Abstract
Bacteriophage P22, a podovirus infecting strains of Salmonella typhimurium, packages a 42 kbp genome using a headful mechanism. DNA translocation is accomplished by the phage terminase, a powerful molecular motor consisting of large and small subunits. Although many of the structural proteins of the P22 virion have been well characterized, little is known about the terminase subunits and their molecular mechanism of DNA translocation. We report here structural and assembly properties of ectopically expressed and highly purified terminase large and small subunits. The large subunit (gp2), which contains the nuclease and ATPase activities of terminase, exists as a stable monomer with an α/β fold. The small subunit (gp3), which recognizes DNA for packaging and may regulate gp2 activity, exhibits a highly α-helical secondary structure and self-associates to form a stable oligomeric ring in solution. For wildtype gp3, the ring contains nine subunits, as demonstrated by hydrodynamic measurements, electron microscopy and native mass spectrometry. We have also characterized a gp3 mutant (Ala 112 → Thr) that forms a ten subunit ring, despite a subunit fold indistinguishable from wildtype. Both the nonameric and decameric gp3 rings exhibit nonspecific DNA binding activity, and gp2 is able to bind strongly to the DNA/gp3 complex but not to DNA alone. We propose a scheme for the roles of P22 terminase large and small subunits in the recruitment and packaging of viral DNA and discuss the model in relation to proposals for terminase-driven DNA translocation in other phages.
Keywords: P22, terminase, structure, assembly, function, gp2, gp3, DNA binding, Raman spectroscopy, mass spectroscopy
INTRODUCTION
The assembly of many tailed dsDNA bacteriophages follows a common strategy in which the viral genome is packaged through a unique (portal) vertex of a preformed shell (procapsid) by a DNA translocase molecular motor.1 A similar packaging mechanism has been proposed for the Herpesviridae.2 A working model for packaging in the tailed phages, where overlength DNA is usually the packaging substrate, is as follows. The force-generating protein (called terminase) binds to a viral DNA concatemer at a specific site and cuts the duplex to create a packaging-competent DNA end that remains bound by terminase. The terminase-DNA complex then docks at the portal vertex, threads DNA into the portal channel and translocates a genome-length of DNA from the concatemer into the procapsid. DNA translocation is fueled by ATP hydrolysis and proceeds until the procapsid shell interior has been filled. The packaging process, which also transforms the architecture of the procapsid into that of the mature capsid, concludes with a second terminase-catalyzed cleavage of the DNA strands.
For some viruses the terminal DNA cleavage site is sequence specific.3 For others, like the Salmonella phage P22, little or no sequence specificity is apparent.4 In either case, terminase utilizes the unpackaged end of the concatemeric DNA in the subsequent cycle of genome packaging. Packaging of P22 DNA follows a prototypical headful mechanism.5-7 Between 2 and 12 sequential cycles of headful packaging can occur processively from a single DNA concatemer, depending upon the specific conditions within the cell.7-9 On average, 103.8 % of a P22 genome is packaged per procapsid, such that the DNA is 3.8% terminally redundant and circularly permuted.4 Thus, the terminal repeats can be utilized for DNA circularization by homologous recombination after injection into the host cell.
Active terminase enzymes are typically composed of small and large subunits. In the well studied cases of phages T4 and λ,1 the terminase small subunit is responsible for both the specific recognition and binding of DNA that initiate the packaging reaction. The requisite enzymatic activities of T4 and λ terminases reside in the large subunit. P22 terminase likewise comprises small and large subunits, which are the products, respectively, of phage genes 3 (gp3, 18.7 kDa) and 2 (gp2, 57.6 kDa).10-12 Both of the P22 terminase subunits are required for in vivo DNA packaging.13 They copurify from infected Salmonella cells and have been found bound to procapsids, although the binding sites are not yet known (M. Adams, E. Gilcrease & S. Casjens, unpublished observations).14,15 Neither subunit is present in the mature virion.16 Mutations that alter the sequence specificity of packaging initiation also alter the amino acid sequence of gp3, indicating that the small subunit is responsible for recognition of a DNA packaging initiation site (pac).17,18 Using directed mutational analysis, it has been shown that the unique pac site of the P22 genome is a 22 bp sequence within gene 3.19 The large subunit of P22 terminase (gp2) has not been studied genetically or biochemically, and the locations of its putative nuclease and ATPase active sites have not been established experimentally.20 Similarly, the small subunit of P22 terminase (gp3) has not been characterized in detail.
In this study we report structural and biophysical properties of the P22 terminase subunits. We also describe the formation of a stable oligomeric state for the small subunit in vitro and demonstrate that it adopts a novel ring morphology. The present results suggest a plausible in vivo molecular mechanism for recognition of the P22 pac site and initiation of packaging by terminase.
RESULTS
The gp2 subunit is a stable monomer in solution
The gp2 subunit of P22 terminase was cloned, expressed and purified as described below (Materials and Methods). To determine the most stable oligomeric state of gp2 in solution, we employed sedimentation velocity and sedimentation equilibrium measurements, which are capable of yielding shape-independent molecular masses. The gp2 peak fraction eluted from the SEC column was diluted to final concentrations of 0.18, 0.34 and 0.41 mg/mL and equilibrated at 4 °C overnight. Figure 2 shows typical results obtained from the sedimentation velocity experiments. The boundary sedimentation exhibits monophasic behavior, and a direct fit of the Lamm equation (eq. 1) reveals a single dominant major component (99.6%) and a single minor component (0.4%). Conversion of the distribution of the apparent sedimentation coefficient to molecular mass indicates that the major component is a 56.6 kDa protein corresponding to monomeric gp2. (The mass predicted from the gene 2 sequence is 57.6 kDa.21) The mass distribution of the minor species is very broad and shows a maximum at about 103 kDa. This minor species profile is reproducible and may correspond to a trace contaminant or a small fraction of gp2 dimers in equilibrium with the predominant monomer.
Figure 2.

Sedimentation velocity profile of gp2 in 0.1 M NaCl and 4 °C. Absorbances as a function of radial position were acquired at intervals of 90 s. (A) Data at intervals of 15 min (open circles) and corresponding residuals are shown for sedimentation at 42,000 rpm. The monophasic sedimentation boundaries suggest a single gp2 species. (B) Fitted distributions of the sedimentation coefficient (left) and molecular mass (right) using eqs. (1)-(4) indicate a gp2 monomer as the major species (99.6% of protein mass). The minor species (0.4%) is presumed to be a dimer.
The sedimentation velocity measurements were performed at 4, 20 and 30 °C and at different salt concentrations (including 0.1 M and 0.4 M NaCl) to assess possible changes in protein shape. The apparent sedimentation coefficients were transformed to standard conditions yielding s020,w = 4.0 ± 0.1 S (Table 1), consistent with previous estimates from sucrose gradient sedimentation measurements.15
Table 1.
Hydrodynamic parameters of P22 terminase large (gp2) and small (gp3) subunits from sedimentation velocity
| Subunit | State | T (°C) | [NaCl] | s* (S) | f/f0 | s020,w (S) |
|---|---|---|---|---|---|---|
| gp2 (wt) | Monomer | 4 | 0.1 | 2.4 | 1.4 | 3.9 |
| 10 | 0.4 | 2.9 | 1.3 | 4.1 | ||
| 20 | 0.1 | 3.9 | 1.4 | 4.0 | ||
| 30 | 0.1 | 5.0 | 1.3 | 4.1 | ||
| gp3 (wt) | Nonamer | 4 | 0.1 | 5.4 | 1.2 | 8.5 |
| gp3 (A112T) | Monomer | 4 | 0.1 | 1.1 | 1.5 | 1.8 |
| Decamer | 4 | 0.1 | 5.9 | 1.2 | 9.1 | |
| gp3 (A112T) | Decamer | 4 | 0.1 | 5.9 | 1.2 | 9.1 |
| n-mera | 4 | 0.1 | 6.7 | 1.2 | 10.5 | |
Dimer of the gp3 (A112T) decamer. See text.
The oligomeric state of gp2 was also determined by sedimentation equilibrium measurements. The gp2 peak fraction noted above was diluted to 0.16, 0.24 and 0.32 mg/mL and spun until equilibrium was achieved at three successively increasing speeds. The equilibrium was checked by comparison of the measured profiles at three hour intervals. The last profile was then used for fitting to individual species models. Typical sedimentation equilibrium results and fits are shown in Figure 3. A single species model was used to fit the equilibrium profiles of the three samples at the three speeds. A fitted molecular mass of 57.4 ± 0.4 kDa with a confidence limit of 95% was obtained, which is in excellent agreement with the calculated monomer mass (57.6 kDa). If two species are included in the model, the distribution of fitted residuals does not improve significantly. Thus, both approaches indicate a stable monomeric state for the plasmid expressed gp2 subunit in solution.
Figure 3.

