKP-1 contains several evolutionally conserved putative AREs in the 3′ untranslated region [35, 48]. It has been well-established that mRNA species containing AREs are highly labile, and these transcripts can be stabilized through a complex process involving a number of RNA-binding proteins, including TTP and HuR [49, 50]. However, earlier studies using a human cervical cell line SiHa did not detect a significant change in mRNA half-life upon a number of extracellular stimulation [51], suggesting that post-transcriptional mechanisms might not play a significant role in the induction of MKP-1 mRNA expression. As with most studies, observations made in a specific system may not be generalized to apply to other system. Very recently, Gorospe and colleagues found that in response to hydrogen peroxide, MKP-1 mRNA undergoes a dramatic increase in half-life, changing from ∼50 min in unstimulated HeLa cells to ∼ 6 h (Myriam Gorospe, personal communication). Interestingly, they also found that a number of RNA-biding proteins, including HuR and NF90, can interact with the AU-rich motif of MKP-1 mRNA in hydrogen peroxide-treated HeLa cells. Such findings are like to contribute considerably to the understanding of the regulation of MKP-1 expression.
At the post-translational level, MKP-1 activity can be modulated through stabilization of the protein and catalytic activation. The MKP-1 protein can be phosphorylated by ERK both in vivo and in vitro at two serine sites at the carboxyl terminal and potentially also at a threonine site in the middle of the protein [52]. Notably, the carboxyl terminus of MKP-1 also contains a Phe-Asn-Phe-Pro (FNFP) sequence motif. The FXFP motif has been shown to serve as a docking site through which ERK binds many of its substrates [53, 53]. Interestingly, phosphorylation of MKP-1 by ERK substantially increases the half-life of MKP-1 protein while it has no effect on its intrinsic phosphatase activity. MKP-1 protein is degraded by the ubiquitin-proteasome system [52], but the ubiquitin-mediated degradation is inhibited through phosphorylation of MKP-1 by ERK [52]. In addition to increasing protein stability, the interaction of MKP-1 with its substrate MAP kinases also modulates the catalytic activity of MKP-1 [54, 55]. We and others have shown that the binding of substrate MAP kinases, including ERK, JNK, and p38, with recombinant MKP-1 protein results in a 6-to 8-fold increase in MKP-1 activity [54, 55]. The interaction between MAP kinases and MKPs is dependent on a kinase-interaction domain at the amino terminus of the phosphatases and an acidic domain at the carboxyl terminus of the kinases [54]. The kinase-interaction domain of MKP-1 contains the consensus sequence ΨΨXRRΨXXG (where Ψ represents a hydrophobic residue and X is any amino acid), flanked by two Cdc25-homology domains [15]. Analysis of the crystal structure of a functionally related phosphatase, MKP-3, has suggested that binding of catalytically activatable MKP with its substrate MAP kinases may enable the phosphatase to adopt a more efficient conformational configuration at the catalytic site [56]. Stewart et al. have shown that the critical conserved aspartic acid in the MKP-3 catalytic domain is nearly 10 Å away from the nucleophilic cysteine and arginine at the catalytic site [56], suggesting that substrate binding-induced catalytic activation of MKP-3 is due to the movement of the aspartic acid residue closer to the catalytic site. Movement of the aspartate residue toward the active site enables the aspartate to serve as a general acid to donate a proton to the tyrosine or threonine leaving group oxygen, thus facilitating dephosphorylation [56]. The strong similarity with MKP-3 and the catalytic activation of MKP-1 upon substrate binding suggest that MKP-1 likely undergoes a similar conformational change upon encountering its substrate MAP kinases.
4. Role of MKP-1 in the Regulation of Microbial-Induced Host Inflammatory Response
4.1 Regulation of Cytokine Biosynthesis in Innate Immune Cells by MKP-1
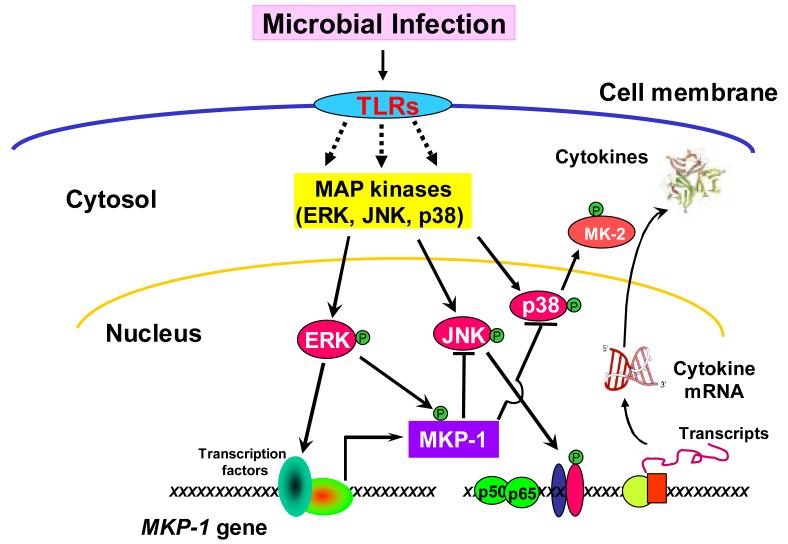
Given the critical role of MAP kinases in the regulation of innate immune responses, MKP-1 was widely expected to play an important role in innate immune regulation. Immortalized murine macrophages were reported to undergo a robust MKP-1 induction following exposure to Listeria monocytogenes, a Gram-positive bacterium [57]. Overexpression of MKP-1 in immortalized macrophages significantly attenuated the phagocytosis of L. monocytogenes, suggesting that MKP-1 negatively regulated innate immune function [58]. Using bone marrow-derived murine macrophages, Celadar and colleagues demonstrated that MKP-1 was highly induced by LPS through a transcriptional mechanism mediated by protein kinase Cε and a tyrosine kinase(s) [59]. They found that the induction of MKP-1 coincided with ERK inactivation, raising the possibility that MKP-1 might be responsible for the deactivation of ERK in this system [59]. To understand the regulation of cytokine expression in