Abstract
The preS1 surface antigen of hepatitis B virus (HBV) is known to play an important role in the initial attachment of HBV to hepatocytes. We have characterized structural features of the full-length preS1 using heteronuclear NMR methods and discovered that this 119-residue protein is inherently unstructured without a unique tertiary structure under a nondenaturing condition. Yet, combination of various NMR parameters shows that the preS1 contains “pre-structured” domains broadly covering its functional domains. The most prominent domain is formed by residues 27–45 and overlaps with the putative hepatocyte-binding domain (HBD) encompassing residues 21–47, within which two well-defined pre-structured motifs, formed by Pro32–Ala36 and Pro41–Phe45 are found. Additional, somewhat less prominent, pre-structured motifs are also formed by residues 11–18, 22–25, 37–40, and 46–50. Overall results suggest that the preS1 is a natively unstructured protein (NUP) whose N-terminal 50 residues, populated with multiple pre-structured motifs, contribute critically to hepatocyte binding.
Keywords: hepatitis B virus, preS1, NMR, natively unstructured protein, pre-structured motif
Hepatitis B virus (HBV) is a member of the hepadnaviridae family (Ganem and Varmus 1987; Chisari et al. 1989) that poses a significant health threat to millions of people worldwide, particularly in Africa, Asia, and certain parts of Europe (Fung and Lok 2004; Wright 2006). Whereas HBV infection itself may not appear so fatal as AIDS or SARS chronic hepatitis B infection often develops into more serious symptoms such as cirrhosis, liver failure, and hepatocelluar carcinoma (Blumberg et al. 1989). While successful administration of HBV vaccines led to a significant reduction of HBV-related casualties (Stephenne 1990; Mahoney et al. 1993; Jilg 1998; Poland and Jacobson 2004), emergence of escape mutants or nonresponders to current vaccines argues for attempts to develop vaccines with better efficacy. In addition, more specific anti-HBV therapeutics are needed in order to completely eradicate HBV infection, since nucleoside drugs currently in use for HBV-related symptoms are not HBV specific, often causing side effects or inducing drug resistance. Efforts to discover anit-HBV-specific agents based upon natural products or siRNAs have been met with only partial success (Saag 2006; Shin et al. 2006).
Even though it is generally accepted that HBV infection begins with an attachment of HBV surface antigens to a HBV-specific hepatocyte receptor, followed by subsequent entry of viral DNA into hepatocyte (Neurath et al. 1992; De Meyer et al. 1997), little structural knowledge is available regarding the critical step of HBV binding to hepatocyte. More fundamentally, an ambiguity exists if there is a hepatocyte receptor protein specific for HBV (Neurath et al. 1992; Pontisso et al. 1992; Ryu et al. 2000; De Falco et al. 2001). Hence, detailed structural information for the HBV receptor is not likely to be available in the foreseeable future. The surface antigen of HBV consists of three envelope glycoproteins called the large, middle, and small proteins, all of which are translated from a single open reading frame divided into preS1, preS2, and S regions, respectively (Heermann et al. 1984). The large protein, including all three regions, is the most abundant form found in the surface of infectious viral particles (Stibbe and Gerlich 1983; Heermann et al. 1984). As the preS1 region in the large protein is the outermost part of HBV particles, it is believed to play the most critical role in the binding of HBV to hepatocytes (Neurath et al. 1986; Pontisso et al. 1989; Ryu et al. 2000; De Falco et al. 2001).
One way of developing more efficient HBV vaccines is to add an immunodominant domain from the preS1 into the current immunogen (Jilg 1998). A prerequisite for such an attempt is to know which fragment to use (Barrera et al. 2005; Glebe et al. 2005; Hu et al. 2005). One may simply use the putative heptatocyte-binding domain encompassing residues 21–47 (Neurath et al. 1986). But, a recent study has suggested that a broader region encompassing residues 13–59 (adr numbering) is important for receptor recognition, which in turn can be subdivided into an essential domain composed of residues 13–29 and an accessory domain of residues 39–59 (Glebe et al. 2005). Another investigation, yet, revealed that the residues 16–31 are sufficient for hepatocyte binding (Barrera et al. 2005). Furthermore, a recent antibody screening study argues that residues 34–59 should be involved with hepatocyte binding (Hu et al. 2005).
Delineating the precise HBD is not only necessary for development of more efficient HBV vaccines, but may also provide a useful structural template for designing an anti-HBV fusion inhibitor. An antiviral fusion inhibitor “Pentafuside” was developed against HIV following such a strategy (Chan and Kim 1998). We have been studying structural features of HBV surface antigens to gain insight into interactions between HBV and hepatocyte (Chi et al. 2006, 2007). Preliminary CD studies showed that the preS1 exists in an unstructured state without a three-dimensional structure (Lee et al. 1994; Maeng et al. 2001). NMR spectroscopy is a powerful tool that provides detailed structural characteristics of such natively unstructured proteins (NUPs) at an amino acid residue level (Lee et al. 2000; Chi et al. 2005). Here, we have applied a heteronuclear three-dimensional NMR technique to a 119-residue preS1 surface protein of adr origin (Heermann et al. 1984). We have found that the preS1 protein, albeit devoid of a three-dimensional structure, is not totally unstructured but contains several noncontiguous “pre-structured” motifs overlapping with the putative hepatocyte binding or immunogenic domains. The fact that all the pre-structured motifs are located within the putative hepatocyte binding or potent immunogenic domains suggests that the hepatocyte-binding function or immunogenicity of preS1 is likely to be mediated by such pre-structured motifs.