Sedimentation equilibrium profiles of gp2 acquired at 280 nm. Samples were spun to equilibrium at speeds of 20,000 (red triangles), 25,000 (blue squares) and 30,000 (green circles) rpm in a Beckman An-Ti 60 rotor. The equilibrium profiles were fitted simultaneously by using a single-species, one-state model in Sedphat software using eq. (5).
The gp3 subunit forms a stable oligomer in solution
The wildtype and A112T mutant variants of gp3 were cloned, expressed, purified and shown to be fully functional, as described in the Materials and Methods section. The studies reported in this section were performed on both gp3 variants and were generally indistinguishable, except where specifically noted otherwise.
The final step in gp3 purification was performed on a prepGrade HPLC-SEC column. The protein eluted in two fractions, one corresponding to an apparent monomer and the other to a high-order oligomer (Figure 1B). Both gp3 variants exhibited a salt-dependent oligomerization reaction that favors the oligomer state at physiological salt conditions (∼0.1 M NaCl). Removal of imidazole from the apparently monomeric material leads to rapid gp3 self-association to the more stable oligomer. The rate of oligomerization of the material from the monomer peak at fixed NaCl concentration was also found to increase with increasing gp3 concentration (data not shown). Once formed, the gp3 oligomer (wildtype or A112T) is highly stable and no dissociation was observed when assayed on an SEC column several days after the initial elution. The elution volume of the oligomeric species occurs reproducibly within the range 150-180 mL depending upon the buffer and contains only the gp3 protein (Figure 1B). We also observed that the A112T monomer is significantly more stable than the wildtype monomer, as judged by the relative intensities of their corresponding SEC elution peaks at the same salt conditions. Accordingly, detailed studies of the gp3 monomer were restricted to the A112T mutant.
Figure 1.
(A) Elution of the P22 terminase large subunit (gp2) by size exclusion chromatography (SEC). Labeled arrows near the gp2 peak maximum correspond to lanes 3-7 of the accompanying SDS-PAGE gel (inset). Also shown in the inset are molecular weight standards (lane 1) and the load fraction (lane 2). (B) Elution of the P22 terminase small subunit (gp3) by SEC. Labeled arrows near the oligomer peak correspond to lanes 2-6 and near the monomer peak to lanes 7 and 8 of the accompanying SDS-PAGE gel (inset). Also shown in the inset are molecular weight standards (lane 1).
We employed sedimentation experiments to characterize the hydrodynamic properties of the putative monomeric and oligomeric species and to assess the stoichiometry of the oligomer. Peak fractions were equilibrated overnight prior to sedimentation runs. Sedimentation of the apparent gp3 monomer manifested biphasic behavior and fit a distribution of the sedimentation coefficient and molecular mass that is diagnostic of two species (Figure 4A). The first (s* = 1.1 S, 19 kDa) corresponds to the monomer and the second (s* = 5.5 S, 210 kDa) to the oligomer. The integrated intensity of the sedimentation coefficient distribution is proportional to the protein loading concentration. Comparison of the integrated intensities of monomer and oligomer fractions indicates that 45% of the gp3 monomer oligomerized during overnight storage at 4 °C. Because the algorithm of the fit assumes that the frictional ratio is the same for both species (which is unlikely to be the case), the calculated molecular mass is considered to be approximate and no definitive conclusion can be drawn about the three-dimensional shape of the monomer. Analyses of later sedimentation profiles, which were uncomplicated by contributions from rapidly sedimenting oligomers, gave the same results. Use of the empirical Teller method and a prolate ellipsoid approximation22 to evaluate the hydrodynamic shape of the gp3 monomer from the frictional ratio suggested an elongated shape with axial ratio of 5:1.
Figure 4.

Sedimentation velocity profiles of gp3 in 0.1 M NaCl at 4 °C. Absorbances as a function of radial position were acquired at intervals of 120 s. (A) Data at intervals of 20 min (open circles) and corresponding residuals are shown for sedimentation at 60,000 rpm. The biphasic sedimentation boundaries suggest two distinct gp3 assembly states. In this experiment the 100% monomeric gp3 sample was stored at 4°C for 24 h prior to the start of the sedimentation run. (B) Fitted distributions of the sedimentation coefficient (left) and molecular mass (right) using eqs. (1)-(4) indicate a gp3 monomer and a single multimer as the prevalent species. (C) Data as in (A) for sedimentation of 100% oligomeric gp3 at 35,000 rpm. (D) Data as in (B) for oligomeric gp3.
The sedimentation profile of the gp3 oligomer is virtually monophasic (Figure 4C). The fitted distributions yield a sedimentation coefficient (transformed to standard conditions) of s020,w = 9.1 ± 0.1 S for the dominant oligomer, a very small contribution (∼5%) from a minor oligomeric species (s020,w = 10.5 ± 0.2 S) and a negligible contribution (<1%) from monomer. The parameters determined from the sedimentation velocity experiments on both wildtype and A112T gp3 are summarized in Table 1.
Sedimentation equilibrium analyses were also performed on gp3. The fraction eluting from the SEC column as the oligomer was diluted to several different concentrations and each was dialyzed overnight against 10 mM tris (pH 7.8), 0.1 M NaCl. Samples were spun until equilibrium was reached at different speeds and equilibrium profiles were measured at 235, 250 and 280 nm (Figure 5). For wildtype gp3, satisfactory fitting of the experimental profiles required two distinct species, a major component with global minimum at 174 ± 3 kDa corresponding to nonamer (n = 9) and a minor component with n = 18. Although this global fit favors a nonamer, individual fits of profiles from independently prepared samples suggest oligomer sizes within the range n = 9 ± 1. For A112T gp3, the major component exhibited a global minimum at 188 ± 3 kDa corresponding to a decamer (n = 10 ± 1) and a minor component with n = 20 ± 2.
Figure 5.

Sedimentation equilibrium profiles of gp3 acquired at 280, 250 and 235 nm, as labeled. Samples were spun to equilibrium at speeds of 9,000 (red triangles), 11,000 (blue squares) and 13,000 (green circles) rpm in a Beckman An-Ti 60 rotor. The equilibrium profiles were fitted simultaneously by using a single-species, two-state model in Sedphat software using eq. (5).
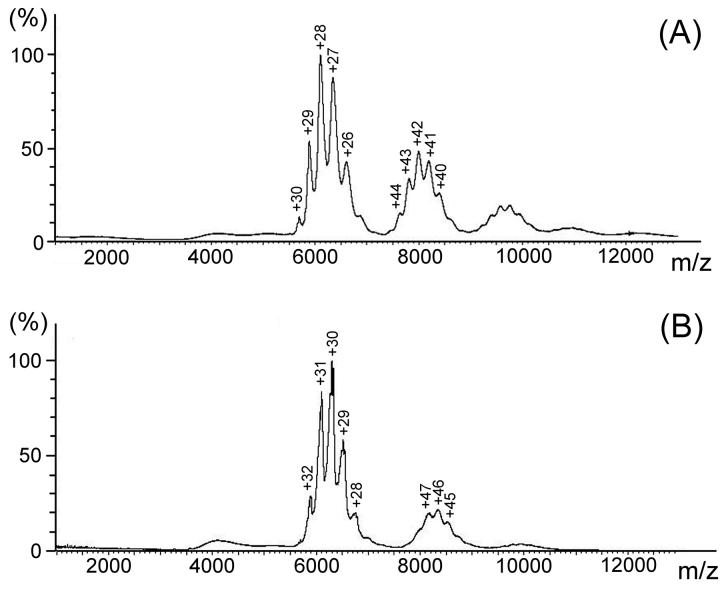
To verify the oligomeric states of the wild type and A112T gp3 oligomers observed in analytical centrifugation experiments we measured the masses of the noncovalent gp3 complexes. Electrospray ionization preserves noncovalent protein complexes and the TOF mass analyzer has a suitably wide mass range.23 The acquired spectra contained major and satellite peak clusters of charged proteins (Figure 6). Data smoothing and deconvolution show that the major clusters correspond to the same oligomers identified in the sedimentation analyses, i.e. masses of 170 ± 1 kDa (n = 9; theoretical value 170.41 kDa) and 189 ± 1 kDa (n = 10; theoretical value 189.61) for wildtype and A112T, respectively. In each case the principal satellite cluster corresponds to a dimer of the major cluster, again consistent with the sedimentation results. Because the masses of the expressed gp3 subunits after thrombin cleavage of His6 are 18,930 and 18,961 Da, respectively, the ESI-TOF spectra demonstrate a nonamer as the major oligomeric state of wildtype gp3 and a decamer as the major oligomeric state of A112T gp3. The dimer of the major oligomer is the prevalent satellite species in each case.
Figure 6.
