Abstract
Background
GPI anchor attachment is catalyzed by the GPI transamidase (GPIT) complex. GAA1, PIG-T and PIG-U are the three of five GPIT subunits. Previous studies demonstrated amplification and overexpression of GPIT subunits in bladder and breast cancer with oncogenic function. We performed an analysis of these subunits in head and neck squamous cell carcinoma (HNSCC).
Results
To evaluate GAA1, PIG-T and PIG-U in HNSCC, we used quantitative PCR (QPCR) and quantitative RT-PCR (QRT-PCR) to determine the copy number of those genes in primary tumors and the matching lymphocytes in 28 patients with HNSCC and quantified RNA expression of those genes in 16 primary HNSCC patients and 4 normal control tissue samples. GAA1 showed a significant increase in normalized mRNA expression, 2.11 (95% CI: 1.43, 2.79), in comparison to that of normal controls, 0.43 (95% CI: -0.76, 1.61), p = 0.014 (Mann-Whitney test). The mean genomic copy number of GAA1 was significantly increased in HNSCC, 0.59 (95% CI: 0.50, 0.79), in comparison to lymphocyte DNA, 0.35 (95% CI: 0.30, 0.50), p = 0.001 (paired t-test).
Conclusion
An increased expression level and elevated copy number for GAA1 suggest a role for this GPI anchor subunit in HNSCC.
Background
HNSCC has an annual incidence of over than 40,000 cases per year in United States and is characterized by local tumor aggressiveness, a high rate of early recurrences, and development of second primary carcinomas [1]. Despite modern therapeutic strategies, overall 5-year survival rate does have only modestly improved.
Glycosylphosphatidylinositol (GPI) anchoring is a membrane attachment mechanism for cell surface proteins widely used in eukaryotes. GPI anchor attachment is catalyzed by GPI transamidase (GPIT) complex, which is composed of at least five subunits: Phosphatidylinositol Glycan Class U (PIG-U), Glycosylphosphatidylinositol Anchor Attachment Protein 1 (GAA1), Phosphatidylinositol Glycan Class K (Gpi8), Phosphatidylinositol Glycan Class S (PIG-S), Phosphatidylinositol Glycan Class T (PIG-T) [2-11]. All of the subunits are required for GPIT to function [3,6-8]. Gpi8 is the likely enzymatic component of the GPIT complex and can be cross-linked to proproteins [5,8,12-14]. GAA1 is able to assemble into PIG-U-containing GPIT complexes that are capable of interacting with a proprotein substrate, and this subunit also is critical in GPI recognition by GPIT [15]. PIG-U contains a sequence motif found in yeast and mammalian fatty acid elongases. This motif has been suggested to play a role in recognizing long chain fatty acids in GPI [3], and PIG-T contains endoplasmic reticulum (ER) localization information.
In a previous study amplification of PIG-U was noted in the bladder cancer, and was found to contribute to an oncogenic phenotype [16]. A recent study showed PIG-T and GPAA1 were overexpressed in breast cancer cell lines and primary tumors and caused malignant transformation in vitro [17]. Genomic amplification and/or DNA copy number gain are common genetic alterations in cancer that lead to the overexpression of oncogenes [18-20]. PIG-U and PIG-T are located at chromosome 20q and GAA1 is located at chromosome 8q, a chromosomal arm with increased copy number in HNSCC [21,22]. To evaluate PIG-U, PIG-T and GAA1 as possible oncogene candidates in HNSCC, we employed real-time PCR and real-time RT-PCR to determine copy number and RNA expression of those genes in primary HNSCCs.