Results
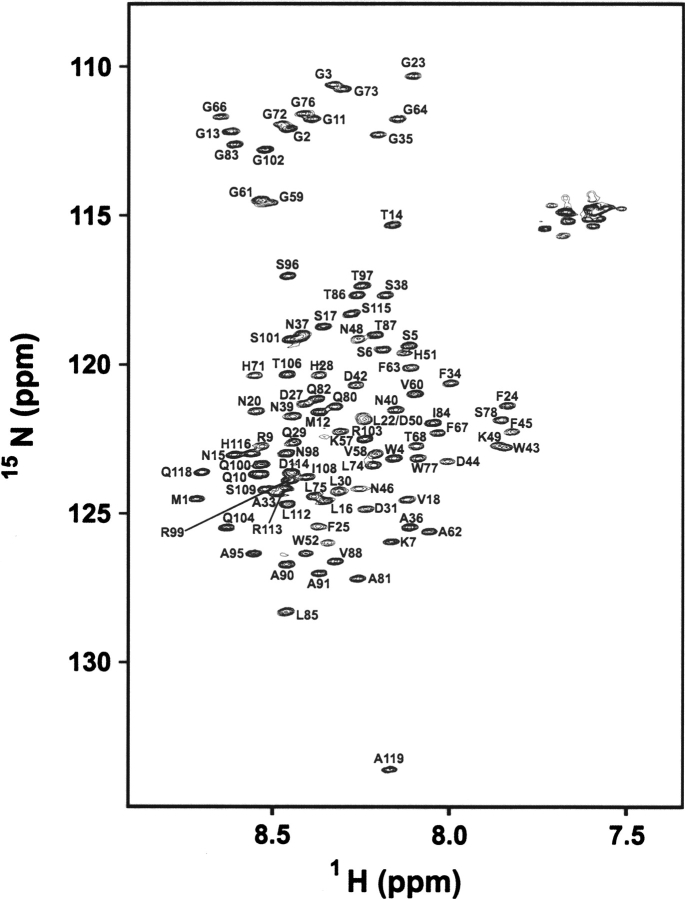
Figure 1 shows a 15N-1H HSQC spectrum of the preS1 with assigned resonances. The spectrum shows narrow chemical-shift dispersion in both dimensions, indicating that it is largely unstructured with no tertiary structure under a nondenaturing experimental condition. Hydrogen–deuterium exchange data show that all backbone amide protons of the preS1 disappear within 10 min after addition of 2H2O, also reflecting absence of a tertiary structure related exchange-retarding effect. The overall unstructured nature of preS1 observed by NMR is consistent with previous CD results (Maeng et al. 2001) and bioinformatics predictions made by AGADIR (Munoz and Serrano 1994) and GLOBPLOT (Linding et al. 2003). The overall helical content (10 of 119 residues) obtained from our NMR experiments is in good agreement with that (∼8%) obtained from CD measurements (Maeng et al. 2001). NMR resonance assignment for the 119-residue HBV preS1 was achieved by following standard triple-resonance NMR assignment procedures (Table 1). The first two N-terminal residues of the recombinant preS1, Met, and Gly originated from the N-terminal glutathione-S-transferase fusion linker. Unambiguous assignment of the backbone 15N and amide protons was possible for all residues except for Asp54–Ile56 and 21 prolines. The level of achieved resonance assignment, however, was sufficient for subsequent structural characterization of the preS1, since the aim of this investigation is not to determine a three-dimensional structure absent in preS1, but to delineate residues that form pre-structured motifs associated with hepatocyte binding or antibody recognition.
Figure 1.
15N-1H HSQC spectrum of the 119-residue full-length preS1. All of the backbone 15N and amide protons are assigned except for Asp54–Ile56 and 21 prolines.