innate immune cells during bacterial infection, we studied the role of MKP-1 using RAW 264.7 macrophages as a model system. We found that RAW264.7 cell stimulation with LPS resulted in a robust but transient activation of JNK and p38 [16]. The activities of these MAP kinases peaked within 15 min, and their activities returned to nearly basal levels within 60 min. The kinetics of p38 and JNK deactivation correlated closely with the accumulation of MKP-1 protein. Unlike JNK and p38, ERK was potently activated in response to LPS stimulation and remained unchanged despite the accumulation of MKP-1. Similarly, stimulation of RAW264.7 cells with peptidoglycan, another bacterial cell wall component, also elicited a transient activation of JNK and p38 that promptly subsided as MKP-1 levels increased [17]. The importance of MKP-1 in the inactivation of p38 and JNK was further demonstrated by pharmacological inhibition of MKP-1 expression by triptolide, a diterpenoid triepoxide. Triptolide blocked MKP-1 induction in LPS-stimulated macrophages and prolonged p38 and JNK activation, but had little effect on ERK activity [16, 17]. These results underscore the central role of MKP-1 in the deactivation of p38 and JNK in these cells. In contrast, modest levels of ectopic MKP-1 expression in RAW264.7 cells shortened the window of p38 and JNK activation in LPS-stimulated cells, and markedly attenuated the production of both TNF-α and IL-6 [16-18]. These studies established the concept that MKP-1 is an important negative regulator of the innate immune response. The very low basal level of MKP-1 in quiescent innate immune cells allows for a window of robust inflammatory response for cytokine production. Yet the rapid induction of MKP-1 allows the system to tune down the inflammatory response, thus achieving homeostasis. Thus, by limiting the strength and duration of p38 and JNK activation, MKP-1 determines the output of inflammatory cytokines. In this sense, MKP-1 serves as a servocontrol mechanism to prevent the overreaction of the innate immune system (Fig. 3).
Figure 3. Restraint of pro-inflammatory cytokine biosynthesis by MKP-1.
In response to microbial infection, TLRs initiate a series of signal transduction pathways, including NF-κB and MAP kinase cascades, leading to production of pro-inflammatory cytokines. Simultaneously, the signals initiated at the TLRs also induce MKP-1 gene transcription. ERK regulates MKP-1 expression by two mechanisms: by enhanceing MKP-1 gene transcription and by phosphorylating MKP-1 and thereby increasing its half-life. The MKP-1 protein in turn dephosphorylates JNK and p38, thus stopping the perpetuation of the inflammatory cascades and terminating cytokine production.
To define the physiological function of MKP-1 in microbial infection, several laboratories, including ours, have studied the effects of MKP-1 knockout mice on host immune responses [18-22]. Compared to primary macrophages isolated from wild-type mice, macrophages originated from MKP-1 knockout mice exhibited prolonged p38 and JNK activation after stimulation with microbial components, while the kinetics of ERK activation was not altered by MKP-1 deficiency. These results confirmed the observation made in immortalized macrophages, and firmly established MKP-1 as a primary phosphatase for p38 and JNK in innate immune cells. Compared to wild-type macrophages, macrophages isolated from MKP-1-deficient mice produced substantially greater quantities of pro-inflammatory cytokines, including TNF-α and IL-6. The profound exaggeration in the host inflammatory responses was evidenced in the considerably higher synthesis of chemokines, including macrophage inflammatory protein (MIP)-1α, MIP-1β, and MIP-2, by MKP-1 knockout macrophages than by the wild-type counterparts [19-22]. It is important to note that MKP-1 deficiency not only substantially altered the dynamics of production of pro-inflammatory cytokines and chemokines, but it also enhanced greatly the production of a powerful anti-inflammatory cytokine, IL-10, in LPS-stimulated macrophages, splenocytes, and bone marrow-derived dendritic cells [19, 22]. The augmented inflammatory responses in MKP-1-deficient innate immune cells are not restricted to the responses to LPS, but are seen in cells exposed to other microbial components, including ligands for TLR2, TLR3, TLR5, TLR7 and TLR9 [17, 22]. However, the expression of two classic TH-1 cytokines, IL-12 and interferon (IFN)-γ, was decreased in MKP-1-deficient splenocytes and dendritic cells [19], suggesting a shift in cytokine production profiles. To understand the function of MKP-1 in the regulation of inflammation, Hammer et al. challenged wild-type and MKP-1 knockout mice with LPS, and compared the gene expression profiles between the spleens of these two strains of mice using microarrays [20]. This analysis demonstrated that approximately 160 transcripts exhibited >2-fold increased levels in the wild-type mice, while some 480 transcripts exhibited >2-fold increased levels in MKP-1 knockout mice.
