Abstract
3-hydroxy-3-methylglutaryl–CoA (HMG-CoA) reductase is the rate-limiting enzyme and the first committed step in the biosynthesis of cholesterol in mammals. We have determined the crystal structures of two nonproductive ternary complexes of HMG-CoA reductase, HMG-CoA/NAD+ and mevalonate/NADH, at 2.8 Å resolution. In the structure of the Pseudomonas mevalonii apoenzyme, the last 50 residues of the C terminus (the flap domain), including the catalytic residue His381, were not visible. The structures of the ternary complexes reported here reveal a substrate-induced closing of the flap domain that completes the active site and aligns the catalytic histidine proximal to the thioester of HMG-CoA. The structures also present evidence that Lys267 is critically involved in catalysis and provide insights into the catalytic mechanism.
The reaction catalyzed by 3-hydroxy-3-methylglutaryl–CoA (HMG-CoA), the conversion of (S)-HMG-CoA to (R)-mevalonate, represents a major point of control for isoprenoid biogenesis (for a review, see ref. 1). Because in mammals this reaction is the first committed step in cholesterol biosynthesis, HMG-CoA reductase is a primary target enzyme for chemotherapy of hypercholesterolemias (2). The crystal structure of the HMG-CoA reductase from Pseudomonas mevalonii previously was solved at 3.0 Å resolution (3). The structure revealed a tightly bound dimer that brings together conserved residues implicated in binding and catalysis at the subunit–interface active site. Each monomer is composed of two major domains. The large domain (residues 1–108 and 220–375) binds HMG-CoA and consists of a central 24-residue α-helix surrounded by three roughly triangular walls. The small NAD(H) binding domain (residues 110–215) has a nonclassical dinucleotide-binding fold. This domain consists of a four-strand antiparallel β-sheet with two crossover helices that lie on one side of the sheet. Connecting the third strand and the second helix in the small domain, there is a highly conserved sequence, the DAMG loop (residues 180–186), which is analogous to the G-rich loop in the classic dinucleotide-binding domain.
Knowledge of the spatial location of catalytic residues is crucial for understanding the mechanism of this enzyme. However, the last 50 residues of the C terminus (377–428), which include the catalytic His381 (4, 5), were not visible in the electron density maps of the HMG-CoA reductase apoenzyme and were presumably disordered. It was proposed that these C-terminal residues form a flap domain that closes over the active site when substrates are bound (3). We have now determined the crystal structures of two nonproductive ternary complexes, HMG-CoA/NAD+ and mevalonate/NADH, at 2.8 Å resolution. The structures demonstrate that the flexible flap domain closes on binding of the substrates and positions the catalytic residue His381 close to the scissile bond of HMG-CoA, completing the active site in its catalytic conformation.
MATERIALS AND METHODS
Crystallization.
P. mevalonii HMG-CoA reductase was crystallized in 1.2 M ammonium sulfate and 100 mM N-(2-acetamino)-2-iminodiacetic acid buffer at pH 6.7, as described (6). Large crystals were produced by microseeding and were grown over several weeks up to 1.0 × 1.0 × 0.5 mm in size. Soaks with substrates were done in two steps. First, HMG-CoA or mevalonate solutions were added to the crystallization drops to a final concentration of 4.5 mM for HMG-CoA (50× the Km value) and 3.3 mM for mevalonate (12× the Km value). The crystals then were transferred to a solution with 4 mM NAD+ or 2 mM NADH (20- and 50× the Km values), were soaked for ≈2 min, and were mounted immediately in a glass capillary for data collection.
Data Collection.
X-ray data were collected at room temperature by using an R-axis II (Molecular Structure Corporation, The Woodlands, TX) with a Rigaku (Tokyo) RU-200 rotating anode source. Full data sets were collected from a single crystal of each ternary complex. The x-ray data were indexed and integrated with denzo (7) and were scaled by using scalepack (7). The scaled data set for the mevalonate/NADH complex (Mev) has 23,498 unique reflections with an Rmerge on intensities (Rm) = 8.2% and is 93% complete (25–2.8 Å). The data set for the HMG-CoA/NAD+ complex (Hmg) has 23,346 unique reflections with an Rm = 7.1% and is 92% complete (25–2.8 Å). The crystal lattice for both complexes is isomorphous with the native crystals and belongs to the cubic space group I4132 with a = 228.5 Å for Mev and a = 228.7 Å for Hmg (native crystals have a = 229.4 Å), with one dimer per asymmetric unit.