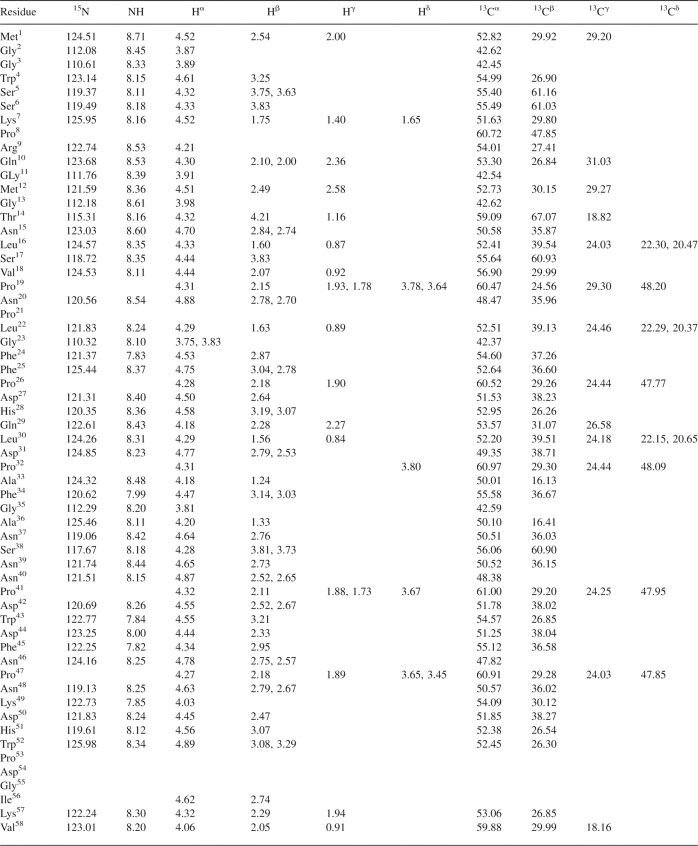
Table 1.
Chemical-shift assignments for the 119-residue HBV preS1 at pH 6.0 and 5°C
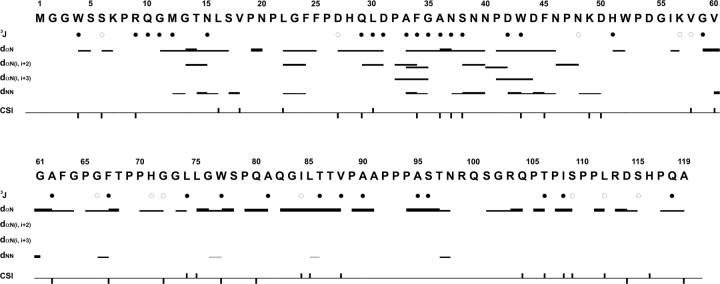
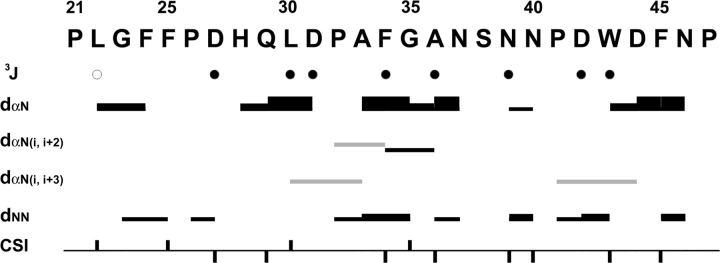
Figure 2 is a summary of interproton NOEs, 3JHNHα coupling constants (Supplemental Table S1) and Hα chemical-shift index (CSI) values of −1, 0, or +1 based on the method of Wishart and Sykes (1994) obtained for the preS1 (1–119) in 100% aqueous solution. Presence of sequential dNN, dαN(i + 2), and dαN(i + 3) NOEs in addition to 3JHNHα coupling constants and Hα CSIs indicates that preS1 is not completely unstructured, but contains transient local structural elements at its N-terminal domain from Met12–Asp50. Beyond the residue Asp50, no noticeable structural ordering is present. Figure 3 describes actual deviation of the Hα and Cα chemical shifts from random coil values (ΔHα and ΔCα) as well as the temperature coefficients of the backbone amide protons (ΔNH). These features indicate that residues 32–50 behave differently from the rest of the preS1 molecule. For Pro32–Ala36 and Pro41–Phe45, ΔHα and ΔNH concur well with interproton NOEs and 3JHNHα in terms of pointing out a local structural order, whereas for other residues, correlation among different parameters are not as clear (Fig. 2). The small (<4 ppb/K) temperature coefficients for the amide NH protons of residues 40–49 (black circles in Fig. 3B), deviating significantly from that of a random coil, suggest that the backbone amide protons of these residues are likely to be involved in hydrogen bonding.
Figure 2.
Summary of interproton NOEs, 3JHNHα, and chemical-shift index (CSI) of Hα protons for preS1 (1–119). Thickness of the bar represents the relative strength of NOEs. Gray bars indicate an ambiguity due to overlapping of NOEs. Black circles are drawn when 3JHNHα is <6 Hz and white circles when 3JHNHα is >8 Hz. The black squares above and below the horizontal line represent CSI values of +1 and −1, respectively.
Figure 3.