4.2. In vivo Function of MKP-1 in the Inflammatory Response to Bacterial Infection
Consistent with the finding that MKP-1 deficiency leads to hyper-responsiveness to LPS stimulation in innate immune cells, MKP-1-deficient mice also produced substantially greater amounts of TNF-α, IL-1β, monocyte chemoattractant protein (MCP)-1/CCL2, GM-CSF, IL-6, IL-10, and IL-12p70, than did wild-type mice upon LPS challenge [19-22]. The excessive production of the pro-inflammatory cytokines and chemokines was associated with a marked increase in physiological sensitivity to endotoxemia. Compared to wild-type mice, MKP-1-deficient mice more readily succumbed to LPS challenge, as indicated by the increased incidence and severity of multi-organ failure and the higher mortality [19-22]. Histological analyses of the lungs of the LPS-challenged MKP-1 knockout mice revealed severe lung edema associated with massive neutrophil infiltration [19]. Reflecting the hepatic damage in the LPS-challenged MKP-1 knockout mice, higher blood alanine aminotransferase activities were observed in these mice [19]. Histological analysis of the livers revealed a marked increase in leukocyte infiltration in the vicinity of the bile ducts in LPS-challenged MKP-1 knockout mice, but not in similarly treated wild-type mice. Moreover, glycogen contents were lower in the livers of LPS-challenged MKP-1 knockout mice than in similarly treated wild-type mice, likely reflecting the dramatic decrease in food intake due to severe distress by these mice [60]. Kidney function was also impaired in the MKP-1 knockout mice challenged with LPS, whereas similarly treated wild-type mice exhibited normal kidney function [19]. Severe hypotension, clinically characteristic of sepsis and implicated in the development of shock and multi-organ dysfunction syndrome [61], was also markedly different between the two strains of mice after LPS challenge [19]. While LPS challenge at a dose of 1.5 mg/kg body weight did not significantly change the systemic blood pressure in wild-type mice, the same challenge caused a substantial and long-lasting decrease in systemic blood pressure in MKP-1 knockout mice. Underlying the severe decrease in blood pressure in MKP-1 knockout mice, circulating NO levels were markedly elevated in these mice [19]. Analysis of the lungs and livers of the mice have indicated that iNOS expression levels were substantially increased in LPS-challenged MKP-1 knockout mice relative to similarly treated wild-type mice (Zhao and Liu, unpublished observations). Additionally, in response to components of Staphylococcus aureus (a Gram-positive bacteria), MKP-1 knockout mice also suffered more severe inflammatory injuries to a number of vital organs compared to wild-type mice [60]. Massive neutrophil infiltration, pulmonary edema, liver injury, and kidney dysfunction were also observed in the treated MKP-1 knockout mice [60]. Consequently, MKP-1 knockout mice exhibited significantly elevated mortality relative to similarly treated wild-type mice [60]. These observations lend strong support to the idea that MKP-1 functions as a critical negative regulator during both Gram-negative and Gram-positive bacterial infection. By limiting the strength and duration of the inflammatory signals, MKP-1 serves to constrain the host inflammatory responses and prevents septic shock (Fig. 3).
Paradoxically, MKP-1-deficient mice also produced substantially more IL-10 than did wild-type mice after LPS challenge. Unlike wild-type mice, which exhibited a transient increase in serum IL-10 levels that quickly returned to basal levels within 2-3 h after LPS administration, MKP-1-deficient mice displayed a robust and long-lasting increase in serum IL-10 levels [19, 20]. This observation suggests that like pro-inflammatory cytokines such as TNF-α, the anti-inflammatory cytokine IL-10 is also controlled in a negative manner, either directly or indirectly, by MKP-1-mediated pathways. Interestingly, either prophylactic administration of IL-10 or administration of IL-10 shortly after LPS challenge can prevent the lethal endotoxic shock syndrome [62, 63]. The fact that a substantial increase in serum IL-10 levels in MKP-1-deficient mice did not prevent endotoxic shock and mortality raises several possibilities. First, it is possible that IL-10 may not be produced early enough, or in sufficient amounts, to suppress the devastating effects of the excessive pro-inflammatory cytokines [62, 63]. It is worth noting that MKP-1 knockout mice exhibit a more robust inflammatory reaction, thus the amount of IL-10 sufficient for protecting wild-type mice from endotoxic shock might not be enough for the prevention of endotoxic shock in MKP-1 knockout mice. Alternatively, the anti-inflammatory function of IL-10 may actually depend on a functional MKP-1 gene. Supporting the latter possibility, Hammer et al. have found that IL-10 boosts MKP-1 expression in LPS-stimulated macrophages, leading to an accelerated and long-lasting p38 deactivation [64]. In the absence of MKP-1, the inhibitory function of IL-10 on the host inflammatory responses may be substantially compromised and thus unable to prevent the onset of the sprawling cytokine storms responsible for endotoxic shock. Further analysis of the effects of MKP-1 knockout on the function of IL-10 on immune effector cells may provide insightful information on the importance of MKP-1 on the function of IL-10.
4.3. Regulation of MKP-1 during the Innate Immune Response
The expression of MKP-1 is primarily regulated through a transcriptional mechanism. However, the mechanism MKP-1 mRNA can be detected within minutes after exposure of macrophages to bacterial components, with maximal mRNA levels > 100-fold above basal levels reached within 1 hour after exposure [16]. In macrophages, MKP-1 can be induced by a variety of TLR ligands, including LPS, peptidoglycan, lipoteichoic acid, CpG DNA, Pam3CSK4, and poly (I-C), as well as mitogenic factor M-CSF [16, 17, 22, 65]. To determine the mediators involved in the induction of MKP-1, Flavell and colleagues examined the expression of MKP-1 in primary macrophages derived from knockout mice lacking MyD88 or TRIF [22]. They found that the induction of MKP-1 by LPS was reduced in both the MyD88-/- and the TRIF mutant macrophages, as compared with wild-type macrophages, indicating that optimal LPS-induced MKP-1 expression requires both the MyD88 and TRIF functions. In response to ligands of TLR9 and TLR2, which only signal through MyD88, MKP-1 induction was completely ablated in MyD88-/- macrophages, but was normal in the TRIF mutant cells. Conversely, in response to poly (I-C), a TLR3 ligand that signals solely through TRIF, MKP-3 induction was abrogated by TRIF mutation, but it was unaffected by MyD88 knockout. To dissect the downstream mediators involved in this process, a number of studies utilized pharmacological inhibitors or suppression of expression approach to investigate the pathways mediating MKP-1 induction. Celada and colleagues found that both PKCε and a tyrosine kinase(s) are involved in the induction of MKP-1 in macrophages stimulated by LPS [59, 65]. They also found that PI-3 kinase, cyclic AMP- and cyclic GMP-dependent protein kinases are not involved in MKP-1 induction in response to LPS. More recently, this group observed that induction of MKP-1 in response to both M-CSF and LPS is mediated by Raf-1 [66], a well established MAP kinase kinase kinase. However, activation of Raf-1 is clearly not sufficient for the induction of MKP-1, as activation of Raf-1 in RAW264.7 macrophages expressing Raf-1:ER fusion protein by estrogen did not significantly induce MKP-1 levels [67]. Thus, it is likely that Raf-1 activation is required but not sufficient for MKP-1 induction in innate immune cells in response to TLR ligands. Furthermore, the role of ERK in the induction of MKP-1 in response to TLR ligands remains controversial. Pretreatment with the MEK1/2 inhibitor PD98059 did not appreciably alter MKP-1 induction in bone marrow-derived macrophages stimulated with LPS, although it almost completely blocked ERK activation in these cells. However, we found that U0126, a different MEK1/2 inhibitor, attenuated the induction of MKP-1 in RAW264.7 macrophages stimulated with LPS [16]. Moreover, such attenuation of MKP-1 induction correlated with lower MKP-1 protein accumulation and protection of the p38 activity in these cells. The cause for the discrepancy between these studies is unclear, but it may arise from differences in the cells used, as it has long been recognized that macrophages from distinct anatomic sites display widely diverse properties [4]. Further research efforts are clearly needed to understand the transcriptional regulation of MKP-1. In light of the recent realization that MKP-1 is a critical negative regulator of cytokine production, the interest on the transcriptional regulation of MKP-1 is likely to grow considerably.