Results and discussion
Analysis of mRNA expression of GAA1, PIG-T and PIG-U by QRT-PCR
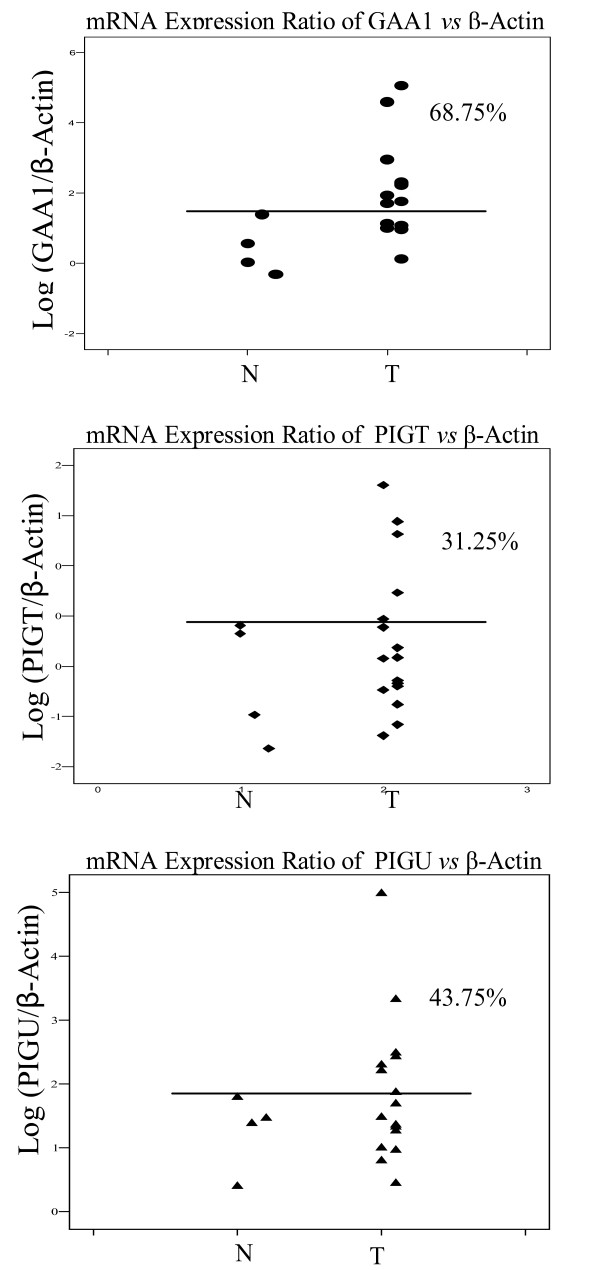
Overexpression of PIG-U, one of the GPIT subunits, was reported previously in primary bladder cancer and cell lines [16]. By comparing the mRNA expression ratios of the oral mucosa of normal subjects and tumor tissues of HNSCC patients, we were able to assess GAA1, PIG-T and PIG-U expression alteration in a quantitative manner. In this study, 68.75%, 31.25% and 43.75% of HNSCC exhibited higher mRNA expression in comparison to normal controls for GAA1, PIG-T and PIG-U, respectively (Fig 1). In particular, GAA1 showed a significant increase in mRNA expression in HNSCC, 2.11 (95% CI: 1.43, 2.79), in comparison to that in normal controls, 0.43 (95% CI: -0.76, 1.61), p = 0.014 (Mann-Whitney test).
Figure 1.
mRNA Expression Ratio of GAA1, PIGT and PIGU vs β-Actin. The expression differential of GAA1, PIGT and PIGU between normal (health subjects mucosa) and HNSCC was measured by calculating relative fluorescence amplification of transcripts of gene of interests normalized by the corresponding β-Actin signal. N, normal; T, HNSCC.