Structure Determination.
Crystallographic refinement was performed by using x-plor 3.1 (8) and the Engh and Huber parameters (9). Initial rigid-body refinement for data in the 15- to 4.5-Å resolution range led to final correlation coefficients of 0.85 (Mev) and 0.83 (Hmg) and Rfactor of 25.5% (Mev) and 26.5% (Hmg). The 2(Fo − Fc), αc density map calculated at this stage showed extra density in one of the two active sites of the dimer for both complexes. This allowed us to model the substrates (HMG-CoA and mevalonate) and the cofactor (NAD+/NADH) using the graphics display program chain (10). Additionally, a large region of extra density beyond residue 377 (the last visible residue in the apoenzyme) of the monomer with bound substrates provided the first hint for the tracing of the flexible flap at the C-terminal region of the protein. Successive cycles of combined conjugate gradient minimization refinement with simulated annealing further improved the phases sufficiently to trace the last 50 residues in the flap domain region of the Hmg complex (377–428) and the first 10 residues of the flap in the Mev complex (377–387). Because of the asymmetry of the two active sites in the dimer (the first one is solvent accessible and occupied by substrates and the second is packed against symmetry-related molecules), noncrystallographic symmetry restraints were applied to both monomers (residues 4–375 in the first monomer and residues 504–875 in the second monomer) during the refinement. Water molecules were modeled by using the 2(Fo − Fc) map contoured at 3.5 σ. Temperature factors were refined according to the restrained individual isotropic B-factor refinement protocol in x-plor, using data from 6–2.8 Å with a resulting Rwork = 19.4% (Rfree =27.4%) for the Hmg complex and Rwork = 18.8% (Rfree = 26.6%) for the Mev complex. The last refined model contains protein residues 4–428 (first monomer) and 504–875 (second monomer) in the Hmg complex plus 1 HMG-CoA and 1 NAD+. The last refined Mev model contains protein residues 4–387 and 504–875, plus 1 mevalonate and 1 NADH. The geometrical parameter analysis gave an rms deviation for bond distance of 0.016 and 0.015 Å (Hmg/Mev) and for bond angles of 1.85 and 1.88°, respectively.
RESULTS AND DISCUSSION
The Active Site and the Flap Domain.
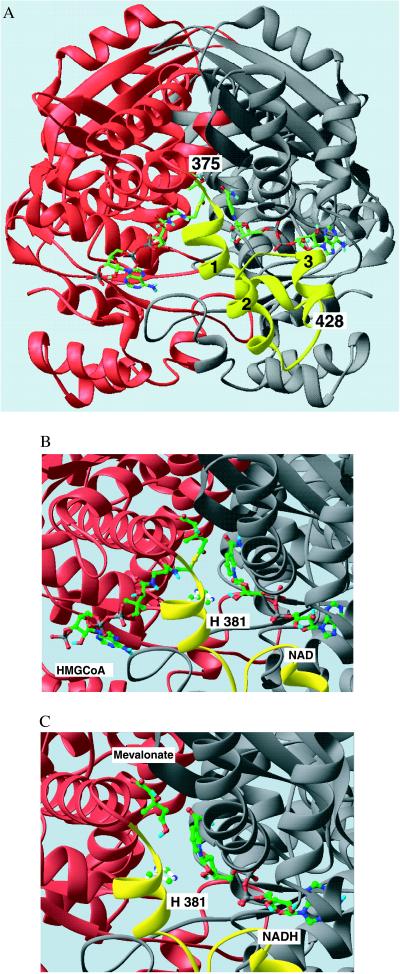
The active site of P. mevalonii HMG-CoA reductase is a large, open cavity 42 Å in length that contains three distinct subsites that bind cofactors and substrates (Fig. 1A). The first site, formed by the large domain of one monomer, binds the CoA portion of HMG-CoA. The small domain of the second monomer forms the NAD(H) binding site. At the interface of the two monomers, there is a smaller, deeper pocket that is the mevalonate binding site. The previously identified catalytic residues, Glu83 and Asp283 (1, 11), are located at the bottom of the mevalonate binding site. The HMG-CoA/NAD+ structure reveals HMG-CoA bound in an extended conformation with the HMG moiety in the central pocket (mevalonate pocket). The NAD+ also binds in an extended conformation with the nicotinamide ring approaching the thioester of HMG-CoA in an anti/pro-S relationship (Fig. 1B). In the mevalonate/NADH structure, the NADH binds in the same conformation as NAD+ (Fig. 1C). Mevalonate sits in the central pocket in an open linear conformation that overlays the HMG moiety of HMG-CoA.