In addition to the elevation in MKP-1 mRNA levels, MKP-1 protein became markedly more stable upon LPS stimulation, with a three-to four-fold increase in half-life [16]. The increase in MKP-1 stability in response to LPS is abolished by a pharmacological inhibitor of the ERK pathway, supporting an additional role of ERK in the regulation of MKP-1. Perhaps it is not a surprise that ERK-mediated stabilization of MKP-1 protein in response to LPS was largely abolished by deletion of 53 amino acids from the carboxyl terminus of MKP-1 [16]. This carboxyl terminal region contains a consensus ERK-docking site [53] and two serine residues phosphorylated by ERK [52]. Previously, Brondello et al. showed that phosphorylation of MKP-1 by ERK attenuates MKP-1 degradation, a process mediate by the ubiquitin-proteasome system [52]. Thus, ERK can regulate MKP-1 expression through at least two mechanisms: by transcriptional induction of the MKP-1 gene and by post-translational stabilization of the MKP-1 protein.
5. MKP-1 and Immunomodulatory Agents
The fact that MKP-1 serves as an important negative regulator of the inflammatory responses raises the interesting question of whether MKP-1 plays a significant role in the action of immunomodulatory agents. A screen of commonly used anti-inflammatory and anti-rheumatic drugs for induction of MKP-1 revealed that MKP-1 was significantly induced by dexamethasone in RAW264.7 macrophages [16]. Such an induction explains an earlier observation by Swantek et al. that JNK activation in response to LPS was inhibited by dexamethasone [68]. Studies to elucidate the mechanisms underlying the inhibitory effect of dexamethasone on cyclooxygenase (COX) 2 expression led Clark and colleague to discover that dexamethasone induced MKP-1 expression in HeLa cells, and that this induction was responsible for the inhibition of p38 and the decreased expression of COX2 [69, 70] [71]. An earlier investigation by Cato and colleagues had shown that dexamethasone induced MKP-1 in mast cells and that such an induction was responsible for the inhibitor effects of glucocorticoids on ERK activity [72]. To delineate the role of MKP-1 in the anti-inflammatory function of glucocorticoids, our laboratory compared a group of synthetic corticosteroids with different anti-inflammatory potencies with regard to their induction of MKP-1 in RAW264.7 macrophages. We found that the ability of these synthetic glucocorticoids to induce MKP-1 was proportional to their relative anti-inflammatory potencies [18]. Very recently, using bone marrow-derived macrophages originated from MKP-1 knockout mice, Clark and colleagues demonstrated that p38 and JNK activation in response to LPS stimulation was no longer inhibited by dexamethasone [73]. Accordingly, many of the inflammatory genes, including cytokines, were less sensitive to dexamethasone in MKP-1-deficient cells. We have also found that dexamethasone accelerates deactivation of p38 and JNK following LPS stimulation in wild-type but not in the MKP-1-deficient peritoneal macrophages elicited by thioglycollate. Moreover, dexamethasone was unable to prevent endotoxic shock in MKP-1 knockout mice while it effectively protected wild-type mice from endotoxin-induced mortality (Wang and Liu, unpublished observations). However, we found that cytokine production by MKP-1-deficient macrophages was still inhibited by dexamethasone and that cytokine production in vivo after LPS stimulation was also inhibited by dexamethasone. The reasons for the discrepancy between the two studies remain unclear at this time. Glucocorticoids have broad effects on a variety of biological systems [74]. In addition to the inhibitory effects on p38 and JNK, glucocorticoids also inhibit the activity of transcription factor NF-κB, and enhance the production of the powerful anti-inflammatory cytokine IL-10 [74]. Furthermore, glucocorticoids can alter the metabolic and cardiovascular systems; therefore, the effects of glucocorticoids observed in the endotoxic shock model may be due to their influence upon the cardiovascular system to prevent shock and multi-organ failure. A challenge in assessing the role of MKP-1 in the anti-inflammatory effects of glucocorticoids is the difficulty in determining the contribution of extra MKP-1 produced as the result of glucocorticoid treatment towards inhibiting the inflammatory response. In theory, this can be accomplished by the generation of knock-in mice that carry an MKP-1 gene devoid of glucocorticoid responsiveness. Further studies on the regulation of MKP-1 by glucocorticoids are needed to shed light on these important questions.