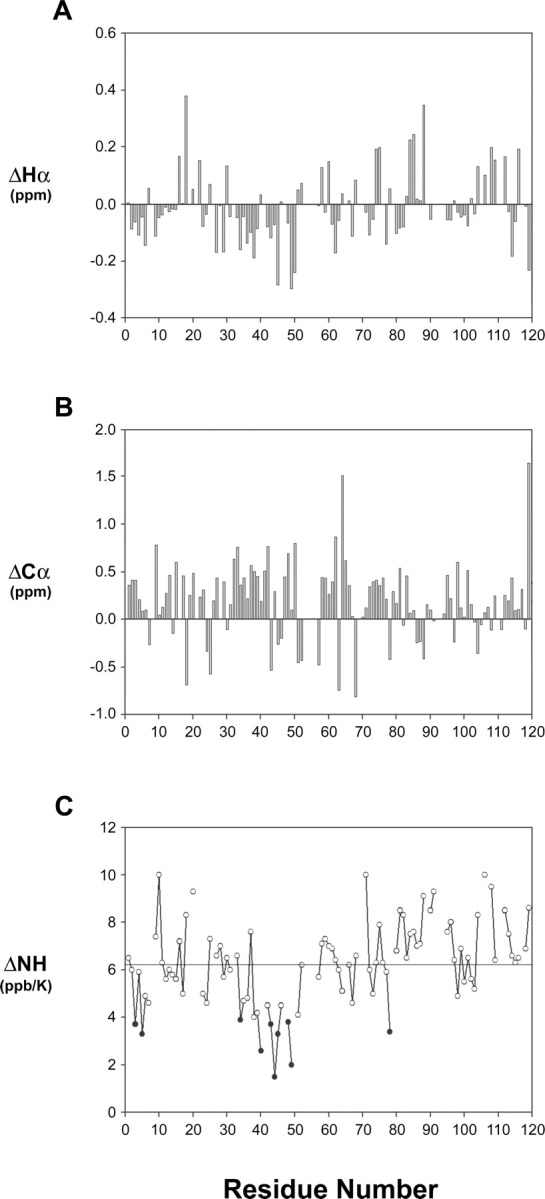
Hα chemical-shift deviation and temperature coefficient of preS1 (1–119). (A) Difference plot showing actual displacement of the measured Hα proton chemical shifts from random coil values. ΔHα = Hα (measured) −Hα (random). (B) Difference plot showing actual displacement of the measured Cα chemical shifts from random coil values. ΔCα = Cα (measured) −Cα (random). (C) Temperature coefficients of amide protons (ΔNH) are shown in ppb/K. To highlight the regions with the temperature coefficients that deviate significantly from those of a random conformation, ΔNH values of <4 ppb/K are displayed as black circles. The horizontal line indicates an average value.
Presence of the above pre-structured motifs in the preS1 can be further confirmed by backbone dynamics data. Figure 4, A and C shows 15N-1H heteronuclear NOEs and T2 relaxation times of preS1 that are useful indicators of local structural ordering. For residues 28–50 (black circles) these two parameters are discernibly different from the rest of the preS1 molecule. The residues that form only transient structural ordering cannot be identified based upon their backbone dynamics data. Figure 5 summarizes interproton NOEs, 3JHNHα coupling constants, and Hα CSIs for a preS1 (21–47) peptide. Several dNN, dαN(i + 2), and dαN(i + 3) NOEs observed in the full preS1 are missing in the NOESY spectrum of preS1 (21–47) peptide, indicating that the propensity to form pre-structured motifs weakens when the putative HBD exists only as a peptide. Nonetheless, the pre-structuring tendency observed in the full-length preS1 persists to a detectable degree, suggesting that the immunogenicity or hepatocyte binding ability of the putative HBD peptide closely correlates with its conformational characteristics.
Figure 4.
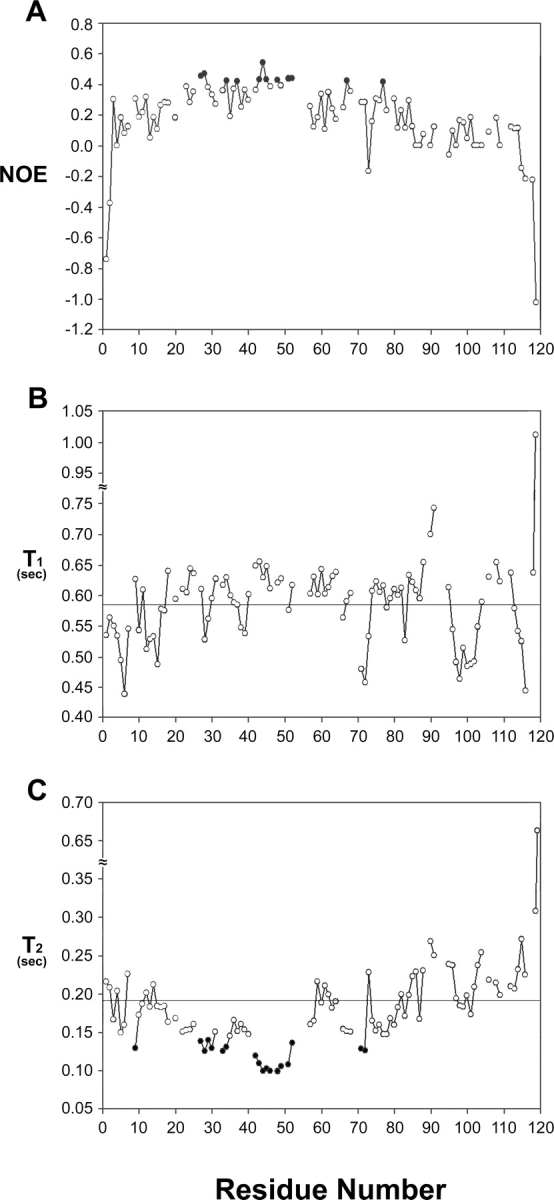
Plots of backbone 15N-1H heteronuclear NOEs and 15N relaxation times and against the residue number of preS1 (1–119). (A) 15N-1H heteronuclear NOEs, (B) T1, (C) T2 relaxation times. Horizontal lines in B and C indicate average values. To highlight the regions that show local structural ordering, heteronuclear NOE values of >0.4 in A and T2 values of <0.14 sec in C are displayed as black circles.