Figure 1.
(A) Ribbon diagram of the HMG-CoA reductase dimer with HMG-CoA and NAD+ bound. The view looks directly into the active site cavity between the two monomers shown in red and gray. The flap domain, in yellow, extends from residue 375 in the red monomer in three helical segments (marked 1–3) to the C-terminal residue of the molecule, residue 428. The substrates are shown as stick models with C = green, N = blue, O = red, P = gray, and S = yellow. Both substrates bind in an extended conformation with, HMG-CoA binding to the large domain of the red monomer on the left and NAD+ binding to the small domain of the gray monomer on the right. (B and C) Close-up views of the active site in the ternary complexes of HMG-CoA reductase with HMG-CoA/NAD+ and mevalonate/ NADH, respectively. The catalytic His381 side chain in the closed flap is shown as a ball and stick model, positioned near the sulfur of the HMG-CoA in B. The similar position of the HMG moiety of HMG-CoA and of mevalonate in these complexes is readily seen. The NAD(H) nicotinamide ring in the anti/pro-S configuration lays side-by-side with the substrate, positioned for hydride transfer. This figure was prepared with the program ribbons (19).
The flap domain was apparent in the initial difference electron density maps of both complexes as an extra density region covering the mevalonate pocket. The first 10 residues of the flap domain were readily built into this extra density as an α-helix. Subsequent cycles of rebuilding and refinement improved the maps sufficiently to allow all 50 residues of the C-terminal region (377–428) to be built in the HMG-CoA/NAD+ complex (Fig. 2). In the mevalonate/NADH complex, only the first 10 residues (377–387) were clearly defined and built in the density. Beyond this point, there was no visible electron density. The flap domain in the HMG-CoA/NAD+ complex contains three α-helices connected by loops: helix 1 from residue 377 to residue 393, helix 2 from residue 399 to residue 409, and helix 3 from residue 415 to residue 428 (Fig. 1A). Helix 1 is connected to the long 24-residue α-helix (351–374) of the large domain by a 2-residue turn (375–377) that may act as a hinge for the flap. These two helices lie almost perpendicular to each other in the closed flap structure.
Figure 2.
The entire flap domain region superimposed on the final electron density 2Fo − Fc map, contoured at the 1.0 σ level. This figure was prepared with the program setor (20).
Ligand-Induced Conformational Changes.
Of the conformational changes that accompany ligand binding, the most interesting is the closure of the flap domain, which positions the catalytic histidine, His381, at the active site. At the same time, juxtaposition of residues Ile213, Leu372, and Ile377 forms a hydrophobic pocket at the inner face of helix 1. This pocket is part of a more extensive hydrophobic region that may serve to shelter the hydride transfer reaction from the solvent and to place the dihydronicotinamide moiety in a hydrophobic environment. Diffusion of substrates into the apoenzyme crystal is accompanied by packing rearrangements in both complexes and causes main-chain shifts of ≈0.7 Å in the region from residues 9 to 37. Most of the flap contacts occur on the inner face of helix 1 and the two helices of the small domain of the second monomer. In the HMG-CoA/NAD+ complex, Ile377 interacts with Glu195 (N…Oɛ1 3.2 Å), Gln378 Oɛ1 interacts with Thr192 Oγ1 (3.2 Å), and Met382 Sδ makes a contact with Asn188 Nδ2 (3.1 Å), all residues on the second helix of the small domain. The catalytic His381 forms a hydrogen bond to Ser85 in the large domain through the main chain O…Oγ 2.7 Å whereas the neighboring His385 Nδ1 forms a hydrogen bond to Asn188 Nδ2 (3.1 Å) and interacts with the pyrophosphate on the NAD+ molecule (Nɛ2…O1PA in NAD+ 3.2 Å). Helix 1 then extends toward the large domain, making a single contact between Asn388 Nδ2 and Thr234 Oγ1 (3.2 Å). Helix 2 makes no close contacts with the main body of the protein. A single contact links helix 3 with the first helix of the small domain (Asp416 Oδ2 is 2.9 Å from Lys145 O). In the mevalonate/NADH complex, only the interactions at the N terminus of helix 1 are seen. In the hinge region, both Ile377 N and Gln378 N interact with Glu195 Oɛ1 (3.2 Å), and Gln378 Oɛ1 forms a hydrogen bond with Thr192 Oγ1 (2.9 Å).