IL-10 also enhances MKP-1 expression induced by LPS, although IL-10 alone does not significantly increase MKP-1 expression [64]. Through microarray analysis, Lang and colleagues found that several MKP genes were induced in macrophages by LPS [64]. They further demonstrated that upregulation of MKP-1 mRNA was transient after stimulation with LPS alone, and IL-10 enhanced the magnitude and duration of the LPS-induced MKP-1 expression. IL-10 also synergized with dexamethasone in the induction of MKP-1 and in the inhibition of IL-6 and IL-12 production. Upregulation of MKP-1 by IL-10 in LPS-stimulated macrophages was correlated with a faster downregulation of p38 activity, suggesting that induction of MKP-1 may constitute an important part of the anti-inflammatory mechanism of IL-10 [64].
Since MKP-1 acts to restrain the inflammatory responses, it is not surprising that cytokines known to enhance inflammation can inhibit MKP-1 expression. It was shown that priming resident peritoneal macrophages with IFN-γ, a TH-1 cytokine that enhances macrophage antimicrobial activity, dramatically increased the production of inflammatory cytokines and NO upon stimulation with LPS [75, 76]. We found that priming of peritoneal macrophages with IFN-γ significantly attenuated the MKP-1 expression induced by LPS, associated with a prolonged activation of p38 and JNK [19]. Interestingly, while LPS does not significantly induce iNOS expression in wild-type resident macrophages without IFN-γ priming, LPS in the absence of IFN-γ substantially elevates iNOS expression in MKP-1-deficient resident macrophages (Zhao and Liu, unpublished observations). These observations suggest that the inhibition of MKP-1 by IFN-γ may be an important part of the mechanism underlying the immune-enhancing properties of IFN-γ.
Macrophage migration inhibitory factor (MIF) is a powerful pro-inflammatory cytokine that enhances the expression of other pro-inflammatory cytokines in macrophages. MIF is tightly linked to lethality associated with both Gram-positive and Gram-negative bacterial sepsis in experimental models. Either knockout of the MIF gene or immnunodepletion of MIF protein confers protection against septic shock [77, 78]. Interestingly, MIF was identified as a physiological counter-regulator of the immunosuppressive effects of glucocorticoids [79]. However, the mechanisms whereby MIF exerts its counter-balancing effect remain largely unknown. Recently, Roger et al. reported that MKP-1 is a critical mediator in the MIF-glucocorticoid crosstalk [80]. They demonstrated that recombinant MIF antagonized the effect of dexamethasone in activated macrophages. They found that MIF inhibited the induction of MKP-1 by LPS and dexamethasone. MIF rescued the production of both TNF-α and IL-8 from dexamethasone-treated macrophages. In contrast, attenuation of MIF expression augmented MKP-1 induction by dexamethasone, leading to decreased TNF-α production. Their studies demonstrate that endogenous MIF acts through attenuating MKP-1 expression to override the inhibition of glucocorticoids on cytokine production in innate immune cells.
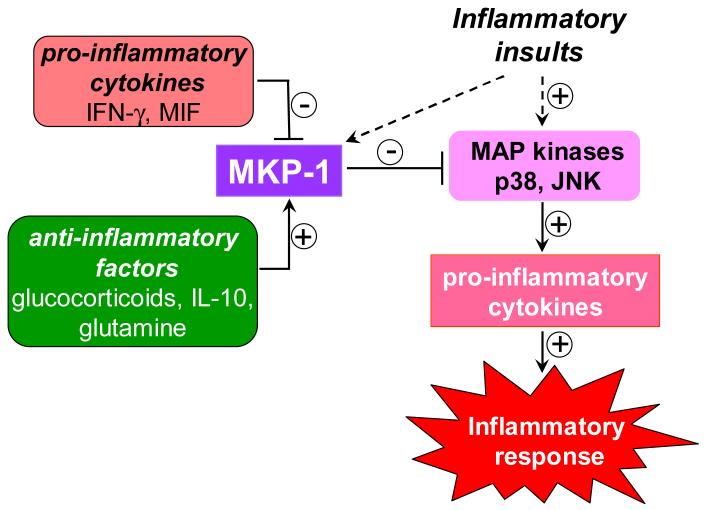
Glutamine was shown to attenuate cytokine release from LPS-stimulated human peripheral blood mononuclear cells, although the mechanism underlying the anti-inflammatory action of glutamine is poorly understood. Using a rat model of polymicrobial sepsis, Singleton et al. examined the effects of glutamine administration on the signaling events mediating the inflammatory responses [81]. They found that glutamine treatment inhibited the phosphorylation of both p38 and ERK, and increased MKP-1 expression in lung tissues, which was correlated with a significant inhibition of the production of pro-inflammatory cytokines, including TNF-α and IL-6. Furthermore, glutamine abrogated the increases in lung iNOS expression and enhanced eNOS during sepsis. These results suggest that induction of MKP-1 by glutamine may be, at least in part, responsible for the anti-inflammatory effect of glutamine. Taken together, it appears that a number of immunomodulatory factors exert their function, at least in part, through modulation of MKP-1 expression, thereby affecting the MAP kinase-mediated inflammatory responses (Fig. 4). Further investigation in this area will provide more insight into the modes of action of various immunomodulatory agents.
Figure 4. Regulation of MKP-1 expression by immunomodulatory agents.
Anti-inflammatory/immunosuppressive agents, such as glucocorticoids, IL-10, and glutamine, induce/augment MKP-1 expression, leading to inhibition of the p38 and JNK cascades and attenuation of inflammatory response. In contrast, pro-inflammatory cytokines, such as IFN-γ and MIF, inhibit MKP-1 expression, thereby perpetuating the p38 and JNK signalling pathways and enhancing the inflammatory response.