Figure 5.
Summary of interproton NOEs, 3JHNHα, and chemical-shift index (CSI) of Hα protons for preS1 (21–47) peptide. Thickness of bar represents relative strength of NOEs. Black circles are drawn when 3JHNHα is <6 Hz and white circles when 3JHNHα is >8 Hz. The black squares above and below the horizontal line represent CSI values of +1 and −1, respectively. Gray bars indicate an ambiguity due to overlapping of NOEs.
Discussion
It has been well documented that residues 21–47 in the preS1 contain the most important residues for hepatocyte binding, so that alanine mutation of these residues decreased the HBV binding to HepG2 cells (Neurath et al. 1992). Over the years, several neutralizing antibodies have been produced against various segments from the N-terminal domain of the preS1 (Neurath et al. 1989; Pontisso et al. 1989). An adx/adw serotype-specific neutralizing antibody, F35.25, was generated against the residues 32–53 in the preS1 (Petit et al. 1989) and an adr serotype specific antibody KR127 against the residues 37–45 (Maeng et al. 2000). On the other hand, a monoclonal antibody MA 18/7 specific for an ay serotype targets the residues 31–34 in the preS1 (Sominskaya et al. 1992). Another ayw-specific 5a-19–1 monoclonal antibody can specifically interact with the residues 36–43 (Budkowska et al. 1995). A short consensus Q29LDPAF34 motif of the preS1 was suggested to be the hepatocyte binding motif, as it is highly conserved in many cellular, bacterial, and viral proteins involved in cell adhesion, attachment, and fusion (Paran et al. 2001).
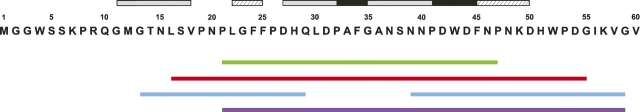
Given the knowledge that the preS1 is unstructured with no tertiary structure (Lee et al. 1994; Maeng et al. 2001), this study aimed at applying heteronuclear three-dimensional NMR methods to delineate the structural features, particularly location of pre-structured motifs, that are likely to be associated with important function of preS1 such as hepatcocyte binding or immunogenicity. Pre-structured motifs identified by NMR in other NUPs were shown to be critically involved in target protein binding (Lee et al. 2000; Ramelot et al. 2000; Sayers et al. 2000; Zitzewitz et al. 2000; Bienkiewicz et al. 2002; Domanski et al. 2004; Bochkareva et al. 2005; Chi et al. 2005; Di Lello et al. 2006). Here we have shown that the preS1 exists in a “mostly unstructured” state, a state where several noncontiguous “pre-structured” motifs are present in the absence of an overall globular three-dimensional structure. Although not all NMR parameters in unison point to the same location of pre-structured regions, we were able to reasonably map out the pre-structured regions or motifs. Figure 6 summarizes the location of the pre-structured motifs delineated in this study, as well as the previously suggested immunodominant or hepatocyte-binding domains (Neurath et al. 1992; Barrera et al. 2005; Glebe et al. 2005; Hu et al. 2005; Engelke et al. 2006). As summarized in Figure 6 there is a general correlation between presence of the pre-structured motifs and immunogenicity or hepatocyte-binding ability in preS1.
Figure 6.
Location of pre-structured regions (this work) and previously suggested functional domains in preS1. Pre-structured regions are shown above the amino acid sequence of preS1 N-terminal 60 residues and are indicated by black, gray, or hatched bars. The most prominently pre-structured region is formed by residues 27–45. The black bars indicate two well-defined helical turn motifs within this region. The gray bars are for residues showing an intermediate degree of structural ordering and the hatched bars for least pre-structuring residues. Shown below the amino acid sequence of preS1 are the previously reported functionally important domains; the putative hepatocyte receptor-binding domain with a green (Neurath et al. 1986) or a red bar (Barrera et al. 2005), hepatocyte attachment domains with blue bars (Glebe et al. 2005), and an immunogenic domain with a violet bar (Hu et al. 2005), respectively.