Additional changes induced by substrate binding are observed in the HMG-CoA and NAD(H) binding sites. In the HMG-CoA site, loop 236–240 shows an rms deviation for main chain atoms of 0.6 Å relative to the apoenzyme structure. Several side chains extend toward hydrogen bond acceptor groups in the HMG-CoA molecule (Arg11, Arg261, Arg370, Lys95, Asn553, and Gln364). In the NAD(H) binding site, the main chain in the loop–helix region 328–339 moves toward the pyrophosphate moiety of NAD(H) (rms deviation of 0.9 Å) for both ternary complexes. The small domain helix 185–195, which contacts helix 1 on the flap, shifts toward the pyrophosphate moiety of NAD(H) by 1.3 Å in the HMG-CoA complex and 0.7 Å in the mevalonate complex. The small domain helix 141–151, which contacts helix 3 on the flap, shifts toward the adenine ring of NAD(H). This helix contains Asp146, a residue that discriminates against NADP(H), the cofactor of biosynthetic HMG-CoA reductases (12). The side chain of Asp146 also moves from its original position in the apoenzyme to form a hydrogen bond to the 2′-hydroxyl group of the adenine ribose of NAD(H).
Mechanism of Catalysis.
HMG-CoA reductase of P. mevalonii reversibly catalyzes both the oxidative conversion of mevalonate to HMG-CoA, in the presence of CoA, and the reduction of HMG-CoA to mevalonate and CoA via consecutive two-electron transfer steps to and from NAD(H). This process also formally requires the transfer of two protons. The reduction of HMG-CoA can be described in three steps (see scheme in Fig. 3). In step 1, hydrogen bond formation of the HMG-CoA thioester oxygen with Lys267 induces partial carbonium ion character in the thioester carbon. This facilitates the first hydride transfer from NADH, forming the thiohemiacetal (mevaldyl-CoA) and NAD+. In step 2, hydrolysis of the C-S bond forms bound mevaldehyde. The CoAS− thioanion then is protonated and released as CoASH. In step 3, protonation of the carbonyl oxygen of bound mevaldehyde facilitates the second hydride transfer from NADH, forming mevalonate.
Figure 3.
Scheme of the reaction mechanism proposed for P. mevalonii HMG-CoA reductase. The three steps of the reaction from HMG-CoA to mevalonate are shown, and the roles proposed for the key catalytic residues Lys267, Asp283, Glu83, and His381 are indicated.
Previous mechanisms proposed for the yeast (5) and hamster enzymes (13) postulated that the chemical mechanism catalyzed by HMG-CoA reductase might follow the general chemical reaction proposed for dehydrogenases in which an acid/base catalyst on the enzyme assists the hydride transfer between substrate and cofactor (14). For the yeast enzyme, Veloso et al. (5) used kinetic analyses to infer participation of a histidine and an acidic residue in catalysis. Extensive mutagenesis and kinetic analysis of the P. mevalonii and hamster enzymes led to the identification of a histidine (4, 15), a glutamate (11), and an aspartate residue (13) involved in catalysis. For the hamster enzyme, Frimpong et al. (13) proposed a catalytic mechanism in which the aspartate serves as a general acid during both reductive stages and as a general base during deacylation of mevaldyl-CoA. The three catalytic residues proposed to be involved in this mechanism were Glu83, Asp283, and His381 in the P. mevalonii enzyme.