6. Concluding Remarks
Although MKP-1 cDNA was cloned 22 years ago [34], only recently the physiological function of this phosphatase began to emerge. Most studies so far have focused on the function of MKP-1 in cytokine regulation during the response to infectious microbial components; however, it is likely that MKP-1 also plays a significant role in cytokine expression evoked by early cytokines such as TNF-α. Since cytokines are critical in the regulation of other immune function, a natural extension of the studies described here will be to inquire whether other aspects of the immune response, eg., the development of adaptive immunity, are also affected by MKP-1 deficiency. On a different front, alterations in cytokine production have been implicated in a wide variety of human diseases, including rheumatoid arthritis, Crohn’s disease, and psoriasis. It is not only intellectually intriguing to study whether the MKP-1 knockout mice are more susceptible to these diseases, as shown by Solajin and colleagues [21], but it is also important to investigate whether potential polymorphisms exist in the populations suffering from these types of diseases. Indeed, efforts to modulate MKP-1 activity in a time-, magnitude-, and tissue-restricted manner could prove to be a fruitful venue for the treatment of many inflammatory diseases.
In addition to the field of immunology, JNK and p38 are implicated in a myriad of physiological and pathophysiological processes, including atherosclerosis [82], diabetes[83], and cancer [84, 85]. As a major negative regulator of these kinases, MKP-1 may also be involved in these processes. It was already shown that MKP-1 is essential for metabolic homeostasis [86]. MKP-1 was found to be overexpressed in a number of human cancer tissue or cell lines [87], and to contribute to the tumorigenicity of pancreatic cancer cells [88]. Recently, Wang et al. have shown that MKP-1-deficient embryonic fibroblasts more readily undergo apoptosis than wild-type cells upon exposure to cisplatin [89], a chemotherapeutic drug widely used in cancer treatment. Thus, the selective inhibition of MKP-1 expression or activity may represent a valuable strategy to overcome drug resistance and improve the efficacy of cancer chemotherapy. Clearly, further studies of the role and regulation of MKP-1 and the development of MKP-1 selective inhibitors will not only enhance our understanding of the function of this phosphatase and its involvement in various human diseases, but will also guide in the treatment of these diseases.
Acknowledgments
This work was supported by a grant (AI 57798) from the National Institutes of Health. The authors want to thank Dr. Myriam Gorospe for editing and sharing unpublished findings.
§Abbreviations used
- MKP
MAP Kinase Phosphatase
- MAP
mitogen-activated protein
- JNK
c-Jun N-terminal kinase
- ERK
extracellular signal-regulated kinase
- MK-2
MAP kinase- activated protein kinase-2
- IKK
IκB kinase
- IRAK
IL-1 receptor-associated kinase
- LPS
lipopolysaccharide
- TNF
tumor necrosis factor
- IL
interleukin
- IFN
interferon
- GM-CSF
granulocyte macrophage colony stimulating factor
- MIF
macrophage migration inhibitory factor
- MIP
macrophage inflammatory protein
- MCP
monocyte chemoattractant protein
- NO
nitric oxide
- iNOS
inducible nitric oxide synthase
- COX
cyclooxygenase
- NF-κB
nuclear factor-κB
- I-κB
inhibitor-κB
- AP-1
activating protein-1
- ARE
AU-rich element
- TTP
tristetraprolin
- TLR
toll-like receptor
- MyD88
myeloid differentiation factor 88
- TRIF
Toll-IL- 1 receptor domain containing adaptor inducing IFN-β
- TRAM
TRIF-related adaptor molecule
- TRAF
TNF receptor-associated factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Janeway CAJ, Travers P, Walport M, Shlomchik MJ. Immunobiology: the immune system in health and disease. Garland Publishing; New York: 2001. [Google Scholar]
- [2].Medzhitov R, Janeway C., Jr. N. Engl. J. Med. 2000;343:338. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- [3].Medzhitov R, Janeway CA., Jr. Curr. Opin. Immunol. 1997;9:4. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- [4].Paulnock DM. Curr. Opin. Immunol. 1992;4:344. doi: 10.1016/0952-7915(92)90087-u. [DOI] [PubMed] [Google Scholar]
- [5].O’Neill L. Biochem. Soc. Trans. 2000;28:557. doi: 10.1042/bst0280557. [DOI] [PubMed] [Google Scholar]
- [6].Inohara N, Ogura Y, Nunez G. Curr. Opin. Microbiol. 2002;5:76. doi: 10.1016/s1369-5274(02)00289-8. [DOI] [PubMed] [Google Scholar]
- [7].Akira S. Curr. Opin. Immunol. 2003;15:5. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- [8].Han J, Thompson P, Beutler B. J. Exp. Med. 1990;172:391. doi: 10.1084/jem.172.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Beutler B, Kruys V. J. Cardiovasc. Pharmacol. 1995;25(Suppl 2):S1–S8. doi: 10.1097/00005344-199500252-00002. [DOI] [PubMed] [Google Scholar]
- [10].Beutler B. Curr. Opin. Immunol. 2000;12:20. doi: 10.1016/s0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- [11].Fan H, Cook JA. J. Endotoxin. Res. 2004;10:71. doi: 10.1179/096805104225003997. [DOI] [PubMed] [Google Scholar]
- [12].Nomura F, Akashi S, Sakao Y, Sato S, Kawai T, Matsumoto M, Nakanishi K, Kimoto M, Miyake K, Takeda K, Akira S. J. Immunol. 2000;164:3476. doi: 10.4049/jimmunol.164.7.3476. [DOI] [PubMed] [Google Scholar]