Analysis of copy number of GAA1, PIG-T and PIG-U by QPCR
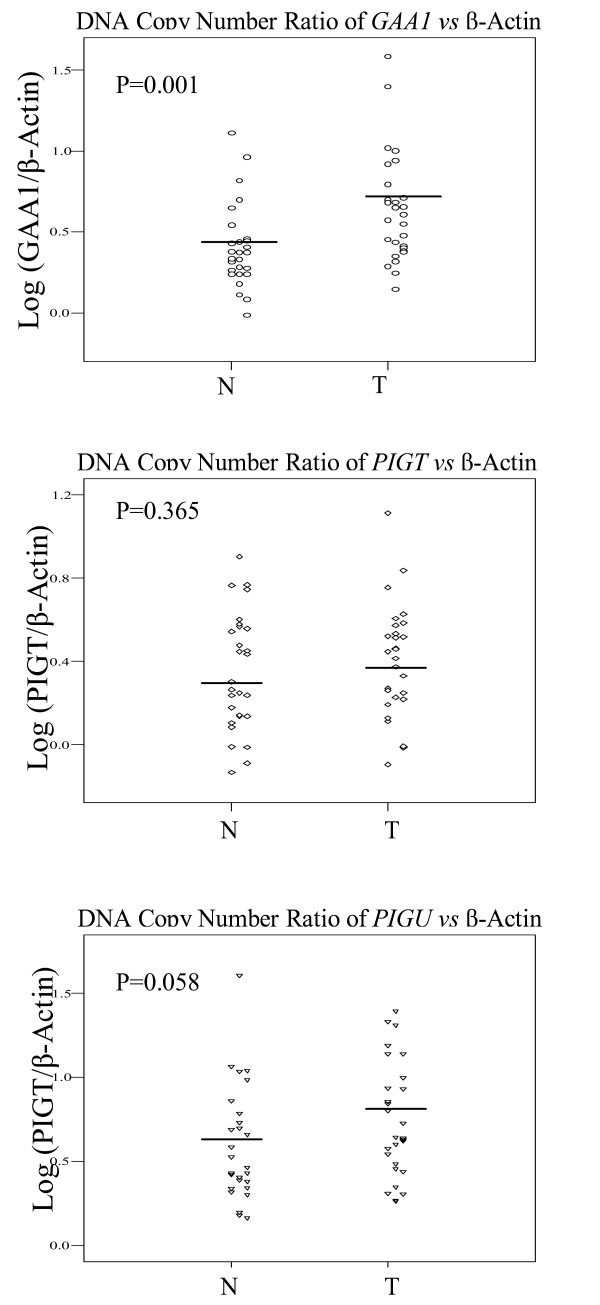
The combination of QPCR and QRT-PCR is a facile approach to detecting gene amplification and over-expression in tumor specimens. To assess whether the copy number of GAA1, PIG-T and PIG-U was altered in HNSCC, we performed QPCR in 28 HNSCC and matched peripheral leukocyte DNA. The relative copy number of each gene vs β-Actin is shown in Table 1. Overall, a trend of increasing copy number was found in HNSCC. GAA1 copy number was significantly increased in primary HNSCCs, 0.59 (95% CI: 0.50, 0.79), in comparison to peripheral leukocyte DNA, 0.35 (95% CI: 0.30, 0.50), p = 0.001, while PIG-U showed a border line difference (paired t-test). (Fig 2) Taken together, the increased mRNA expression of GAA1 and low level copy number increase in HNSCC may be relevant as a possible contributor to oncogenic transformation in HNSCC.
Table 1.
Copy number alteration of GAA1, PIG-T and PIG-U in HNSCC
| DNA Copy Number Ratio | Number | Mean | Median | 95%CI | p (paired-t test) | |
| Log (GAA1/β-Actin) | Control | 28 | 0.40 | 0.35 | (0.30, 0.50) | 0.001 |
| HNSCC | 28 | 0.63 | 0.59 | (0.50, 0.76) | ||
| Log (PIG-T/β-Actin) | Control | 28 | 0.34 | 0.28 | (0.24, 0.45) | 0.365 |
| HNSCC | 28 | 0.40 | 0.43 | (0.30, 0.50) | ||
| Log (PIG-U/β-Actin) | Control | 28 | 0.59 | 0.45 | (0.46, 0.72) | 0.058 |
| HNSCC | 28 | 0.74 | 0.64 | (0.61, 0.87) |
Figure 2.
Relative DNA copy number of GAA1, PIGT and PIGU in HNSCC. The copy number differential of GAA1, PIGT and PIGU between paired lymphocytes and HNSCC was measured by calculating relative fluorescence amplification of gene of interest normalized by the corresponding β-Actin signal using standard curve method. N, normal; T, HNSCC.
Many membranous enzymes, receptors, differentiation antigens and other biologically active proteins proved to be bound to the plasma membrane by GPI. Recent studies report that there are increased levels of the GPI-anchored adhesion molecules CEACAM5, CEACAM6 and mesothelin in response to forced expression of ΔN-TCF-1B, which could contribute to the induction of effecter molecules potentially relevant for tumor invasion in colorectal carcinomas patients [23]. Notably, our data showed that GAA1 copy numbers were much higher in HNSCC than in control white blood cells. This difference was highly significant, which was corresponded to the elevated RNA expression level in HNSCC patients.