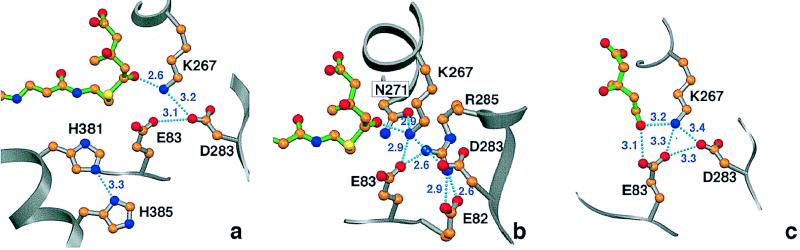
From the two x-ray crystal structures of the ternary complexes of the P. mevalonii enzyme, we have obtained structural evidence for roles of the participating residues and propose a revised mechanism. The ternary complexes show all three of these residues in the active site, with the two acidic residues at the bottom of the mevalonate site and the histidine on the first helix of the closed flap. The structures also reveal that Lys267, not previously implicated in mutagenesis experiments, is critically involved in catalysis by the P. mevalonii enzyme. In the HMG-CoA/NAD+ complex, the thioester oxygen of the substrate points toward the Nζ atom of Lys267 at a distance of 2.6 Å (Fig. 4a). Lys267 is located midway between the substrate and Asp283, which is 5.6 Å away from the thioester oxygen on the opposite side of Lys267. In the light of this structural evidence, the assumption that Asp283 directly transfers a proton to the thioester oxygen, as proposed for the cognate residue of the hamster enzyme, can be ruled out for the P. mevalonii enzyme. Lys267 is at the center of a hydrogen bond network that involves Asn271, Glu83, and Asp283 as the closest neighbors, and Arg285 as a second shell neighbor through hydrogen bonds to Glu83, Glu82, and Asp283 (Fig. 4b). These interactions hold the side chain of Lys267 in place for substrate binding and catalysis. We therefore propose that Lys267 is the residue that polarizes the carbonyl oxygen in steps 1 and 3 of the reaction and provides the proton in step 3.
Figure 4.
Ball and stick diagrams of the HMG-CoA (a and b) and mevalonate (c) substrates in the active site of HMG-CoA reductase. The substrates are shown as stick models with C = orange, N = blue, O = red, and S = yellow. (a) Critical contacts with the enzyme (silver bonds) at the site of cleavage of HMG-CoA (green bonds). The thioester oxygen of the substrate points directly at Lys267, making a strong hydrogen bond with this newly identified catalytic residue. The contact between the catalytic His381 and His385, proposed to aid in the protonation of the CoAS− group, also is shown. (b) The hydrogen bonding network that holds Lys267 in place. Shown are the first shell of hydrogen bond contacts that involve primarily positive and negatively charged residues. (c) Critical contacts of the catalytic residues with the mevalonate substrate. In this ternary complex, the tetrahedral carbon (C-5) points its OH toward one of the Glu83 carboxylic oxygens, suggesting a change in the primary contact of that oxygen after isomerization. This also suggests that Glu83 may be involved in accepting a proton from mevaldyl-CoA, which should have the same tetrahedral configuration for C-5. This figure was prepared with setor (20).
His381 is proximal to the sulfur atom of HMG-CoA, as would be expected for its proposed role in protonating the leaving CoAS− anion (Fig. 4a). A third residue implicated in catalysis, Glu83, is also in contract with Asp283. In Frimpong’s mechanism (13), the cognate of Glu83 was proposed to reprotonate His381 in step 2. However, in both ternary structures, Glu83 and His381 are separated by 4.0–4.3 Å. Instead, His381 probably is reprotonated by His385, which is within hydrogen bonding distance of His381 (2.8 Å in the mevalonate complex and 3.3 Å in the HMG-CoA complex) (Fig. 4a). An alternate role for the glutamate can be proposed from the mevalonate/NADH complex. In this complex, the oxygen of the 5-hydroxyl of mevalonate is 3.1 Å from a carboxylic oxygen of Glu83 (Fig. 4c). The tetrahedral configuration for C-5 of mevalonate is expected to be the same as in the thiohemiacetal intermediate. The displacement of the C-5 oxygen in the mevalonate complex with respect to the carbonyl oxygen in the HMG-CoA complex then mimics the isomerization from the trigonal configuration to the tetrahedral configuration in the thiohemiacetal. Therefore, we propose that Glu83 could participate in proton abstraction from bound mevaldyl-CoA. No residue other than Glu83 is closer to the C-5 oxygen of mevalonate, and the closest histidine, assigned to this role for the yeast enzyme, is 5.3 Å away. The location of Lys267, Asp283, and Glu83 suggests that all three residues participate in proton transfer at different stages of the reaction and thus may constitute a proton relay system (Fig. 3).