ESI-TOF spectra of wildtype (A) and A112T mutant (B) gp3 oligomers. Deconvolutions of the major peak clusters in the respective spectra indicate oligomer masses of 170 ± 1 and 189 ± 1 kDa, respectively, for wildtype and mutant oligomers, which correspond to nonameric and decameric assemblies. Deconvolution of the secondary cluster in each spectrum indicates a dimer of the respective oligomer. Peak labels indicate protonation states.
Visualization of gp3 oligomers by transmission electron microscopy reveals a ring structure
Specimens of gp3 were stained with uranyl acetate or uranyl formate and visualized by transmission electron microscopy (TEM) (Figures 7A and 7B). Refinement of the data by means of in-plane translational and rotational operations24-26 reveals distinct ring morphologies for the two variants (Figures 7C and 7D). The averaged images, which were reproduced in independent selections and exhibited variance within the noise level, confirm the nonameric (wildtype) and decameric (A112T) oligomerization states, demonstrate the symmetries of the rings and provide some detail of subunit domains. In these averaged images, it appears that the subunits project radially and manifest the elongated shape deduced from sedimentation velocity analyses. The images of Figure 7 suggest that the inner annulus comprises subunit oligomerization domains and the spikes at high radius serve other functions. The overall outer diameter of each ring is approximately 11 nm, while the diameter of the central hole is approximately 2 nm.
Figure 7.
Electron micrographs of negatively stained wildtype (A) and A112T mutant (B) gp3 oligomers at minimal dose and 75,000X magnification. Ring morphologies of the wildtype (C) and mutant (D) oligomers revealed in two dimensions by reference-free averaging of 450 single particles in each case. The dimensions of the wildtype ring (∼10 nm outer diameter, ∼2 nm hole diameter) are about 10% smaller than those of the mutant ring. The apparent difference in intensity of the inner annuli of wildtype and mutant rings is attributed to staining artifacts.
Secondary and tertiary structures of the terminase large and small subunits
Purified gp2 was brought to 0.7 mg/mL in aqueous solution using a Centricon molecular filtration device (Millipore) of 50-kDa molecular weight cutoff. At this protein concentration gp2 remains soluble and monomeric, as confirmed by analytical SEC (data not shown). The Raman spectrum of the solution monomer is shown in Figure 8A. The wavenumber values, intensities and half-widths of bands in the amide I (1650-1680 cm−1) and amide III (1230-1320 cm−1) spectral intervals, as well as the intensity of the α-helix marker at 937 cm−1, indicate an α/β fold in the native gp2 molecule.27-29 Decomposition of the amide I band profile using the reference spectra method of Berjot et al.30 provides the following semi-quantitative assessment of secondary structure distribution in gp2: 36% β-strand, 36% α-helix and 28% irregular structure (Table 2). Among the contributions of specific side chains to the Raman signature of gp2,29 the results show that the average tyrosine phenoxyl participates principally as a strong hydrogen bond donor and the average tryptophan indolyl ring occurs in a hydrophilic environment.
Figure 8.
(A) Raman spectrum (532 nm excitation) of the P22 terminase large subunit at 0.7 mg/mL in 10 mM Tris, 0.4 M NaCl. Amide I (1664 cm−1), amide III (1241 cm−1) and related markers (900-950 cm−1) indicate an α/β-fold. See Table 2. (B) Raman spectra (532 nm) of the P22 terminase small subunit [wildtype (top) and A112T (middle), each at 20 mg/mL in 10 mM Tris, 0.1 M NaCl]. Amide marker bands (1657 and 1253 cm−1) indicate α-helix as the principal secondary structure component of gp3. See Table 2. The Raman difference spectrum computed between the wildtype and mutant proteins (bottom) shows bands diagnostic of the Ala → Thr substitution and confirms the invariance of the gp3 subunit fold to the A112T mutation. The inset at upper left shows the CD profile of the gp3 monomer (open circles) and oligomer (open squares).
Table 2.
Secondary structures of P22 terminase large (gp2) and small (gp3) subunitsa
| Subunit | State | α-Helix (%) | β-Strand (%) | Other (%) |
|---|---|---|---|---|
| gp2 | Monomer | 36 ± 4 | 36 ± 2 | 28 ± 2 |
| gp3 (wt) | Nonamer | 40 ± 2 | 22 ± 2 | 38 ± 3 |
| gp3 (A112T) | Monomer | b (38) | b (15) | b (47)c |
| gp3 (A112T) | Decamer | 44 ± 2 (46) | 24 ± 2 (19) | 32 ± 3 (35)d |
Data are from analysis of Raman spectra, except for values given in parentheses, which are from analysis of CD spectra. See text.
Data unavailable from Raman spectra due to rapid self-assembly of monomer.
CD analysis indicates 17% turns and 30% undefined structure.
CD analysis indicates 12% turns and 23% undefined structure.
Purified gp3 was examined for secondary and tertiary structural features by both Raman (oligomer) and CD (monomer) methods. For Raman experiments the oligomer fraction separated by SEC was brought to 20 mg/mL using a Centricon device with a 50-kDa cutoff. The oligomerization states of wildtype and mutant proteins were confirmed by SEC before and after acquiring the spectral data shown in Figure 8B. The Raman signature of gp3 was unchanged over the protein concentration range 3-50 mg/mL. Amide I (1657 cm−1) and amide III (1253 cm−1) peaks demonstrate that the native gp3 fold of the oligomer subunit is principally α-helical. Decomposition of the amide I profile30 yields 42% α-helix, 23% β-strand and 35% irregular structures (Table 2, averages of wildtype and mutant gp3). Raman markers bands of tyrosine and tryptophan residues of gp329,31-34 indicate that these aromatics are distributed roughly equally between solvent accessible and inaccessible sites.
To assess the secondary structure of gp3 in the monomeric state we obtained the CD spectrum of the protein at very low concentration (∼0.1 mg/mL in 10 mM phosphate, 0.1 M NaF, pH 7.8). Spectral data were recorded on both the monomer and oligomer in the interval 190-240 nm and converted to mean residue ellipticity as shown in the inset of Figure 8B. Deconvolution of these CD profiles by the CDSSTR method using the largest available database (http://www.cryst.bbk.ac.uk/cdweb/html/home.html35-38 gives the secondary structure distributions listed in Table 2. The results suggest a small increase of ordered secondary structure with gp3 ring formation (cf. last two rows of Table 2), as has been observed for oligomerizations of other viral proteins.39-41 It has been proposed that the small subunits of other phage terminases may utilize a region in the center of the peptide sequence to effect intersubunit association via a coiled-coil.42 Enhanced α-helicity with gp3 oligomerization is consistent with coiled-coil formation. Although the locus of the conformational change is not known, structure prediction algorithms suggest that residues 85-105 of the gp3 sequence may have a propensity for coiled-coil formation.42
Preliminary assessment of terminase subunit interactions and binding to DNA
The P22 terminase small subunit is responsible for recognition of a 22-bp pac site of the viral DNA concatemer and for selection of the specific residues within the pac site at which packaging is initiated. Conversely, the large subunits of other phage terminases carry nuclease and ATPase activities.18,19 Both subunits might therefore be expected to interact with DNA. We studied the DNA binding capability of gp3, alone and in combination with gp2, to a 50-bp double-stranded (ds) target sequence incorporating the pac site. As a control, DNA-binding capability of gp3 was also studied using a 50-bp dsDNA target incorporating a base permutation within the pac site. (See Materials and Methods.) The results of such an electromobility shift experiment are shown in Figure 9. When no DNA is present, the gp3 oligomer (lane 8) and gp2 (lane 9) run as rather wide bands. However, when incubated together at room temperature for 30 min prior to electrophoresis, the gp3 and gp2 bands both become less intense and a large amount of material remains at the top of the resolving gel (lane 10). This indicates that the subunits interact with one another and form a larger complex that cannot enter the gel. When the DNA target is mixed with protein in binding buffer, incubated at room temperature for 30 min and loaded onto a 4-20% TBE polyacrylamide electrophoresis gel, some of the DNA shows retarded mobility that coincides with a protein band. Comparison of lanes 2 and 11 indicates that gp2 alone co-migrates with a small amount of stained DNA, and comparison of lanes 3 and 11 shows a strong band of DNA co-migrating with gp3. These results suggest that both gp2 and gp3 have DNA binding capability in vitro.
Figure 9.

Native gel electromobility shift assay of gp2 and gp3 binding to 50-bp DNA. DNA and proteins are stained in green and red, respectively. (A) Robust DNA binding occurs only for monomeric (lane 3) and oligomeric (lane 5) gp3. When gp2 is added concomitantly with gp3 (lanes 4 and 6) or prior to mixing with DNA (lane 8), virtually no DNA binding is observed. Oligomeric gp3 appears to have a higher affinity than monomeric gp3 for DNA (cf. lanes 3 and 5). (B) Binding of gp2 with gp3 is demonstrated by the high stain intensity at the entry to the gel (lane 10). Minimal DNA binding is seen for free gp2 (lanes 2 and 5), while oligomeric gp3 manifests significant binding to DNA. Addition of gp2 to the DNA/gp3 complex results in the formation of a higher order (ternary) complex (lanes 4 and 7). No differences in terminase subunit binding are observed between DNAs either containing or lacking the pac site.