Clinical associations with copy number alterations in GAA1, PIG-T and PIG-U
To analyze whether clinical factors are associated with GAA1, PIG-T and PIG-U as potential markers, therefore, we further sub-grouped HNSCC patients by gender, race, stage and tumor site. We found that female HNSCC patients had a higher copy number of GAA1, 0.77 (95% CI: 0.63, 0.91), in comparison to male patients 0.58 (95% CI: 0.40, 0.75), p = 0.025 (Mann-Whitney test) (Table 2). In comparison to GAA1, PIG-T showed significant difference of copy number in paired lymphocytes DNA between male and female patients but not in tumor tissue DNA (Table 3b). Neither race nor tumor site was independently associated with GAA1 (Table 2), PIG-T (Table 3) and PIG-U (Table 4) copy number alterations. One might, therefore, speculate that gender might be a factor in the catalysis of GPI anchor attachment. Five out of 28 HNSCCs were located in larynx. We noticed that the PIG-T DNA copy number of laryngeal HNSCC, showed a relatively lower value in comparison to that in the other sites (Table 3), however, we were unable to draw definite conclusions due to limited case numbers. Additionally, no significances were shown among different stages for each gene.
Table 2.
The effects of clinical parameters on copy number of GAA1
| Log (Gaa1/β-Actin) | Number | Mean | Median | 95%CI | p (Mann-Whitney test) | |
| Control | Male | 20 | 0.36 | 0.33 | (0.25, 0.48) | 0.242 |
| Female | 8 | 0.49 | 0.39 | (0.29, 0.70) | ||
| HNSCC | Male | 20 | 0.58 | 0.44 | (0.40, 0.75) | 0.025* |
| Female | 8 | 0.77 | 0.75 | (0.63, 0.91) | ||
| Control | White | 17 | 0.35 | 0.32 | (0.23, 0.46) | 0.165 |
| Other | 11 | 0.48 | 0.43 | (0.29, 0.68) | ||
| HNSCC | White | 17 | 0.52 | 0.48 | (0.42, 0.63) | 0.095 |
| Other | 11 | 0.81 | 0.68 | (0.52, 1.09) | ||
| Control | OC | 11 | 0.35 | 0.33 | (0.18, 0.53) | |
| HP | 3 | 0.46 | 0.43 | (0.05, 0.88) | 0.392 | |
| L | 5 | 0.42 | 0.28 | (0.04, 0.80) | 0.865 | |
| OP | 9 | 0.43 | 0.38 | (0.22, 0.64) | 0.518 | |
| HNSCC | OC | 11 | 0.67 | 0.68 | (0.39, 0.94) | |
| HP | 3 | 0.55 | 0.11 | (-0.26, 1.37) | 0.697 | |
| L | 5 | 0.65 | 0.61 | (0.37, 0.92) | 0.955 | |
| OP | 9 | 0.61 | 0.57 | (0.36, 0.86) | 0.676 |
Table 3.
The effects of clinical parameters on copy number of PIG-T
| Log (PIG-T/β-Actin) | Number | Mean | Median | 95%CI | p (Mann-Whitney test) | |
| Control | Male | 20 | 0.28 | 0.25 | (0.15, 0.41) | 0.042* |
| Female | 8 | 0.51 | 0.56 | (0.30, 0.72) | ||
| HNSCC | Male | 20 | 0.37 | 0.39 | (0.24, 0.49) | 0.222 |
| Female | 8 | 0.48 | 0.54 | (0.25, 0.71) | ||
| Control | White | 17 | 0.33 | 0.44 | (0.18, 0.47) | 0.621 |
| Other | 11 | 0.37 | 0.26 | (0.18, 0.57) | ||
| HNSCC | White | 17 | 0.40 | 0.45 | (0.29, 0.51) | 0.621 |
| Other | 11 | 0.40 | 0.41 | (0.16, 0.63) | ||
| Control | OC | 11 | 0.39 | 0.43 | (0.18, 0.61) | |
| HP | 3 | 0.46 | 0.54 | (-0.40, 1.33) | 0.938 | |
| L | 5 | 0.35 | 0.25 | (-0.01, 0.72) | 0.692 | |
| OP | 9 | 0.24 | 0.24 | (0.08, 0.40) | 0.239 | |
| HNSCC | OC | 11 | 0.52 | 0.52 | (0.34, 0.70) | |
| HP | 3 | 0.44 | 0.10 | (-0.34, 1.22) | 0.586 | |
| L | 5 | 0.20 | 0.19 | (-0.08, 0.48) | 0.027* | |
| OP | 9 | 0.35 | 0.41 | (0.17, 0.53) | 0.271 |
Table 4.