The most prominent pre-structured region encompasses residues 27–45 and contains two helical turn motifs (black bars in Fig. 6) composed of residues Pro32–Ala36 and Pro41–Phe45, both of which are located within the putative HBD (residues 21–47) proposed by other techniques. As preS1 peptides encompassing residues 40–45 are actively hepatocyte binding (Budkowska et al. 1995; Barrera et al. 2005; Glebe et al. 2005; Engelke et al. 2006), the specific conformation of this pre-structured motif must be responsible for the hepatocyte-binding activity of such peptides. This observation is consistent with the general concept that pre-structured motifs in NUPs serve as “recognition antennae” for target protein interactions (Lee et al. 2000; Ramelot et al. 2000; Sayers et al. 2000; Zitzewitz et al. 2000; Bienkiewicz et al. 2002; Domanski et al. 2004; Bochkareva et al. 2005; Chi et al. 2005; Di Lello et al. 2006). The second most stable pre-structured motif in the preS1 is formed by residues Pro32–Ala36 and partially overlaps with the putative hepatocyte binding stretch (Q29LDPAF34) (Paran et al. 2001), which is an immunodominant epitope recognized by several neutralizing monoclonal antibodies (Kuttner et al. 1999). The P32AFG35 segment forms a β-turn when it is bound to neutralizing monoclonal antibodies such as PC282, PC283, and PC287 (Nair et al. 2002). The fact that this β-turn motif is similar to a helical turn we have observed in an unbound state of preS1 indicates that, before binding to antibodies, the helical turn conformation for the Pro32–Ala36 is sufficiently populated and can be easily converted into a β-turn upon binding.
Our investigation reveals that there are functionally important pre-structured motifs outside of the putative HBD such as the one involving residues 11–18 (Fig. 6). While the pre-structuring tendency of this motif is not as strong as that of the Pro32–Ala36 and Pro41–Phe45 motifs, the result that peptides containing residues 11–18 are involved in hepatocyte binding (Glebe et al. 2005) also agrees with the general concept that pre-structured motifs in NUPs are important for target protein binding. The Q29LDPAF34 motif shows an inconsistency with such a general trend, in that its absence does not seem to affect hepatocyte binding to a significant degree (Glebe et al. 2005). One should not exclude a possibility that these residues make a subtle conformational contribution to receptor binding of the preS1. The region encompassing residues 20–29 was shown to be important for hepatocyte binding, but shows only a weak tendency for pre-structural ordering in our NMR analysis. In summary, our results should help to determine which regions are to be used for production of more efficient HBV vaccines and to design structure-based inhibitors against HBV attachment to hepatocytes. Such an effort could be expedited by elucidation of the binding surface of preS1 to hepatocytes through NMR titration experiments once an unambiguous HBV receptor is identified.
Materials and Methods
Protein preparation
A recombinant preS1 construct corresponding to residues 1–119 (adr subtype) was expressed in pGEX-2T vector. Transformed Escherchia coli DH5α cells were grown at 37°C to an OD600 of 0.6 and the culture was induced with 0.2 mM isopropyl thio-β-D-thiogalactopyranoside (IPTG). Then, the cells were further cultivated at 20°C for 16 h. The harvested cell suspension was sonicated in 50 mM TrisHCl (pH 8), 0.2 M NaCl, 1 mM PMSF, 10 mM β-mercaptoethanol, and centrifuged for 30 min at 30,000g. The 15N-labeled or 13C, 15N-labeled preS1 (1–119) were purified using Glutathione–Sepharose column, SP–Sepharose column, and Hiprep 26/60 Sephacryl S-200 FPLC column (Amersham Pharmacia Biotech). The molecular weights of the purified proteins were confirmed by MALDI-TOF mass spectrometry.
Peptide preparation
The preS1 (21–47) peptide was synthesized by a solid-phase method with Multiple Peptide Synthesizer APEX 348Ω (Advanced Chemtech). The C-termini of the synthesized peptide was amidated. The peptide was purified by reverse-phase HPLC using Vydac C18 columns and the peptide masses confirmed by MALDI-TOF mass spectrometry.