In addition, when both proteins are present, a substantial amount of DNA co-migrates with the gp2/gp3 complex at the top of the gels (compare lanes 4 and 11). The fact that bound DNA does not substantially change protein mobilities (compare lanes 2, 3 and 4 to lanes 8, 9 and 10) is not surprising because of the presumably large sizes of the complexes and the difficulty in resolving high-molecular-weight complexes using this type of gel. The observed DNA binding does not appear to be sequence-specific, since identical results are found with dsDNAs containing pac and permuted pac sequences (compare lanes 2, 3 and 4 with lanes 5, 6 and 7). Again, this is not unexpected owing to the facts that the gp2 nuclease activity is not sequence-specific4 and that the reactants were likely present at concentrations high enough to enable non-specific binding.
This assay illustrates two interesting properties of P22 terminase that have not been reported previously. First, mixing of the purified terminase subunits leads spontaneously to the formation of a large complex. Second, both subunits appear to have non-sequence specific DNA-binding capabilities in vitro, even though sequence-specific binding is a requisite for packaging initiation in vivo.19 These properties are being further explored.
DISCUSSION
All tailed dsDNA phages package a genome by the action of protein complexes called terminases, which are built from large and small subunits. Generally, these molecular machines exploit their small subunit for DNA recognition and their large subunit for DNA translocation via ATP hydrolysis.1 In the case of phage P22, where the substrate for DNA packaging is the overlength concatemer generated by rolling circle replication, the terminase large subunit (gp2) employs a headful mechanism6,43,44 to package the DNA into a preformed procapsid structure.12 As in most phages, the P22 terminase large subunit also exhibits the DNase activity that is required to cleave the unpackaged portion of the concatemer from the DNA-filled capsid (D. Nemecek, S. R. Casjens & G. J. Thomas, Jr., unpublished results).
Terminases of different phages vary greatly in their DNA recognition and cleavage modes. For example, different kinds of packaging initiation mechanisms and chromosome end cleavages are known.12 Nevertheless, the large subunits of different phage terminases exhibit some sequence homology, consistent with the overall similarities of their enzymatic activities.45,46 Among terminases bearing nuclease activity, two types have been studied in some detail: those with a cohesive end mechanism (e.g. phage λ) and those with a headful packaging mechanism (e.g. phages T4 and SPP1). Unfortunately, in all cases the stoichiometric and structural aspects of the translocase and nuclease activities during in vivo packaging remain poorly understood.1
The small subunits of different phage terminases are even more highly diverse than their corresponding large subunits, and accordingly, may function with even greater diversity.47 The terminase large subunit of phage P22 belongs to a “sequence type” subfamily that differs from that of phage T4 and is considered only a distant relative of the phage SPP1 terminase (about 14% amino acid sequence identity, despite being in the same subfamily).45 Terminases of the P22 and SPP1 subfamily are very common among known phages, yet very limited information is available on their structures and molecular mechanisms of function. In this report we have characterized key properties of the P22 terminase small and large subunits using ectopically expressed proteins purified to better than 99%.
The large subunit - gp2
Our sedimentation experiments show that gp2 remains monomeric over a wide range of temperature (4-30 °C) and salt concentration (0.1-0.4 M NaCl). The calculated frictional ratio is very similar at all conditions. We conclude that the protein fold of monomeric gp2 is quite stable to both temperature and ionic strength. Raman spectral analyses of gp2 indicate a secondary structure comprising about 36% α-helix and 36% β-strand. It is interesting to note that although gp2 and the terminase large subunit of phage T4 (gp17) have very little sequence similarity, the available structure information on gp17 (ATPase domain) indicates also a stable monomer with a prevalence of β-strand over α-helix secondary structure.48 The same applies to the terminase large subunits of phage λ (gpA)49 and SPP1 (G2P).50 Thus, although more than one large subunit may be required for terminase function during the packaging event,48-50 in every phage so far examined the terminase large subunit is monomeric in the absence of its small subunit and DNA substrate.
Previously studied terminase large subunits contain two domains. These exhibit the two distinct enzymatic activities – DNA cleavage (DNase) and ATP hydrolysis (ATPase) – that are required at different stages of DNA packaging.48,51,52 Because purified gp2 also exhibits these activities (D. Nemecek, S. R. Casjens & G. J. Thomas, Jr., unpublished results), it is presumed to contain similar corresponding domains. The fold of the putative ATPase domain of gp2 is also likely to be similar to that of gp17 of phage T4, for which an X-ray crystal structure has recently been determined.48 Unfortunately, the complete 3D structure is not known for any full-length terminase large subunit; nor is the 3D structure known for the isolated DNA cleavage domain of any terminase large subunit. It seems likely, however, that the two distinct domains are connected by a hinge-like region to permit switching between the different conformations required at different stages of DNA packaging. In phage SPP1 for example, it has been proposed that DNA packaging is mediated by a conformational change within the terminase large subunit (G2P) that selectively controls DNase and ATPase functions.53
The small subunit - gp3
The previously studied terminase small subunits of phages T4 (gp16), SPP1 (G1P), SF6 (G1P) and λ (gpNu1) generally lack sequence similarity with one another or with the small subunit (gp3) of phage P22 terminase. Nevertheless, all share the interesting characteristics of oligomerization, DNA binding and terminase large subunit binding. T4 gp16 forms ∼8 nm diameter rings estimated to contain about 8 subunits;54 SPP1 G1P forms short ∼9 nm diameter cylinders of about 10 subunits55 and λ gpNu1 forms dimers and perhaps larger oligomers56,57 as well as a more complex large-plus-small subunit hetero-oligomer.58 These properties are considered here in relation to our findings on gp3.
(1) Oligomerization
We have found that P22 gp3 spontaneously and completely assembles into stable oligomeric rings of either 9 (wildtype) or 10 (A112T) subunits. Despite the different number of subunits, the rings of the two gp3 variants are similar in overall morphology, stability and hydrodynamic properties. The rings exhibit distinct symmetries with outwardly projecting spikes (Figure 7). Together with the supporting data of mass spectrometry and sedimentation analysis, these results represent the first definitive demonstration of both the stoichiometry and gross structure of terminase small subunit assembly. The findings here are reminiscent of the SPP1 G1P and T4 gp16 structures, although in those cases the subunit stoichiometries and arrangements are not precisely known.
Both the wildtype and mutant gp3 rings exhibit a central hole that is compatible with the cross-sectional diameter of double-stranded B DNA (∼2 nm). Both also exhibit a spike-to-spike spacing at high subunit radius that is compatible with the helical pitch of B DNA (∼3.5 nm).59 These findings immediately suggest two possible modes of gp3/DNA recognition: (i) threading of the duplex through the central channel of the annulus, and (ii) alignment of DNA grooves with the spike-like density at the outer circumference of the ring. It is not possible at this stage of analysis to cite direct evidence favoring one or the other mode of gp3/DNA recognition, although there is circumstantial evidence to support both options. Thus, the large footprint of SPP1 G1P on its DNA binding site55 and the tandem binding of λ gpNu158,60,61 imply a role for circumferential DNA binding. On the other hand, the similarity in approximate hole diameters of the gp3 ring (∼2.0 nm) and P22 portal entrance (∼2.5 nm)62 suggest at least the plausibility of central channel binding.
The in vitro assembly of monomeric gp3 into a ring is not dependent on the presence of either DNA or gp2, although the kinetics of ring formation can be retarded by high salt (data not shown). This implies that electrostatic interactions play a role in stabilizing the subunit/subunit interface of the gp3 ring.
It is also interesting to note that the self-assembly of gp3 is dramatically affected by the A112T mutation, which leads not only to a different order oligomer but also to a retardation of the rate of ring formation vis-à-vis the wildtype protein. This suggests that hydrophobic bonding may also play a role at the intersubunit interface. At present, it is not known whether the subunit interface includes residue 112 or is altered indirectly through the effect of the mutation on a neighboring domain. It is clear, however, that the A112T mutant is fully functional in vivo in spite of its distinctive oligomer size in vitro. We cannot exclude the possibility that the wildtype (nonameric) and mutant (decameric) gp3 rings achieve their distinct sizes as a consequence of factors, such as assembly kinetics, that have not been controlled in vitro but are subject to precise regulation in vivo. Importantly, the oligomeric size of the wildtype gp3 protein in vivo remains an open question. It may well be possible that the A112T mutant gp3 is less susceptible than the wildtype gp3 to an artifactual nonameric assembly in vitro, although there is no reason at present to suspect this is the case.