The effects of clinical parameters on copy number of PIG-U
| Log (PIG-U/β-Actin) | Number | Mean | Median | 95%CI | p (Mann-Whitney test) | |
| Control | Male | 20 | 0.56 | 0.43 | (0.39, 0.72) | 0.476 |
| Female | 8 | 0.66 | 0.68 | (0.41, 0.92) | ||
| HNSCC | Male | 20 | 0.65 | 0.62 | (0.51, 0.80) | 0.053 |
| Female | 8 | 0.95 | 0.97 | (0.66, 1.24) | ||
| Control | White | 17 | 0.56 | 0.43 | (0.40, 0.71) | 0.655 |
| Other | 11 | 0.64 | 0.59 | (0.38, 0.89) | ||
| HNSCC | White | 17 | 0.69 | 0.68 | (0.53, 0.86) | 0.466 |
| Other | 11 | 0.81 | 0.64 | (0.56, 1.06) | ||
| Control | OC | 11 | 0.57 | 0.41 | (0.29, 0.86) | |
| HP | 3 | 0.57 | 0.43 | (-0.05, 1.19) | 0.484 | |
| L | 5 | 0.62 | 0.59 | (0.27, 0.96) | 0.533 | |
| OP | 9 | 0.59 | 0.43 | (0.36, 0.82) | 0.470 | |
| HNSCC | OC | 11 | 0.83 | 0.80 | (0.57, 0.10) | |
| HP | 3 | 0.68 | 0.54 | (-0.45, 1.81) | 0.586 | |
| L | 5 | 0.67 | 0.57 | (0.19, 1.16) | 0.396 | |
| OP | 9 | 0.69 | 0.64 | (0.51, 0.86) | 0.425 |
Conclusion
Our data indicate that increasing of copy number and mRNA expression of GAA1 is characteristic for HNSCC and GAA1 play a role for this GPI anchor subunit in HNSCC.
Methods
Subjects
Genomic DNA samples from 28 HNSCC tissues and matching lymphocytes were subjected to QPCR analysis. RNA of tissue samples from 16 microdissected HNSCCs and 4 oral mucosal biopsies of healthy people were subjected to QRT-PCR. All samples were collected at the Department of Otolaryngology-Head and Neck Surgery, School of Medicine, The Johns Hopkins University, after appropriate approval was obtained from the Johns Hopkins institutional review board (IRB approval # 03-04-11-131d).
Cell lines and culture conditions
DNA and RNA from a virally transformed human cutaneous keratinocyte cell line (HaCaT) were used as standards for QPCR and QRT-PCR. The HaCaT cell line was grown in DMEM media supplemented with 10% FBS and 1% Penicillin-Streptomycin. Media components were obtained from Gibco Invitrogen Corporation (Carlsbad, CA) and cells were incubated at 37°C in an atmosphere of 5% CO2/95% relative humidity.
Quantitative PCR
A Perkin-Elmer/ABI 7900 thermocycler (Applied Biosystems, CA) was used to perform QPCR amplification for β-actin, PIG-U, PIG-T and Gaa1. β-actin primer and probe sequences used previously were employed in this study [24]. PIG-U[16], PIG-T and GAA1 primer and probe sequences were listed in Table 5. Primers were custom made and obtained from Invitrogen (Carlsbad, CA). All TaqMan probes (Applied Biosystems, Foster City, CA) were 5'-FAM and 3'-TAMRA labeled. PCR amplifications were carried out in buffer containing: 16.6 mM ammonium sulfate, 67 mM Trizma (pH 8.8), 2.5 mM MgCl2, 10 mM β-mercaptoethanol, 0.1% DMSO, 600 nM each of forward and reverse primers, 200 nM TaqMan probe, 0.6 units Platinum Taq polymerase, and 2% ROX reference dye. 500 picograms of DNA were used to amplify the mitochondrial regions whereas 10 ng were used to amplify β-actin. The real-time PCR reactions were performed in triplicate for each gene. Data analysis was performed using Microsoft Excel software.PIG-U, PIG-T and GAA1 were normalized using the corresponding β-actin signal.