Based on our revised mechanism and the structures of the ternary complexes, we can hypothesize that the flap closes on binding of the substrates and remains closed during the reaction. We further propose that NAD(H) can be bound and released as NAD+ without requiring the flap to be opened. Similarly, CoASH can be released after protonation by His381. Conversely, aldehyde-level intermediates would be trapped in the central pocket while the flap remains closed. This might explain why no free intermediates in catalysis by HMG-CoA reductase have been isolated (16, 17). After completion of the reaction, the flap would open, permitting departure of mevalonate and regenerating free enzyme.
Mutagenesis of the Catalytic Residues.
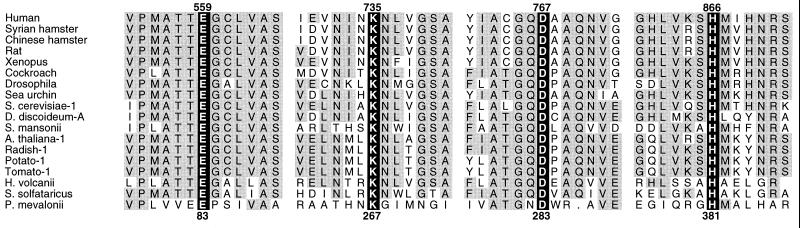
The active site histidine, glutamate, and aspartate are conserved in all known HMG-CoA reductases (Fig. 5), and the changes in activity that accompany their mutagenesis support their proposed roles in catalysis (4, 11, 13, 15). After Lys267 was identified in the P. mevalonii structure as a catalytic residue, site-directed mutagenesis studies were performed. Mutation of Lys267 to alanine crippled the P. mevalonii enzyme, as would be expected for a catalytically critical residue. For oxidation of mevalonate to HMG-CoA, the specific activity of the enzyme K267A was <0.1% that of the wild-type enzyme. All HMG-CoA reductases catalyze, in addition to the reversible interconversion of HMG-CoA and mevalonate, two reactions of free mevaldehyde. For catalysis of mevaldehyde reduction and mevaldehyde oxidation, enzyme K267A had <0.5% wild-type activity. Low activity did not appear to reflect a nonnative structure, as judged by size-exclusion HPLC and far UV-circular dichroism. Neither changing the pH over the range 6–9 nor raising the concentrations of mevalonate, CoASH, or NAD+ to 50- to 100-fold above their Km values for the wild-type enzyme enhanced activity (18). These observations are consistent with the role proposed for Lys267 in catalysis by the P. mevalonii enzyme.
Figure 5.
Alignment of HMG-CoA reductase sequences around the critical catalytic residues. Four regions of sequence around the catalytic residues of P. mevalonii HMG-CoA reductase (Glu83, Lys267, Asp283, and His381) are shown in comparison to selected sequences from a variety of biosynthetic HMG-CoA reductases. The numbering for P. mevalonii is shown in bold at the bottom. The numbering at the top of the figure is that for the human enzyme, given as a reference. Catalytic residues are indicated by white on black lettering. Conserved residues are shaded in gray. Site-directed mutagenesis of the P. mevalonii and Syrian hamster enzymes have confirmed that the aligned residues are catalytic homologues.
For the proposed mechanism to be general, all HMG-CoA reductases must possess an active site lysine. Sequence comparisons indicated that only three lysines are conserved in all known biosynthetic HMG-CoA reductases. For the hamster enzyme, these are Lys690, Lys691, and Lys734. An unambiguous decision as to which of these was the cognate residue of Lys267 could not be drawn from sequence alignment alone. Site-directed mutagenesis and kinetic analysis of the hamster enzyme showed that mutant enzyme K734A lacked detectable activity and identified Lys734 as the probable cognate of Lys267 (data not shown).