The subunits of the gp3 ring assembly are rich in α-helical secondary structure, irrespective of the A112T mutation (Table 2). Nevertheless, about one-fourth of the amino acid residues of the ring subunit and about one-third of those of the monomer subunit do not participate in the formation of repetitive secondary structure (either α-helix or β-strand), implying ample opportunity for gp3 pleiomorphism. Plasticity in the gp3 structure may in fact be essential to its involvement in such diverse functional roles as self-assembly, gp2 recognition and binding, pac site recognition, procapsid portal docking, non-specific DNA recognition and DNA translocation.
In phage λ it has been shown that the terminase large and small subunit heteromultimer as well as the large subunit can bind to the procapsid-incorporated portal.53,63-66 At present it is not known whether the gp3 ring assembly can bind to the portal vertex of the P22 procapsid. We have found no evidence of gp3 binding to in vitro assembled portal dodecamers in the absence of the procapsid (data not shown). Future experiments will address directly the procapsid binding affinity of the gp3 ring alone and in the presence of gp2. If binding were to occur, an apparent mismatch of symmetries of the gp3 ring (9- or 10-fold) and portal (12-fold) would be required.
(2) DNA binding
In other phages the terminase small subunits have been implicated in DNA binding in vitro, including λ gpNu1 with the cos site,67 SPP1 G1P and SF6 G1P with pac,55,68 and T4 gp16 with the packaging initiation site.54 Genetic analysis strongly implicates P22 gp3 in pac site recognition and initiation of packaging in vivo.17,18,69,70 We show here (Figure 9) that gp3 binds DNA irrespective of the pac site, thus demonstrating only sequence nonspecific binding. Specificity for pac site binding has not yet been demonstrated. It should be noted, however, that sequence-specific DNA recognition occurs in vivo only once in a series of 4 ± 1 genome packaging events per concatemer substrate.12 In addition, gp3 is required in the in vitro packaging reaction developed by Schmieger and colleagues, even though a pac site is not required for the DNA to be packaged.71,72 Thus, it seems reasonable that gp3 might have both pac-specific and nonspecific DNA binding capabilities.
(3) Large subunit binding
All terminase small subunits interact with their corresponding large subunit. Thus, T4 gp16 modulates the ATPase activity and DNA packaging of gp1754,73-76 and λ gpNu1 forms a complex with gpA to regulate packaging activity.77,78 We show here that gp3 binds the P22 terminase large subunit gp2 and this interaction will be further characterized in future studies.
Because atomic level structural information is presently available only for the DNA binding domain of λ gpNu1,58 it is not known whether the various terminase small subunits are highly diverged (beyond the point of having recognizable amino acid sequence similarity) or whether they may have different evolutionary origins. Despite the similarities noted above among previously examined terminase small subunits, it is not yet clear whether they perform each distinct role in a similar or disparate manner. The apparent failure of gpNu1 to form homo-multimeric rings in vitro, the differences in the locations of putative DNA-binding and homo-assembly domains, as well as the differences in large subunit binding properties all point to significant mechanistic differences. More detailed structure-function relationships of both large and small subunits of terminases are required. These objectives are being addressed for the P22 terminase subunits in our ongoing studies.
In summary, we propose in Figure 10 a pathway for terminase mediated packaging of the P22 viral genome. This scheme is based upon the results of the present biochemical and biophysical studies of the recombinantly expressed P22 terminase large and small subunits and existing genetic studies.12 After P22 infection, a viral DNA concatemer is synthesized in the host cell by rolling circle replication and the individual viral proteins are expressed. Procapsid assembly (not shown in Figure 10) requires the coat (gp5), scaffolding (gp8), portal (gp1) and ejection (gp7, gp16, gp20) factors.10-12 The expressed gp3 recognizes and binds the DNA at or near the pac site, either as a monomer or as a preassembled ring. The gp3/DNA complex subsequently binds the terminase large subunit (gp2), and the ternary gp2/gp3/DNA complex cleaves DNA at the pac site. The resulting activated terminase complex (DNA translocase), which is presumed to accommodate the DNA double strand through the central hole of the gp3 ring, docks at the portal vertex of the procapsid where DNA packaging ensues via the phosphohydrolysis of ATP. After headful packaging and shell expansion have been completed, the terminase complex again cleaves DNA, undocks from the portal, and becomes activated for a subsequent round of headful packaging. The cycle continues until either the concatemer is depleted of sufficient genomic DNA to accomplish headful packaging or procapsids become unavailable for docking. The present studies provide new insights into the steps of gp3 self-assembly, gp3/DNA binding and gp2/gp3/DNA complex formation. The results imply two feasible DNA recognition schemes for the gp3 ring assembly, both of which will be further evaluated in future work.
Figure 10.

Proposed pathway for in vivo recognition, binding and packaging of the P22 viral genome under the action of the terminase molecular motor. Roles of the terminase large (gp2) and small (gp3) subunits in the packaging mechanism are based on the present findings and the body of previous work summarized in the review of Casjens & Weigele.12 Clockwise from upper left: gp3 monomer (a), or its pre-assembled ring (b), binds to concatemeric DNA to generate a complex consisting of a gp3 ring at or near the pac site. Two possible pac-site recognition schemes are depicted, viz. central annulus binding (1a), and peripheral ring binding (1b). One or several gp2 molecules are recruited to the gp3/DNA complex, resulting in cleavage of the concatemer and activation of the terminase complex, gp2/gp3/DNA (2). The activated complex docks at the procapsid portal vertex and initiates the ATPase-driven DNA packaging reaction (3). Headful packaging (∼104% of the genome length) and capsid expansion ensue (4), followed by cutting of DNA strands and release of the gp2/gp3/DNA complex from the filled capsid (5). The capsid portal is rendered competent for portal closure and tailspike attachment (not shown), while the gp2/gp3/DNA complex is reactivated (6) for another cycle of procapsid docking and headful DNA packaging (7).
MATERIALS AND METHODS
Plasmid and phage constructs for expression of gp2 and gp3 subunits
P22 gene 2 was cloned into the pET-21b vector (Novagen, Darmstadt, Germany)79 between its NdeI and BamHI sites after PCR amplification with the oligonucleotides 5′-CACGATGCAT ATGGTGGAACTGGACGCGATTC and 5′-CGGAGGATCCTCTTATCGTCCTCTGCGTACTGG to give plasmid PP293. The expressed gp2 subunit has no His6 tag and contains an additional valine residue immediately after the N-terminal methionine. The subunit mass predicted from the gene sequence (57,691 Da) was confirmed by mass spectrometry (57,693 Da) of the denatured protein.
P22 gene 3 was similarly cloned into the pET-15b vector between NdeI and BamHI sites after PCR amplification with the oligonucleotides 5′-CACGATGCATATGGCGGCACCAAAGGGCAACCG and 5′-CGGAGGATCCTCAAGAATCGCGTCCAGTTCC. The resulting plasmid PP295 carried a single fortuitous mutation that resulted in an A112T amino acid change relative to wildtype gp3. Codon 112 was altered to the wildtype gene 3 sequence (plasmid PP353) by the Quickchange® (Stratagene, La Jolla, CA) site directed mutagenesis strategy using the oligonucleotides 5′-AAATTCTCTGGCGCAGCCGCTGACC and 5′-GGTCAGCGGCTGCGCCAGAGAATTTC. The sequences and masses of the gp3 mutant (measured 18,965 ± 5 vs. theoretical 18,961 Da) and wildtype subunits (18,934 ± 5 vs. 18,931 Da) were confirmed by denaturing mass spectroscopy after thrombin cleavage of the His6tags. Plasmid construction was carried out in E. coli NF182980 and E. coli XL2-Blue MRF (Stratagene, La Jolla, CA).
The gene 3 double nonsense mutant phage strain P22 c1-7 3−amH317 3−amN6 was constructed by co-infecting cells with the two c1-7, single amber mutant strains (the nonsense mutations are in codons 45 and 53, respectively)10 and screening progeny plaques on Salmonella DB700481 for inability to recombine to wildtype with either parental phage. P22 c1-7 3−A112T was constructed as follows. P22 c1-7 3−amH317 3−amN6 was used to infect Salmonella DB7000 carrying plasmid PP295 (above) and DB7000 carrying no plasmid.81 Less than 103/mL amber+ phage (measured by their ability to make plaques on the sup° strain DB7000) were present in the lysate of the host with no plasmid, while 2.5 × 109/mL amber+ phage were present in the lysate of the host carrying PP295, indicating that essentially all of these amber+ phage had lost their nonsense mutations through recombination with the PP295 plasmid. The resulting amber+ phages from the DB7000/PP295 infection made normal sized plaques. Four of these were single plaque purified and propagated on DB7000, and screened by direct sequencing for the codon 112 mutation (GCC → ACC) using purified whole phage DNA as template. The mutation was carried by one out of four amber+ plaques that were tested.