Table 5.
PIG-U, PIG-T and GAA1 primer and probe sequences
| Gene | Forward Primer | Probe | Reverse Primer |
| PIG-U | AGCCCTCCAGCCAGAGTTA | CAGGCGAGTGCTTGGGCAGAAGA | ACTTGTGACCCTGGACTCGAA |
| PIG-T | GATCTGCCTCACGTGCACTGT | TGGCCGTGTGCTATGGCTCCTTC | AGGTTCGGGTGAGGAGATTGT |
| Gaa1 | CCGGGCTGGGACAGAGA | TCCCCAAGGACCCCATTCTGCC | CAGACACTCATTTATTTCCCCA |
Statistical analysis
The major statistical first endpoint in this study was the comparison of the difference in PIG-U, PIG-T and GAA1 quantitative DNA ratio between lymphocytes DNA and tumor tissue DNA in the HNSCC patients. The second endpoint in this study was PIG-U, PIG-T and GAA1 RNA expression differences between HNSCC patients and normal control subjects. Distributions of PIG-U, PIG-T and GAA1 DNA and RNA quantitative ratios were examined graphically using scatter plots and bar plots with logarithmic scales. Log transformation was chosen for these values and the difference taken for statistical analyses. Paired t-tests were used to determine if these changes were significantly different from each other. All statistical computations were performed using the SPSS system (SPSS, Chicago) and all p values reported are two-sided.
Authors' contributions
WWJ carried out cell cultures, QRT-PCR, QPCR, conceived the study, participated in its design, and drafted the manuscript. MZ analyzed the data and performed the statistical analysis. ZTZ and HLP critically revised the manuscript. WGJ and ZMG participated in the design of the study. DS conceived the study. BT conceived the study, participated in its design. JAC conceived the study, participated in its design and coordination, and contributed to the final drafting and critical revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This investigation was supported by Maryland Cigarette Restitution Fund and NIDCR 1R01DE015939-01 [JC]; NIH Head and Neck SPORE [WHW, WK, DS, and JC]; the Damon Runyon Cancer Research Foundation CI-#9 [JC].
Contributor Information
Wei-Wen Jiang, Email: wwjiang33@hotmail.com.
Marianna Zahurak, Email: mzahur1@jhmi.edu.
Zeng-Tong Zhou, Email: zhouzengtong@hotmail.com.
Hannah Lui Park, Email: hlui1@jhmi.edu.
Zhong-Min Guo, Email: zmguo2000@yahoo.com.
Guo-Jun Wu, Email: gwu10@jhmi.edu.
David Sidransky, Email: dsidrans@jhmi.edu.
Barry Trink, Email: btrink@jhmi.edu.
Joseph A Califano, Email: jcalifa@jhmi.edu.
References
- Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–18900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Ohishi K, Inoue N, Kinoshita T. PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. Embo J. 2001;20:4088–4098. doi: 10.1093/emboj/20.15.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Ohishi K, Kang JY, Tanaka S, Inoue N, Nishimura J, Maeda Y, Kinoshita T. Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol Biol Cell. 2003;14:1780–1789. doi: 10.1091/mbc.E02-12-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Egerton M, Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. Embo J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Yu J, Nagarajan S, Knez JJ, Udenfriend S, Chen R, Medof ME. The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc Natl Acad Sci USA. 1997;94:12580–12585. doi: 10.1073/pnas.94.23.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y, Komuro I, Chen R, Hosoda T, Mizuno T, Kudoh S, Georgescu SP, Medof ME, Yazaki Y. Molecular cloning of human homolog of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett. 1998;421:252–258. doi: 10.1016/S0014-5793(97)01576-7. [DOI] [PubMed] [Google Scholar]
