Abstract
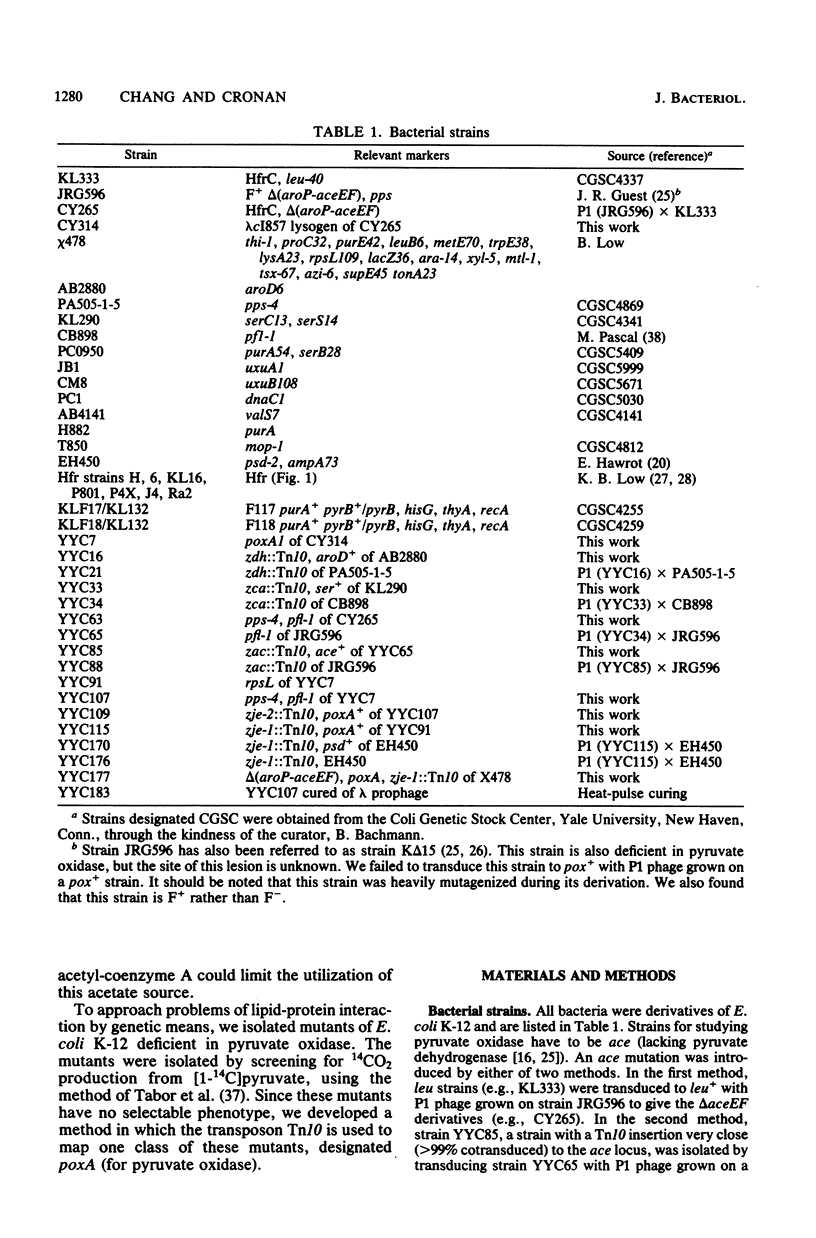
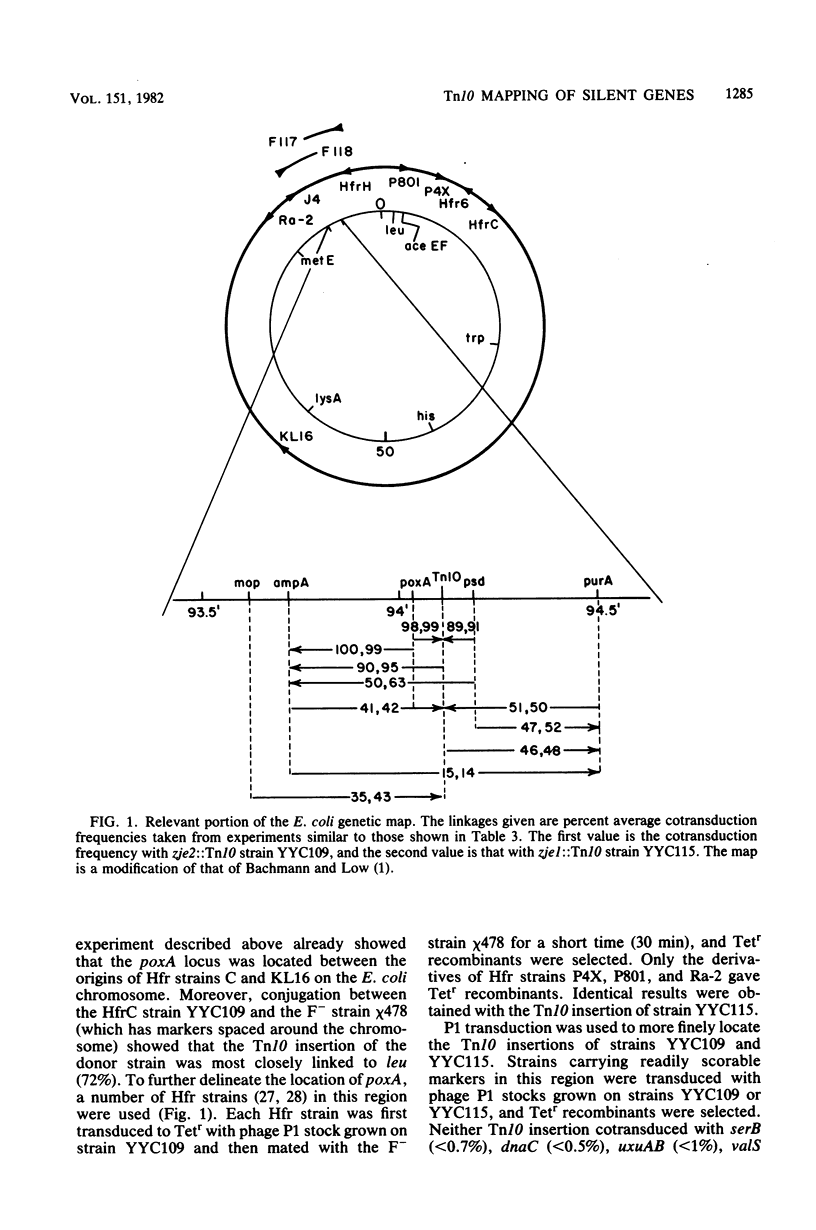
Mutants of Escherichia coli K-12 deficient in pyruvate oxidase were isolated by screening for the production of 14CO2 from [1-14C]pyruvate by the method of Tabor et al. (J. Bacteriol. 128:485-486, 1976). One of these lesions (designated poxA) decreased the pyruvate oxidase activity to 10 to 15% of the normal level but grew well. To map this nonselectable mutation, we isolated strains having transposon Tn10 inserted into the chromosome close to the poxA locus and mapped the transposon. These insertions were isolated by the following procedure: (i) pools of Tn10 insertions into the chromosomes of two different Hfr strains were prepared by transposition from a lambda::Tn10 vector; (ii) these Tn10-carrying strains were then mated with a poxA recipient strain, and tetracycline-resistant (Tetr) recombinants were selected; (iii) the Tetr recombinants were then screened for 14CO2 production from [1-14C]pyruvate. This method was shown to give a greater than 40-fold enrichment of insertions of Tn10 near the poxA gene as compared with transduction. Calculations indicate that a similar enrichment should be expected for other genes. The enrichment is due to the much greater map interval over which strong linkage between selected and unselected markers is found in conjugational crosses as compared with transductional crosses. The use of Hfr conjugative transfer allows isolation of transposon insertions closely linked to a nonselectable gene by scoring hundreds rather than thousands of colonies. Using a Tn10 insertion greater than 98% cotransduced with the poxA locus, we mapped the poxA gene on the E. coli genetic map. The poxA locus is located at 94 min, close to the psd locus. The clockwise gene order is ampA, poxA, psd, purA. The poxA mutation is recessive and appears to be a regulatory gene.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaven M. A., Wilcox G., Terpstra G. K. A microprocedure for the measurement of 14CO2 release from [14C]carboxyl-labeled amino acids. Anal Biochem. 1978 Feb;84(2):638–641. doi: 10.1016/0003-2697(78)90089-1. [DOI] [PubMed] [Google Scholar]
- Blake R., Hager L. P. Activation of pyruvate oxidase by monomeric and micellar amphiphiles. J Biol Chem. 1978 Mar 25;253(6):1963–1971. [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice C. B., Kornberg H. L. Location on the chromosome of Escherichia coli of a gene specifying phosphopyruvate synthase activity. Biochim Biophys Acta. 1967 Mar 22;136(2):412–414. doi: 10.1016/0304-4165(67)90094-3. [DOI] [PubMed] [Google Scholar]