NMR spectroscopy
Isotopically labeled protein samples with concentration of 0.75 mM were prepared in 90% H2O/10% 2H2O or 100% 2H2O containing 25 mM sodium acetate buffer (pH 6.0), 100 mM NaCl, 0.1 mM PMSF, 0.1 mM EDTA, and 0.1 mM benzamide. All NMR experiments were done at 5°C using a Varian Unity INOVA 600 and Bruker Avance II 900 spectrometers equipped with cryogenic probes. 13C chemical shifts are referenced to DSS and 15N chemical shifts were referenced indirectly using the gyromagnetic ratio of 15N/1H (0.101329118) (Wishart et al. 1995). Sequence-specific resonance assignment of HBV preS1 was obtained using standard multidimensional double- and triple-resonance NMR techniques as previously described (Bax and Grzesiek 1993). The sequence-corrected random coil chemical-shift values of Schwarzinger et al. (2001) were used to calculate the secondary shifts of Hα and 13Cα (Fig. 3). The three-bond 3JHNHα coupling constants were measured by a three-dimensional HNHA experiment. Temperature coefficients for the backbone amide protons (ΔNH) were calculated using the 1H resonance assignments obtained at three temperatures (5°C, 10°C, and 25°C). 15N T1 values were measured from spectra recorded with seven relaxation delays (20, 40, 80, 160, 320, 640, and 1280 msec). 15N T2 values were measured from spectra recorded using a CPMG sequence (Kay et al. 1992) with eight relaxation delays (10, 30, 50, 70, 90, 130, 190, and 250 ms). The 15N-1H heteronuclear steady-state NOEs were measured from a pair of spectra recorded with and without a proton presaturation.
The resonance assignment of preS1 (21–47) peptide was obtained by standard two-dimensional NMR experiments such as total-shift correlation spectroscopy (TOCSY), double-quantum filtered correlation spectroscopy (DQF-COSY), and NOESY (NOE spectroscopy). Mixing times of 100 ms for NOESY and 70 ms for TOCSY experiments were used. The three-bond coupling constants, 3JHNHα, for backbone torsion angles were measured using phase-sensitive DQF-COSY experiments. The two-dimensional NMR data consist of 2048 complex points in the T2 dimension with 256 complex T1 increments. Spectral widths were 8 kHz in both dimensions. All data were processed and analyzed on a Sun SPARCstation (Sun Microsystems, Inc.) using a Varian Vnmr software and an nmrPipe/nmrDraw software.
Acknowledgments
We thank H.J. Hong for the preS1 plasmid. This work has been supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MOST) (R01-2006-000-10905-0) (K.H.), a grant from the Korean Ministry of Health and Welfare and Korea Health Industry Development Institute (A060017) (K.H.), and National Research Laboratory Program to I.C. (No. M104-333-06J-33310) funded by the MOST of Korea. This study made use of the NMR facility at Korea Basic Science Institute, which is supported by Bio-MR Research Program of the Korean Ministry of Science and Technology (E26070).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Kyou-Hoon Han, Molecular Cancer Research Center, Division of Molecular Therapeutics, KRIBB, Daejeon 305-806, Korea; e-mail: khhan600@kribb.re.kr; fax: 82-42-860-4259.
Abbreviations: HBV, hepatitis B virus; NMR, nuclear magnetic resonance; NUP, natively unstructured protein; CSI, chemical-shift index.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072983507.
References
- Barrera A., Guerra, B., Notvall, L., and Lanford, R.E. 2005. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J. Virol. 79: 9786–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A. and Grzesiek, S. 1993. Methodological advances in protein NMR. In NMR of proteins (eds. G.M. Clore and A.M. Gronenborn), pp. 33–52. MacMillan, London, UK.
- Bienkiewicz E.A., Adkins, J.N., and Lumb, K.J. 2002. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1). Biochemistry 41: 752–759. [DOI] [PubMed] [Google Scholar]
- Blumberg B.S., Millman, I., Venkateswaran, P.S., and Thyagarajan, S.P. 1989. Hepatitis B virus and hepatocellular carcinoma–treatment of HBV carriers with Phyllanthus amarus. Cancer Detect. Prev. 14: 195–201. [PubMed] [Google Scholar]
- Bochkareva E., Kaustov, L., Ayed, A., Yi, G.S., Lu, Y., Pineda-Lucena, A., Liao, J.C., Okorokov, A.L., Milner, J., Arrowsmith, C.H., et al. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. 102: 15412–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkowska A., Bedossa, P., Groh, F., Louise, A., and Pillot, J. 1995. Fibronectin of human liver sinusoids binds hepatitis B virus: Identification by an anti-idiotypic antibody bearing the internal image of the pre-S2 domain. J. Virol. 69: 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C. and Kim, P.S. 1998. HIV entry and its inhibition. Cell 93: 681–684. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Lee, S.H., Kim, D.H., Ahn, M.J., Kim, J.S., Woo, J.Y., Torizawa, T., Kainosho, M., and Han, K.H. 2005. Structural details on mdm2-p53 interaction. J. Biol. Chem. 280: 38795–38802. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Kim, D.H., Kim, J.S., Lee, M.K., and Han, K.H. 2006. Solution conformation of an immunodominant epitope in the hepatitis B virus preS2 surface antigen. Antiviral Res. 72: 207–215. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Maeng, C.Y., Kim, S.J., Oh, M.S., Ryu, C.J., Kim, S.J., Han, K.H., Hong, H.J., and Ryu, S.E. 2007. Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism. Proc. Natl. Acad. Sci. 104: 9230–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari F.V., Ferrari, C., and Mondelli, M.U. 1989. Hepatitis B virus structure and biology. Microb. Pathog. 6: 311–325. [DOI] [PubMed] [Google Scholar]