Because we had expression clones of both wildtype and mutant (A112T) gp3, we wished to determine whether the A112T protein was functional. To test its functionality, we recombined the A112T mutation into the chromosome of P22 c1-7 as described above and found that the resulting phage produce normal appearing plaques. Parallel infections of Salmonella DB7000 in logarithmic growth phase at 2 × 108 cells/mL (37°C in LB broth) with P22 c1-7 and P22 c1-7 3−A112T at m.o.i. = 5 gave lysate titers of 1.5 × 1011/mL and 4.3 × 1011/mL, respectively. We conclude that the A112T mutation in gp3 does not negatively impact its function in P22 lytic growth. (Both the wildtype protein and the A112T mutant of gp3 were examined in parallel in most of the studies described above. See Results section. Unless indicated otherwise, the reported results refer specifically to the wildtype protein.)
Expression and purification of gp2 and gp3 subunits
BL21(DE3) pLysS E. coli cells82 were transformed with the expression plasmids and grown at 37 °C overnight on LB agar plates with 50 μg/mL ampicilin and 34 μg/mL chloramphenicol. A single colony was inoculated and grown overnight in LB with both antibiotics and 3 mg/mL glucose. The overnight culture (40 mL) was inoculated into fresh LB (2 L) containing both antibiotics and 3 mg/mL glucose and shaken at 37 °C to OD600 ∼ 0.6. Expression of gp2 or gp3 was induced with 0.5 mM IPTG, and the cell cultures were incubated for 5 hours at 14 °C (gp2) or 28 °C (gp3). Finally, the cells were pelleted and stored at −80°C.
Frozen cell pellets from the 2 L culture (above) were thawed on ice in 20 mL of 10 mM Tris, 5 mM imidazole, pH 8.0 buffer. Lysozyme, PMSF, CHAPS and MgCl2 were added to the homogenized cell solution to total concentrations of 1 mg/mL, 0.5 mM, 10 mM and 2 mM, respectively. Cellular nucleic acids were digested in a mixture of nucleases (Benzonase®, Merck Chemicals Ltd, Darmstadt, Germany). Proteases were inhibited using a standard cocktail tablet (Roche, Basel, Switzerland). The cells were sonicated (eight 20 s bursts) and the resulting lysate was centrifuged at 17,400 × g for 1 h. The initial pellet of lysate contained more than half of the over-expressed protein, as estimated by SDS-PAGE. This pellet was again resuspended in buffer and spun at 17,400 × g for 1 h. The final supernatant was loaded onto a Ni-affinity HPLC column pre-equilibrated with buffer N (10 mM Tris, 0.5 M NaCl, pH 8.0). The column was then washed with buffer N containing 20 mM imidazole and gp2 was eluted with buffer N containing 100 mM imidazole. The latter fraction was loaded onto a HiLoad 26-60 Superdex200 prepGrade SEC column (GE Healthcare, Chalfont St. Giles, United Kingdom), which had been equilibrated with buffers containing either high (10 mM Tris, 0.4 M NaCl, pH 7.6) or low salt (10 mM Tris, 0.1 M NaCl, 10 mM MgCl2, pH 7.6). We found that gp2, despite the absence of a His6 tag, bound sufficiently strongly to the Ni-affinity column to separate it from most other proteins. On the other hand, chromatography separations based upon either ion exchange (Q-column) or hydrophobic interaction (FF-column) were ineffective in separating gp2 from contaminating proteins. A final purification step consisting of gel filtration chromatography yielded greater than 99% pure gp2 (Figure 1A).
His6-tagged gp3 was extracted from stored cells and separated from debris as described above for gp2. The supernatant was loaded onto a Ni-affinity column preequilibrated with buffer N. Other proteins were washed out with buffer N containing 100 mM imidazole, while gp3 eluted with buffer N containing 1 M imidazole. The His6 tag was removed by incubation of a solution of one unit of biotinylated thrombin (Novagen, San Diego, CA) per mg of gp3 for 15 h at 22°C. The thrombin was removed by streptavidine agarose and the cleaved gp3 subunits were isolated from His6 and unreacted gp3 by Ni-affinity column chromatography. Gel filtration chromatography of the cleaved protein yielded greater than 99% pure gp3 (Figure 1B). The same purification protocols were followed for wildtype and mutant (A112T) variants of gp3, each expressed from its corresponding plasmid.
Sedimentation velocity
Sedimentation velocity experiments were performed on a Beckman XL-A ultracentrifuge using the An60-Ti rotor. The selected rotor speed and absorbance wavelength varied according to the molecular mass and concentration of the sample. Typically, data were collected at 90 s intervals for 10 h, until complete sedimentation was achieved. The protein sample and a corresponding reference were loaded into the assembled cells of a two-channel Epon (charcoal-filled) centerpiece. To avoid thermal convection, the sample-loaded rotor was incubated at the desired temperature prior to each experiment and the temperature was maintained constant until the end of the sedimentation run. The experimental data were analyzed and fitted using SEDFIT software (http://www.analyticalultracentrifugation.com/sedfit.htm), which allows direct boundary modeling of the Lamm equation (eq. 1) and provides the distribution of apparent sedimentation coefficients (eq. 2) by maximum entropy regularization.83
| (1) |
| (2) |
Here, A(r,t) is the absorbance at radius r and time t, c(s) is the concentration of species with sedimentation coefficient s ∈ (s,s + ds), χ(s,D(s),r,t) is the Lamm equation solution and ε represents background and noise contributions. The sedimentation coefficient distribution c(s) is converted into a molar mass distribution c(M) by use of the Stokes-Einstein formula (eq. 3) and Svedberg equation (eq. 4), which are interrelated through the average friction ratio f/f0.
| (3) |
| (4) |
Rs denotes the radius of a sphere with given molar mass M and partial specific volume ν̃, η is the buffer viscosity, ρ is the buffer density, kB is the Boltzmann constant, R is the gas constant and T is the absolute temperature. The average friction ratio has minimal effect on the sedimentation coefficient distribution, but significantly affects the molecular mass distribution. Therefore this analysis provides only estimates of molecular mass. Input parameters such as the partial specific volume, buffer density and viscosity were determined using Sednterp software (http://www.jphilo.mailway.com/). It also allows the equivalent radius and protein asymmetry to be calculated. A detail description and an overview of the derived equations can be found in the Sednterp documentation on-line.
Sedimentation equilibrium
Protein and corresponding reference samples were loaded into the sedimentation equilibrium cell with a six-channel Epon centerpiece. Three different sample concentrations were loaded simultaneously with decreasing concentration farther from the rotation axis. Data were collected either at 233 nm, 250 nm or 280 nm, depending on the sample concentration. Each experiment was conducted at three consecutively increasing rotor speeds, according to the determined apparent sedimentation coefficient from sedimentation velocity. Equilibrium was reached in 12 to18 hours and then 3 subsequent scans were acquired in 3 h intervals to confirm equilibrium. Only the last scan at a given speed was used for the data analysis in Sedphat software (http://www.analyticalultracentrifugation.com/sedphat/sedphat.htm). A species analysis model was used to fit the sedimentation equilibrium data to a sum of N exponential terms, each corresponding to one species n, according to eq. 5
| (5) |
where A(r) is the total absorbance at radius r, c(r0)n is the concentration at a reference radius r0, εn is the extinction coefficient, Mn is the molar mass, νn is the partial specific volume of species n, d is the optical path, ρ is the buffer density and ω is the angular velocity.
Transmission electron microscopy
Oligomeric gp3 (from the prepGrade SEC separation; see RESULTS) was diluted to an equivalent subunit concentration of ∼2 μM and negatively stained with 2% uranyl acetate or uranyl formate solution. Electron micrographs were taken on a JEOL 1200-EX transmission electron microscope using a minimal dose procedure and 75,000 magnification. The negative films were scanned at 5 Å/pixel resolution and about 600 symmetric particles selected in Eman/Boxer software. The particles were aligned and averaged in a reference-independent method using Spider/Web (http://www.wadsworth.org/spider_doc/spider/docs/spider.html). Thus, the initial reference was randomly chosen and iteratively improved from the image set. The final image of the gp3 oligomer was refined from a selection of 450 particles with correlation coefficient better than 0.4 with respect to the first average image.