- Ohishi K, Inoue N, Maeda Y, Takeda J, Riezman H, Kinoshita T. Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol Biol Cell. 2000;11:1523–1533. doi: 10.1091/mbc.11.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraering P, Imhof I, Meyer U, Strub JM, van Dorsselaer A, Vionnet C, Conzelmann A. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol Biol Cell. 2001;12:3295–3306. doi: 10.1091/mbc.12.10.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Nagamune K, Maeda Y, Kinoshita T. Two subunits of glycosylphosphatidylinositol transamidase, GPI8 and PIG-T, form a functionally important intermolecular disulfide bridge. J Biol Chem. 2003;278:13959–13967. doi: 10.1074/jbc.M300586200. [DOI] [PubMed] [Google Scholar]
- Nagamune K, Ohishi K, Ashida H, Hong Y, Hino J, Kangawa K, Inoue N, Maeda Y, Kinoshita T. GPI transamidase of Trypanosoma brucei has two previously uncharacterized (trypanosomatid transamidase 1 and 2) and three common subunits. Proc Natl Acad Sci USA. 2003;100:10682–10687. doi: 10.1073/pnas.1833260100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Vidugiriene J, Bangs JD, Menon AK. A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes. J Biol Chem. 1999;274:16479–16486. doi: 10.1074/jbc.274.23.16479. [DOI] [PubMed] [Google Scholar]
- Meyer U, Benghezal M, Imhof I, Conzelmann A. Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry. 2000;39:3461–3471. doi: 10.1021/bi992186o. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J, Vainauskas S, Johnson AE, Menon AK. Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment: photocrosslinking studies in a cell-free system. Eur J Biochem. 2001;268:2290–2300. doi: 10.1046/j.1432-1327.2001.02106.x. [DOI] [PubMed] [Google Scholar]
- Vainauskas S, Menon AK. A conserved proline in the last transmembrane segment of Gaa1 is required for glycosylphosphatidylinositol (GPI) recognition by GPI transamidase. J Biol Chem. 2004;279:6540–6545. doi: 10.1074/jbc.M312191200. [DOI] [PubMed] [Google Scholar]
- Guo Z, Linn JF, Wu G, Anzick SL, Eisenberger CF, Halachmi S, Cohen Y, Fomenkov A, Hoque MO, Okami K, Steiner G, Engles JM, Osada M, Moon C, Ratovitski E, Trent JM, Meltzer PS, Westra WH, Kiemeney LA, Schoenberg MP, Sidransky D, Trink B. CDC91L1 (PIG-U) is a newly discovered oncogene in human bladder cancer. Nat Med. 2004;10:374–381. doi: 10.1038/nm1010. [DOI] [PubMed] [Google Scholar]
- Wu G, Guo Z, Chatterjee A, Huang X, Rubin E, Wu F, Mambo E, Chang X, Osada M, Sook Kim M, Moon C, Califano JA, Ratovitski EA, Gollin SM, Sukumar S, Sidransky D, Trink B. Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 2006;66:9829–36. doi: 10.1158/0008-5472.CAN-06-0506. [DOI] [PubMed] [Google Scholar]
- Albertson DG. Profiling breast cancer by array CGH. Breast Cancer Res Treat. 2003;78:289–98. doi: 10.1023/A:1023025506386. [DOI] [PubMed] [Google Scholar]
- Ethier SP. Identifying and validating causal genetic alterations in human breast cancer. Breast Cancer Res Treat. 2003;78:285–7. doi: 10.1023/A:1023078722316. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Bergamo NA, da Silva Veiga LC, dos Reis PP, Nishimoto IN, Magrin J, Kowalski LP, Squire JA, Rogatto SR. Classic and molecular cytogenetic analyses reveal chromosomal gains and losses correlated with survival in head and neck cancer patients. Clin Cancer Res. 2005;11:621–631. [PubMed] [Google Scholar]
- Huang Q, Yu GP, McCormick SA, Mo J, Datta B, Mahimkar M, Lazarus P, Schaffer AA, Desper R, Schantz SP. Genetic differences detected by comparative genomic hybridization in head and neck squamous cell carcinomas from different tumor sites: construction of oncogenetic trees for tumor progression. Genes Chromosomes Cancer. 2002;34:224–233. doi: 10.1002/gcc.10062. [DOI] [PubMed] [Google Scholar]
- Liebig B, Brabletz T, Staege MS, Wulfanger J, Riemann D, Burdach S, Ballhausen WG. Forced expression of deltaN-TCF-1B in colon cancer derived cell lines is accompanied by the induction of CEACAM5/6 and mesothelin. Cancer Lett. 2005;223:159–167. doi: 10.1016/j.canlet.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Califano AJ. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–2491. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]