- Clark D. P., Cronan J. E., Jr Acetaldehyde coenzyme A dehydrogenase of Escherichia coli. J Bacteriol. 1980 Oct;144(1):179–184. doi: 10.1128/jb.144.1.179-184.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D., Cronan J. E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980 Jan;141(1):177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr, Silbert D. F., Wulff D. L. Mapping of the fabA locus for unsaturated fatty acid biosynthesis in Escherichia coli. J Bacteriol. 1972 Oct;112(1):206–211. doi: 10.1128/jb.112.1.206-211.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C. C., Hager L. P. Crystalline pyruvate oxidase from Escherichia coli. 3. Phospholipid as an allosteric effector for the enzyme. J Biol Chem. 1971 Mar 25;246(6):1583–1589. [PubMed] [Google Scholar]
- Cunningham C. C., Hager L. P. Crystalline pyruvate oxidase from Escherichia coli. II. Activation by phospholipids. J Biol Chem. 1971 Mar 25;246(6):1575–1582. [PubMed] [Google Scholar]
- Cunningham C. C., Hager L. P. Reactivation of the lipid-depleted pyruvate oxidase system from Escherichia coli with cell envelope neutral lipids. J Biol Chem. 1975 Sep 25;250(18):7139–7146. [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harb Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- HENNING U. EIN REGULATIONSMECHANISMUS BEIM ABBAU DER BRENZTRAUBENSAEURE DURCH ESCHERICHIA COLI. Biochem Z. 1963 Jul 26;337:490–504. [PubMed] [Google Scholar]
- HENNING U., HERZ C. EIN STRUKTURGEN-KOMPLEX FUER DEN PYRUVAT-DEHYDROGENASE-KOMPLEX VON ESCHERICHIA COLI K 12. Z Vererbungsl. 1964 Nov 11;95:260–275. [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Havekes L. M., Hoekstra W. P. Characterization of an Escherichia coli K-12 F-Con-mutant. J Bacteriol. 1976 May;126(2):593–600. doi: 10.1128/jb.126.2.593-600.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrot E., Kennedy E. P. Conditional lethal phosphatidylserine decarboxylase mutants of Escherichia coli. Mapping of the structural gene for phosphatidylserine decarboxylase. Mol Gen Genet. 1976 Nov 17;148(3):271–279. doi: 10.1007/BF00332901. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol. 1978 May;106(1):103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and biochemical properties of deletion mutants. J Gen Microbiol. 1977 Apr;99(2):263–276. doi: 10.1099/00221287-99-2-263. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNKRES K. D., RICHARDS F. M. THE PURIFICATION AND PROPERTIES OF NEUROSPORA MALATE DEHYDROGENASE. Arch Biochem Biophys. 1965 Mar;109:466–479. doi: 10.1016/0003-9861(65)90391-7. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B. J., Masters M. The variation in frequency with which markers are transduced by phage P1 is primarily a result of discrimination during recombination. Mol Gen Genet. 1980;180(3):585–589. doi: 10.1007/BF00268064. [DOI] [PubMed] [Google Scholar]
- Russell P., Hager L. P., Gennis R. B. Characterization of the proteolytic activation of pyruvate oxidase. Control by specific ligands and by the flavin oxidation-reduction state. J Biol Chem. 1977 Nov 10;252(21):7877–7882. [PubMed] [Google Scholar]
- Russell P., Schrock H. L., Gennis R. B. Lipid activation and protease activation of pyruvate oxidase. Evidence suggesting a common site of interaction on the protein. J Biol Chem. 1977 Nov 10;252(21):7883–7887. [PubMed] [Google Scholar]
- Schrock H. L., Gennis R. B. High affinity lipid binding sites on the peripheral membrane enzyme pyruvate oxidase. Specific ligand effects on detergent binding. J Biol Chem. 1977 Sep 10;252(17):5990–5995. [PubMed] [Google Scholar]
- Tabor H., Tabor C. W., Hafner E. W. Convenient method for detecting 14CO2 in multiple samples: application to rapid screening for mutants. J Bacteriol. 1976 Oct;128(1):485–486. doi: 10.1128/jb.128.1.485-486.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Casse F., Chippaux M., Pascal M. C. A mutant of Escherichia coli deficient in pyruvate formate lyase. Mol Gen Genet. 1975 Nov 24;141(2):181–184. doi: 10.1007/BF00267683. [DOI] [PubMed] [Google Scholar]
- Verhoef C., de Haan P. G. Genetic recombination in Escherichia coli. I. Relation between linkage of unselected markers and map distance. Mutat Res. 1966 Apr;3(2):101–110. doi: 10.1016/0027-5107(66)90023-6. [DOI] [PubMed] [Google Scholar]
- Williams F. R., Hager L. P. Crystalline flavin pyruvate oxidase from Escherichia coli. I. Isolation and properties of the flavoprotein. Arch Biochem Biophys. 1966 Sep 26;116(1):168–176. doi: 10.1016/0003-9861(66)90025-7. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan P. G., Verhoef C. Genetic recombination in Escherichia coli. II. Calculation of incorporation frequency and relative map distance by recombinant analysis. Mutat Res. 1966 Apr;3(2):111–117. doi: 10.1016/0027-5107(66)90024-8. [DOI] [PubMed] [Google Scholar]