Raman spectroscopy
Solution Raman spectra were excited at 532 nm using a NdY:VO4 laser (Coherent Verdi 5, Palo Alto, CA) with ∼100 mW of radiant power at the sample. Raman scattering was collected in the 90° geometry and analyzed using a Spex 500M single spectrograph (JY Horiba, Edison, NJ) equipped with a 1200 groove/mm grating. The scattered light was detected using a liquid nitrogen cooled charge-coupled-device with 2000 × 800 pixels. Samples were contained in sealed 1 mm (inner diameter) glass capillaries, which were thermostated during data collection protocols.84 Raman spectra were collected for up to 12 h from the lowest concentration solutions (∼2 mg/mL) or 2 h from the highest concentration solutions (∼50 mg/mL). The Raman spectrum of crystalline gp3 was excited at 785 nm using an Invictus NIR diode laser (Kaiser Optical, Ann Arbor, MI). The single crystal was mounted in the thermostated platform of a Raman microprobe system (Kaiser) incorporating a Holospec VPT spectrograph with transmissive grating and liquid nitrogen cooled detector. Further details of instrumentation and sample handling for Raman spectroscopy have been described.85
UV spectroscopy
UV spectra were recorded on a Cary 3E UV-Vis spectrometer (Varian, Palo Alto, CA). Protein concentrations were determined with an estimated accuracy of 5% using calculated extinction coefficients (ε280) for gp2 and gp3 (100270 and 33460 M−1·cm−1, respectively).86 Concentrations of synthesized oligonucleotides were determined using A260 conversion factors provided by Sigma Genosys, The Woodlands, TX.
CD spectroscopy
Far UV circular dichroism spectra were recorded on a PiStar-180 spectrometer (Applied Photophysics, Leatherhead, Surrey, U.K.) from samples in 1 mm quartz cuvettes thermostated at 25 °C. Protein concentration was typically 0.1 mg/mL in 10 mM sodium phosphate buffer (pH 7.8), 0.1 M NaF. Data were collected from 180 nm to 260 nm at 0.2 nm intervals with entrance and exit slit-widths of 2 nm. Collection time at each spectral point was 12.5 s. The raw spectra in millidegrees [θ]obs were converted to mean residue ellipticity (θ) using
| (6) |
where M denotes protein molar mass, Nr is the number of residues in the protein, c is the protein concentration in mg/mL and l is the optical path. Secondary structure was determined by spectral deconvolution based on the CDSSTR algorithm and a basis data set of 48 spectra in the 190-240 nm region.35-38
Polyacrylamide gel electrophoresis (PAGE) of proteins and nucleic acids
Aliquots of gp2 and gp3 from various steps in the purification protocols were suspended in denaturing buffer (66 mM Tris, pH = 6.8, 26% glycerol, 2% SDS, 0.011% bromphenol blue and 5 % β-mercaptoethanol). The samples were separated by electrophoresis on 15% Tris-Glycine PAGEr® Gold Precast polyacrylamide gels (Cambrex, Rockland, ME) and stained with Coomassie Blue R-250.
In the electromobility shift assays, purified gp2 and gp3 were mixed with annealed 50-bp dsDNA to a final volume of 60 μL in 10 mM Tris (pH 7.8), 0.1 M NaCl, 10 mM MgCl2 and 1 mM ATP. In these experiments we used the following DNA sequences: 5′-ATCGCCTGAGAGAAGATTTATCTGAAGTCGTTACGCGAGCAGAACAGGTC (the 22 bases of the pac site are underlined) and 5′-GTTACATGGCGAACTAACCGAGGCCGGCTCCGGATAGTGATTATATAGAA (a control sequence lacking the pac site) with their respective complementary strands. The latter sequence, which was designed by permutation of the pac site, contains the same overall base composition as the former. Final concentrations of DNA (0.4 μM), gp2 (3.5 μM) and gp3 (7.0 μM) were the same in all mixtures. DNA/gp2 and DNA/gp3 reaction mixtures were incubated for 30 min at 22 °C. A mixture of DNA with both subunits was made by successively adding gp2 to DNA/gp3 and incubating at 30 min intervals. All mixtures were loaded into prerun 4-20% TBE PAGEr® Gold Precast gels (Cambrex). Separated DNA and proteins were stained with SYBR® Green and SYPRO® Ruby fluorescent dyes (Invitrogen, Eugene, OR). The stained gels were scanned on a Typhoon 9400 Scanner (Amersham Biosciences, Piscataway, NJ) using 532 nm excitation.
Mass spectrometry
Masses of the recombinant subunits in the denatured form were determined using a liquid-chromatography coupled ESI-TOF mass spectrometer equipped with a Z-spray source (Waters Micromass LCT, Framingham, MA). The samples were injected into a 20 μL loop and separated using a linear gradient (5-95% acetonitrile containing 0.1% formic acid) with a flow rate of 36 μL/min. Spectra were acquired and calibrated in the range 200 < m/z < 1650. Mass spectroscopy of native gp3 assemblies was carried out on samples in 50 mM ammonium acetate (pH 7.8) buffer. Spectra were acquired in the range 1000 < m/z < 11500 by directly infusing samples (∼50 μM monomer concentration) into the spectrometer at a flow rate of 0.5 μL/min via a 39 μm (inner diameter) fused silica capillary. Source temperature was 60 °C. Collisional cooling, which facilitated the focusing and transmission of ionized gp3 assemblies in vacuo, was achieved by increasing the source pressure to approximately 7.0 mbar.87,88 The capillary voltage and the sample cone voltage were 2600V and 200V, respectively. Pressure in the TOF tube was maintained at 2.0 × 10−6 mbar.23 Mass calibration was accomplished by directly infusing aqueous CsI (50 mg/mL) at 0.5 μL/min. Charge states and masses were obtained from the spectral data using MassLynx version 4.0 software (Waters). All assigned masses were verified manually.
ACKNOWLEDGMENTS
D.N. thanks UMKC colleagues Drs. James Benevides and Stacy Overman for assistance in all stages of this work. The authors thank Dr. Edward Gogol (UMKC) for assistance with electron microscopy and Drs. Gabe Lander and John E. Johnson (Scripps Research Institute) for sharing experimental results prior to publication. This work was supported by NIH Grant GM50776 to GJT and Grant GM47980 to PEP.
ABBREVIATIONS AND SYMBOLS
- IPTG
isopropyl-β-D-thiogalactopyranoside
- Tris
tris[hydroxymethyl]aminomethane
- PMSF
phenylmethylsulfonyl fluoride
- EDTA
ethylenediaminetetraacetic acid disodium salt
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- HPLC-SEC
high performance liquid chromatography-size exclusion chromatography
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TEM
transmission electron microscopy
- CD
circular dichroism
- ds
double stranded
- TBE buffer
89 mM tris-borate and 2 mM EDTA, pH 8.3
- m.o.i.
multiplicity of infection
- ESI-TOF
electrospray ionization time-of-flight
- m/z
mass to charge ratio of ion detected in mass spectrometer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Catalano CE. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Landes Bioscience / Eurekah.com and Kluwer Academic / Plenum Publishers; Georgetown, TX and New York: 2005. [Google Scholar]
- 2.Brown JC, McVoy MA, Homa FL. Packaging DNA into Herpesvirus Capsids. In: Holzenburg A, Bogner E, editors. Structure-Function Relationships of Human Pathogenic Viruses. Springer US; Cambridge, MA: 2002. pp. 111–153. [Google Scholar]
- 3.Wieczorek DJ, Feiss M. Defining cosQ, the site required for termination of bacteriophage lambda DNA packaging. Genetics. 2001;158:495–506. doi: 10.1093/genetics/158.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol. 1988;199:467–474. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- 5.Streisinger G, Emrich J, Stahl MM. Chromosome structure in phage T4, III. Terminal redundancy and length determination. Proc. Natl. Acad. Sci. U. S. A. 1967;57:292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tye BK, Chan RK, Botstein D. Packaging of an oversize transducing genome by Salmonella phage P22. J. Mol. Biol. 1974;85:485–500. doi: 10.1016/0022-2836(74)90311-8. [DOI] [PubMed] [Google Scholar]
- 7.Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- 8.Casjens S, Huang WM. Initiation of sequential packaging of bacteriophage P22 DNA. J. Mol. Biol. 1982;157:287–298. doi: 10.1016/0022-2836(82)90235-2. [DOI] [PubMed] [Google Scholar]
- 9.Adams MB, Hayden M, Casjens S. On the sequential packaging of bacteriophage P22 DNA. J. Virol. 1983;46:673–677. doi: 10.1128/jvi.46.2.673-677.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 11.Backhaus H. DNA packaging initiation of Salmonella bacteriophage P22: determination of cut sites within the DNA sequence coding for gene 3. J. Virol. 1985;55:458–465. doi: 10.1128/jvi.55.2.458-465.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens S, Weigele P. DNA packaging by bacteriophage P22. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Landes Bioscience / Eurekah.com and Kluwer Academic / Plenum Publishers; Georgetown, TX and New York: 2005. pp. 80–88. [Google Scholar]
- 13.Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- 14.Poteete AR, King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977;76:725–739. doi: 10.1016/0042-6822(77)90254-9. [DOI] [PubMed] [Google Scholar]
- 15.Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–573. doi: 10.1016/0042-6822(79)90509-9. [DOI] [PubMed] [Google Scholar]
- 16.Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- 17.Casjens S, Huang WM, Hayden M, Parr R. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J. Mol. Biol. 1987;194:411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- 18.Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992;227:1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell MS, Rao VB. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J. Biol. Chem. 2006;281:518–527. doi: 10.1074/jbc.M507719200. [DOI] [PubMed] [Google Scholar]
- 21.Casjens S, Eppler K, Parr R, Poteete AR. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology. 1989;171:588–598. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 22.Teller DC. Accessible area, packing volumes and interaction surfaces of globular proteins. Nature. 1976;260:729–731. doi: 10.1038/260729a0. [DOI] [PubMed] [Google Scholar]