The structures of P. mevalonii HMG-CoA reductase nonproductive ternary complexes present compelling evidence for an enzymatic mechanism that requires a hinge closure of the C-terminal flap domain. Concomitant binding of the substrate and cofactor induces closing of the flap domain to complete the active site, positioning the catalytic histidine close to the scissile bond of HMG-CoA. These complexes provide important structural information that contributes to an understanding of the molecular details of the catalytic mechanism. The roles of the catalytic residues His381, Glu83, and Asp283 are established, and a revised mechanism is proposed in which an additional residue, Lys267, is essential in catalysis by P. mevalonii HMG-CoA reductase.
Acknowledgments
We thank Martin Lawrence for his discussion on the potential role of the lysine in catalysis. We thank Haresh Patel for helpful assistance with crystallization and data collection and John W. Burgner for critical revision of the mechanism. We thank V. Munshi and K. Li for their kinetic data on P. mevalonii mutant enzymes. This work has been supported by National Institutes of Health Grant HL 52115 to C.V.S. and Grant HL 47113 to V.W.R. and by a grant from the Lucille P. Markey Foundation to the Structural Biology Group. D.A.B. was a Predoctoral Fellow of the American Heart Association, Indiana Affiliate.
ABBREVIATION
- HMG
3-hydroxy-3-methylglutaryl
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QAX and 1QAY).
References
- 1.Bochar D A, Friesen J A, Stauffacher C V, Rodwell V W. In: Comprehensive Natural Products: Isoprenoids Including Carotenoids and Steroids. Cane D E, editor. New York: Elsevier Pergamon; 1999. pp. 15–44. [Google Scholar]
- 2.Endo A. J Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 3.Lawrence C M, Rodwell V W, Stauffacher C V. Science. 1995;268:1758–1762. doi: 10.1126/science.7792601. [DOI] [PubMed] [Google Scholar]
- 4.Darnay B G, Wang Y, Rodwell V W. J Biol Chem. 1992;267:15064–15070. [PubMed] [Google Scholar]
- 5.Veloso D, Cleland W, Porter J. Biochemistry. 1981;20:887– 894. doi: 10.1021/bi00507a036. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence C M, Chi Y-I, Rodwell V W, Stauffacher C V. Acta Crystallogr D. 1995;51:386–389. doi: 10.1107/S0907444994009819. [DOI] [PubMed] [Google Scholar]
- 7.Otwinowski Z. Proceedings of the CCP4 Study Weekend. Warington, U.K.: Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 8.Brünger A T. x-plor 3.1Manual. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 9.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 10.Sack J S. J Mol Graphics. 1988;6:224–225. [Google Scholar]
- 11.Wang Y, Darnay B G, Rodwell V W. J Biol Chem. 1990;265:21634–21641. [PubMed] [Google Scholar]
- 12.Friesen J A, Lawrence C M, Stauffacher C V, Rodwell V W. Biochemistry. 1996;35:11945–11950. doi: 10.1021/bi9609937. [DOI] [PubMed] [Google Scholar]
- 13.Frimpong K, Rodwell V W. J Biol Chem. 1994;269:11478–11483. [PubMed] [Google Scholar]
- 14.Oppenheimer N J, Handlon A L. Mechanism of NAD-Dependent Enzymes: The Enzymes. Vol. 20. San Diego: Academic; 1992. pp. 453–505. [Google Scholar]
- 15.Darnay B G, Rodwell V W. J Biol Chem. 1993;268:8429–8435. [PubMed] [Google Scholar]
- 16.Siddiqi M A, Rodwell V W. J Bacteriol. 1967;93:207–214. doi: 10.1128/jb.93.1.207-214.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Retey J, von Stetten E, Coy U, Lynen F. Eur J Biochem. 1970;15:72–76. doi: 10.1111/j.1432-1033.1970.tb00977.x. [DOI] [PubMed] [Google Scholar]
- 18.Bochar, D. A., Tabernero, L., Stauffacher, C. V. & Rodwell, V. W. (1999) Biochemistry, in press. [DOI] [PubMed]
- 19.Carson M, Bugg C E. J Mol Graphics. 1986;4:121–122. [Google Scholar]
- 20.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]