- [13].Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr., Medzhitov R, Flavell RA. Cell. 2002;110:191. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- [14].Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, Akira S, Yamanishi K, Kawase I, Nakanishi K, Kishimoto T. Immunity. 2002;17:677. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- [15].Keyse SM. Curr. Opin. Cell Biol. 2000;12:186. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- [16].Chen P, Li J, Barnes J, Kokkonen GC, Lee JC, Liu Y. J. Immunol. 2002;169:6408. doi: 10.4049/jimmunol.169.11.6408. [DOI] [PubMed] [Google Scholar]
- [17].Shepherd EG, Zhao Q, Welty SE, Hansen TN, Smith CV, Liu Y. J. Biol. Chem. 2004;279:54023. doi: 10.1074/jbc.M408444200. [DOI] [PubMed] [Google Scholar]
- [18].Zhao Q, Shepherd EG, Manson ME, Nelin LD, Sorokin A, Liu Y. J. Biol. Chem. 2005;280:8101. doi: 10.1074/jbc.M411760200. [DOI] [PubMed] [Google Scholar]
- [19].Zhao Q, Wang X, Nelin LD, Yao Y, Matta R, Manson ME, Baliga RS, Meng X, Smith CV, Bauer JA, Chang CH, Liu Y. J Exp. Med. 2006;203:131. doi: 10.1084/jem.20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hammer M, Mages J, Dietrich H, Servatius A, Howells N, Cato AC, Lang R. J Exp. Med. 2006;203:15. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Salojin KV, Owusu IB, Millerchip KA, Potter M, Platt KA, Oravecz T. J Immunol. 2006;176:1899. doi: 10.4049/jimmunol.176.3.1899. [DOI] [PubMed] [Google Scholar]
- [22].Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, Flavell RA. Proc. Natl. Acad. Sci U. S. A. 2006;103:2274. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lang R, Hammer M, Mages J. J. Immunol. 2006;177:7497. doi: 10.4049/jimmunol.177.11.7497. [DOI] [PubMed] [Google Scholar]
- [24].Liu Y, Shepherd EG, Nelin LD. Nat. Rev. Immunol. 2007;7:202. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- [25].Salojin K, Oravecz T. J. Leukoc. Biol. 2007 doi: 10.1189/jlb.1006639. [DOI] [PubMed] [Google Scholar]
- [26].Liu Y, Shanley TP. J. Organ Dysfunct. 2007 doi: 10.1080/17471060701200444. in press.
- [27].Nishida E, Gotoh Y. Trends Biochem. Sci. 1993;18:128. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- [28].Dong C, Davis RJ, Flavell RA. Annu. Rev. Immunol. 2002;20:55. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- [29].Whitmarsh AJ, Davis RJ. J. Mol. Med. 1996;74:589. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- [30].Carballo E, Lai WS, Blackshear PJ. Science. 1998;281:1001. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- [31].Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mol. Cell Biol. 2001;21:6461. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Mol. Cell Biol. 2006 doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bhalla US, Ram PT, Iyengar R. Science. 2002;297:1018. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- [34].Lau LF, Nathans D. EMBO J. 1985;4:3145. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Charles CH, Abler AS, Lau LF. Oncogene. 1992;7:187. [PubMed] [Google Scholar]
- [36].Keyse SM, Emslie EA. Nature. 1992;359:644. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- [37].Charles CH, Sun H, Lau LF, Tonks NK. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5292. doi: 10.1073/pnas.90.11.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alessi DR, Smythe C, Keyse SM. Oncogene. 1993;8:2015. [PubMed] [Google Scholar]
- [39].Zheng CF, Guan KL. J Biol Chem. 1993;268:16116. [PubMed] [Google Scholar]
- [40].Sun H, Charles CH, Lau LF, Tonks NK. Cell. 1993;75:487. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- [41].Liu Y, Gorospe M, Yang C, Holbrook NJ. J. Biol. Chem. 1995;270:8377. doi: 10.1074/jbc.270.15.8377. [DOI] [PubMed] [Google Scholar]
- [42].Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. J. Biol. Chem. 1995;270:7420. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- [43].Franklin CC, Kraft AS. J. Biol. Chem. 1997;272:16917. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- [44].Kwak SP, Hakes DJ, Martell KJ, Dixon JE. J. Biol. Chem. 1994;269:3596. [PubMed] [Google Scholar]
- [45].Li J, Gorospe M, Hutter D, Barnes J, Keyse SM, Liu Y. Mol. Cell Biol. 2001;21:8213. doi: 10.1128/MCB.21.23.8213-8224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Peng SS, Chen CY, Shyu AB. Mol. Cell Biol. 1996;16:1490. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wilson T, Treisman R. Nature. 1988;336:396. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- [48].Emslie EA, Jones TA, Sheer D, Keyse SM. Hum. Genet. 1994;93:513. doi: 10.1007/BF00202814. [DOI] [PubMed] [Google Scholar]
- [49].Dean JL, Sully G, Clark AR, Saklatvala J. Cell Signal. 2004;16:1113. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- [50].Chen CY, Shyu AB. Trends Biochem. Sci. 1995;20:465. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- [51].Laderoute KR, Mendonca HL, Calaoagan JM, Knapp AM, Giaccia AJ, Stork PJ. J. Biol. Chem. 1999;274:12890. doi: 10.1074/jbc.274.18.12890. [DOI] [PubMed] [Google Scholar]
- [52].Brondello JM, Pouyssegur J, McKenzie FR. Science. 1999;286:2514. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- [53].Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Genes Dev. 1999;13:163. [PMC free article] [PubMed] [Google Scholar]