- 23.Poliakov A, van Duijn E, Lander G, Fu CY, Johnson JE, Prevelige PE, Jr., Heck AJ. Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J. Struct. Biol. 2007;157:371–383. doi: 10.1016/j.jsb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, Shimkin B, Dowse H. SPIDER-A modular software system for electron image processing. Ultramicroscopy. 1981;6:343–358. [Google Scholar]
- 25.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 26.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 27.Thomas GJ., Jr. Raman spectroscopy of protein and nucleic acid assemblies. Annu. Rev. Biophys. Biomol. Struct. 1999;28:1–27. doi: 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Tuma R, Thomas GJ., Jr. Raman spectroscopy of viruses. In: Chalmers JM, Griffiths PR, editors. Handbook of Vibrational Spectroscopy, Volume 5, Applications in Life, Pharmaceutical and Natural Sciences. Vol. 5. John Wiley & Sons; Chichester, U.K.: 2002. pp. 3519–3535. [Google Scholar]
- 29.Benevides JM, Overman SA, Thomas GJ., Jr. Raman spectroscopy of proteins. In: Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. John Wiley & Sons; New York: 2005. pp. 17.8.1–17.8.35. [DOI] [PubMed] [Google Scholar]
- 30.Berjot M, Marx J, Alix AJP. Determination of the secondary structure of proteins from the Raman amide I band: The reference intensity profiles method. J. Raman Spectrosc. 1987;18:289–300. [Google Scholar]
- 31.Siamwiza MN, Lord RC, Chen MC, Takamatsu T, Harada I, Matsuura H, Shimanouchi T. Interpretation of the doublet at 850 and 830 cm−1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry. 1975;14:4870–4876. doi: 10.1021/bi00693a014. [DOI] [PubMed] [Google Scholar]
- 32.Arp Z, Autrey D, Laane J, Overman SA, Thomas GJ., Jr. Tyrosine Raman signatures of the filamentous virus Ff are diagnostic of non-hydrogen-bonded phenoxyls: Demonstration by Raman and infrared spectroscopy of p-cresol vapor. Biochemistry. 2001;40:2522–2529. doi: 10.1021/bi0023753. [DOI] [PubMed] [Google Scholar]
- 33.Harada I, Miura T, Takeuchi H. Origin of the doublet at 1360 and 1340 cm−1 in the Raman spectra of tryptophan and related compounds. Spectrochim. Acta. 1986;42A:307–312. [Google Scholar]
- 34.Miura T, Takeuchi H, Harada I. Tryptophan Raman bands sensitive to hydrogen bonding and side-chain conformation. J. Raman Spectrosc. 1989;20:667–671. [Google Scholar]
- 35.Manavalan P, Johnson WC., Jr. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 36.Compton LA, Johnson WC., Jr. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 38.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuma R, Prevelige PE, Jr., Thomas GJ., Jr. Structural transitions in the scaffolding and coat proteins of P22 virus during assembly and disassembly. Biochemistry. 1996;35:4619–4627. doi: 10.1021/bi952793l. [DOI] [PubMed] [Google Scholar]
- 40.Tuma R, Tsuruta H, Benevides JM, Prevelige PE, Jr., Thomas GJ., Jr. Characterization of subunit structural changes accompanying assembly of the bacteriophage P22 procapsid. Biochemistry. 2001;40:665–674. doi: 10.1021/bi001965y. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Casado A, Moore SD, Prevelige PE, Jr., Thomas GJ., Jr. Structure of bacteriophage P22 portal protein in relation to assembly: investigation by Raman spectroscopy. Biochemistry. 2001;40:13583–13591. doi: 10.1021/bi0110488. [DOI] [PubMed] [Google Scholar]
- 42.Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: insights into DNA packaging initiation and assembly of packaging motor. J. Mol. Biol. 2006;358:67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 43.Tye BK, Huberman JA, Botstein D. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 1974;85:501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- 44.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol. Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005;187:1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell MS, Rao VB. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology. 2004;321:217–221. doi: 10.1016/j.virol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Maluf NK, Feiss M. Virus DNA translocation: progress towards a first ascent of mount pretty difficult. Mol. Microbiol. 2006;61:1–4. doi: 10.1111/j.1365-2958.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- 50.Gual A, Camacho AG, Alonso JC. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 2000;275:35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 51.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage T4 terminase. J. Biol. Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 52.Gual A, Camacho AG, Alonso JC. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. Journal of Biological Chemistry. 2000;275:35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 53.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J. Biol. Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 54.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 55.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 56.Parris W, Rubinchik S, Yang YC, Gold M. A new procedure for the purification of the bacteriophage λ terminase enzyme and its subunits. Properties of gene product A, the large subunit. J. Biol. Chem. 1994;269:13564–13574. [PubMed] [Google Scholar]
- 57.Yang Q, de Beer T, Woods L, Meyer JD, Manning MC, Overduin M, Catalano CE. Cloning, expression, and characterization of a DNA binding domain of gpNu1, a phage λ DNA packaging protein. Biochemistry. 1999;38:465–477. doi: 10.1021/bi981271d. [DOI] [PubMed] [Google Scholar]
- 58.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 59.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. The anatomy of A-, B-, and Z-DNA. Science. 1982;216:475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 60.Feiss M, Kobayashi I, Widner W. Separate sites for binding and nicking of bacteriophage λ DNA by terminase. Proc. Natl. Acad. Sci. U. S. A. 1983;80:955–959. doi: 10.1073/pnas.80.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shinder G, Gold M. The Nul subunit of bacteriophage λ terminase binds to specific sites in cos DNA. J. Virol. 1988;62:387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 63.Frackman S, Siegele DA, Feiss M. A functional domain of bacteriophage λ terminase for prohead binding. J. Mol. Biol. 1984;180:283–300. doi: 10.1016/s0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- 64.Sippy J, Feiss M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage λ terminase. J. Bacteriol. 1992;174:850–856. doi: 10.1128/jb.174.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 1995;245:141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 66.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 67.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 68.Chai S, Kruft V, Alonso JC. Analysis of the Bacillus subtilis bacteriophages SPP1 and SF6 gene 1 product: a protein involved in the initiation of headful packaging. Virology. 1994;202:930–939. doi: 10.1006/viro.1994.1415. [DOI] [PubMed] [Google Scholar]
- 69.Raj AS, Raj AY, Schmieger H. Phage genes involved in the formation generalized transducing particles in Salmonella--Phage P22. Mol. Gen. Genet. 1974;135:175–184. doi: 10.1007/BF00264784. [DOI] [PubMed] [Google Scholar]
- 70.Laski F, Jackson EN. Maturation cleavage of bacteriophage P22 DNA in the absence of DNA packaging. J. Mol. Biol. 1982;154:565–579. doi: 10.1016/s0022-2836(82)80015-6. [DOI] [PubMed] [Google Scholar]
- 71.Strobel E, Behnisch W, Schmieger H. In vitro packaging of mature phage DNA by Salmonella phage P22. Virology. 1984;133:158–165. doi: 10.1016/0042-6822(84)90434-3. [DOI] [PubMed] [Google Scholar]
- 72.Behnisch W, Schmieger H. In vitro packaging of plasmid DNA oligomers by Salmonella phage P22: independence of the pac site, and evidence for the termination cut in vitro. Virology. 1985;144:310–317. doi: 10.1016/0042-6822(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 73.Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J. Mol. Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 74.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 75.Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 76.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 2006;281:21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 77.Tomka MA, Catalano CE. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage λ. J. Biol. Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 78.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage λ terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 79.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 80.Schwarz JJ, Berget PB. The isolation and sequence of missense and nonsense mutations in the cloned bacteriophage P22 tailspike protein gene. Genetics. 1989;121:635–649. doi: 10.1093/genetics/121.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winston F, Botstein D, Miller JH. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 1979;137:433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 83.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas GJ, Jr., Barylski J. Thermostating capillary cells for a laser-Raman spectrophotometer. Appl. Spectrosc. 1970;24:463–464. [Google Scholar]
- 85.Tsuboi M, Benevides JM, Thomas GJ., Jr. The complex of ethidium bromide with genomic DNA: structure analysis by polarized Raman spectroscopy. Biophys. J. 2007;92:928–934. doi: 10.1529/biophysj.106.093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data [published erratum appears in Anal Biochem 1990 Sep;189(2):283] Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 87.Tahallah N, Pinkse M, Maier CS, Heck AJ. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 2001;15:596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- 88.Chernushevich IV, Thomson BA. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]