- [54].Hutter D, Chen P, Barnes J, Liu Y. Biochem. J. 2000;352(Pt 1):155. [PMC free article] [PubMed] [Google Scholar]
- [55].Slack DN, Seternes OM, Gabrielsen M, Keyse SM. J. Biol. Chem. 2001;276:16491. doi: 10.1074/jbc.M010966200. [DOI] [PubMed] [Google Scholar]
- [56].Stewart AE, Dowd S, Keyse SM, McDonald NQ. Nat. Struct. Biol. 1999;6:174. doi: 10.1038/5861. [DOI] [PubMed] [Google Scholar]
- [57].Schwan WR, Kugler S, Schuller S, Kopecko DJ, Goebel W. Infect. Immun. 1996;64:91. doi: 10.1128/iai.64.1.91-99.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kugler S, Schuller S, Goebel W. FEMS Microbiol. Lett. 1997;157:131. doi: 10.1111/j.1574-6968.1997.tb12763.x. [DOI] [PubMed] [Google Scholar]
- [59].Valledor AF, Xaus J, Comalada M, Soler C, Celada A. J Immunol. 2000;164:29. doi: 10.4049/jimmunol.164.1.29. [DOI] [PubMed] [Google Scholar]
- [60].Wang X, Meng X, Kuhlman JR, Nelin LD, Nicol KK, English BK, Liu Y. J. Immunol. 2007 doi: 10.4049/jimmunol.178.8.5312. [DOI] [PubMed] [Google Scholar]
- [61].Parrillo JE. N. Engl. J. Med. 1993;328:1471. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- [62].Howard M, Muchamuel T, Andrade S, Menon S. J. Exp. Med. 1993;177:1205. doi: 10.1084/jem.177.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gerard C, Bruyns C, Marchant A, Abramowicz D, Vandenabeele P, Delvaux A, Fiers W, Goldman M, Velu T. J. Exp. Med. 1993;177:547. doi: 10.1084/jem.177.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hammer M, Mages J, Dietrich H, Schmitz F, Striebel F, Murray PJ, Wagner H, Lang R. Eur. J Immunol. 2005;35:2991. doi: 10.1002/eji.200526192. [DOI] [PubMed] [Google Scholar]
- [65].Valledor AF, Xaus J, Marques L, Celada A. J. Immunol. 1999;163:2452. [PubMed] [Google Scholar]
- [66].Sanchez-Tillo E, Comalada M, Farrera C, Valledor AF, Lloberas J, Celada A. J. Immunol. 2006;176:6594. doi: 10.4049/jimmunol.176.11.6594. [DOI] [PubMed] [Google Scholar]
- [67].Hambleton J, McMahon M, DeFranco AL. J. Exp. Med. 1995;182:147. doi: 10.1084/jem.182.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Swantek JL, Cobb MH, Geppert TD. Mol. Cell Biol. 1997;17:6274. doi: 10.1128/mcb.17.11.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Mol. Cell Biol. 2000;20:4265. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lasa M, Brook M, Saklatvala J, Clark AR. Mol. Cell Biol. 2001;21:771. doi: 10.1128/MCB.21.3.771-780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lasa M, Abraham SM, Boucheron C, Saklatvala J, Clark AR. Mol. Cell Biol. 2002;22:7802. doi: 10.1128/MCB.22.22.7802-7811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kassel O, Sancono A, Kratzschmar J, Kreft B, Stassen M, Cato AC. EMBO J. 2001;20:7108. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Abraham SM, Lawrence T, Kleiman A, Warden P, Medghalchi M, Tuckermann J, Saklatvala J, Clark AR. J. Exp. Med. 2006;203:1883. doi: 10.1084/jem.20060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rhen T, Cidlowski JA. N. Engl. J. Med. 2005;353:1711. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- [75].Collart MA, Belin D, Vassalli JD, de Kossodo S, Vassalli P. J. Exp. Med. 1986;164:2113. doi: 10.1084/jem.164.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gordon S. Bioessays. 1995;17:977. doi: 10.1002/bies.950171111. [DOI] [PubMed] [Google Scholar]
- [77].Calandra T, Echtenacher B, Roy DL, Pugin J, Metz CN, Hultner L, Heumann D, Mannel D, Bucala R, Glauser MP. Nat. Med. 2000;6:164. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- [78].Roger T, David J, Glauser MP, Calandra T. Nature. 2001;414:920. doi: 10.1038/414920a. [DOI] [PubMed] [Google Scholar]
- [79].Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. Nature. 1995;377:68. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- [80].Roger T, Chanson AL, Knaup-Reymond M, Calandra T. Eur. J Immunol. 2005;35:3405. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- [81].Singleton KD, Beckey VE, Wischmeyer PE. Shock. 2005;24:583. doi: 10.1097/01.shk.0000185795.96964.71. [DOI] [PubMed] [Google Scholar]
- [82].Ricci R, Sumara G, Sumara I, Rozenberg I, Kurrer M, Akhmedov A, Hersberger M, Eriksson U, Eberli FR, Becher B, Boren J, Chen M, Cybulsky MI, Moore KJ, Freeman MW, Wagner EF, Matter CM, Luscher TF. Science. 2004;306:1558. doi: 10.1126/science.1101909. [DOI] [PubMed] [Google Scholar]
- [83].Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. Nature. 2002;420:333. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- [84].Bradham C, McClay DR. Cell Cycle. 2006;5:824. doi: 10.4161/cc.5.8.2685. [DOI] [PubMed] [Google Scholar]
- [85].Heasley LE, Han SY. Mol. Cells. 2006;21:167. [PubMed] [Google Scholar]
- [86].Wu JJ, Roth RJ, Anderson EJ, Hong EG, Lee MK, Choi CS, Neufer PD, Shulman GI, Kim JK, Bennett AM. Cell Metab. 2006;4:61. doi: 10.1016/j.cmet.2006.05.010. [DOI] [PubMed] [Google Scholar]
- [87].Vicent S, Garayoa M, Lopez-Picazo JM, Lozano MD, Toledo G, Thunnissen FB, Manzano RG, Montuenga LM. Clin. Cancer Res. 2004;10:3639. doi: 10.1158/1078-0432.CCR-03-0771. [DOI] [PubMed] [Google Scholar]
- [88].Liao Q, Guo J, Kleeff J, Zimmermann A, Buchler MW, Korc M, Friess H. Gastroenterology. 2003;124:1830. doi: 10.1016/s0016-5085(03)00398-6. [DOI] [PubMed] [Google Scholar]
- [89].Wang Z, Xu J, Zhou JY, Liu Y, Wu GS. Cancer Res. 2006;66:8870. doi: 10.1158/0008-5472.CAN-06-1280. [DOI] [PubMed] [Google Scholar]