- De Falco S., Ruvoletto, M.G., Verdoliva, A., Ruvo, M., Raucci, A., Marino, M., Senatore, S., Cassani, G., Alberti, A., Pontisso, P., et al. 2001. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J. Biol. Chem. 276: 36613–36623. [DOI] [PubMed] [Google Scholar]
- De Meyer S., Gong, Z.J., Suwandhi, W., van Pelt, J., Soumillion, A., and Yap, S.H. 1997. Organ and species specificity of hepatitis B virus (HBV) infection: A review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J. Viral Hepat. 4: 145–153. [DOI] [PubMed] [Google Scholar]
- Di Lello P., Jenkins, L.M., Jones, T.N., Nguyen, B.D., Hara, T., Yamaguchi, H., Dikeakos, J.D., Appella, E., Legault, P., and Omichinski, J.G. 2006. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22: 731–740. [DOI] [PubMed] [Google Scholar]
- Domanski M., Hertzog, M., Coutant, J., Gutsche-Perelroizen, I., Bontems, F., Carlier, M.F., Guittet, E., and van Heijenoort, C. 2004. Coupling of folding and binding of thymosin β4 upon interaction with monomeric actin monitored by nuclear magnetic resonance. J. Biol. Chem. 279: 23637–23645. [DOI] [PubMed] [Google Scholar]
- Engelke M., Mills, K., Seitz, S., Simon, P., Gripon, P., Schnolzer, M., and Urban, S. 2006. Characterization of a hepatitis B and hepatitis δ virus receptor binding site. Hepatology 43: 750–760. [DOI] [PubMed] [Google Scholar]
- Fung S.K. and Lok, A.S. 2004. Viral hepatitis in 2003. Curr. Opin. Gastroenterol. 20: 241–247. [DOI] [PubMed] [Google Scholar]
- Ganem D. and Varmus, H.E. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56: 651–693. [DOI] [PubMed] [Google Scholar]
- Glebe D., Urban, S., Knoop, E.V., Cag, N., Krass, P., Grun, S., Bulavaite, A., Sasnauskas, K., and Gerlich, W.H. 2005. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 129: 234–245. [DOI] [PubMed] [Google Scholar]
- Heermann K.H., Goldmann, U., Schwartz, W., Seyffarth, T., Baumgarten, H., and Gerlich, W.H. 1984. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 52: 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.G., Wei, J., Xia, H.C., Yang, X.X., Li, F., Li, G.D., Wang, Y., and Zhang, Z.C. 2005. Identification of the immunogenic domains in HBsAg preS1 region using overlapping preS1 fragment fusion proteins. World J. Gastroenterol. 11: 2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg W. 1998. Novel hepatitis B vaccines. Vaccine 16: S65–S68. doi: 10.1016/50264-410X(98)00300-4. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Nicholson, F.D., Bax, A., and Torchia, D.A. 1992. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 97: 359–375. [Google Scholar]
- Kuttner G., Kramer, A., Schmidtke, G., Giessmann, E., Dong, L., Roggenbuck, D., Scholz, C., Seifert, M., Stigler, R.D., and Schneider-Mergener, J. 1999. Characterization of neutralizing anti-pre-S1 and anti-pre-S2 (HBV) monoclonal antibodies and their fragments. Mol. Immunol. 36: 669–683. [DOI] [PubMed] [Google Scholar]
- Lee H., Mok, K.H., Muhandiram, R., Park, K.H., Suk, J.E., Kim, D.H., Chang, J., Sung, Y.C., Choi, K.Y., and Han, K.H. 2000. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 275: 29426–29432. [DOI] [PubMed] [Google Scholar]
- Lee M.K., Kim, K.L., Hahm, K.S., and Yang, K.H. 1994. Structure-antigenicity relationship of peptides from the pre-S2 region of the hepatitis B virus surface antigen. Biochem. Mol. Biol. Int. 34: 159–168. [PubMed] [Google Scholar]
- Linding R., Russell, R.B., Neduva, V., and Gibson, T.J. 2003. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31: 3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng C.Y., Ryu, C.J., Gripon, P., Guguen-Guillouzo, C., and Hong, H.J. 2000. Fine mapping of virus-neutralizing epitopes on hepatitis B virus PreS1. Virology 270: 9–16. [DOI] [PubMed] [Google Scholar]
- Maeng C.Y., Oh, M.S., Park, I.H., and Hong, H.J. 2001. Purification and structural analysis of the hepatitis B virus preS1 expressed from Escherichia coli . Biochem. Biophys. Res. Commun. 282: 787–792. [DOI] [PubMed] [Google Scholar]
- Mahoney F.J., Woodruff, B.A., Erben, J.J., Coleman, P.J., Reid, E.C., Schatz, G.C., and Kane, M.A. 1993. Effect of a hepatitis B vaccination program on the prevalence of hepatitis B virus infection. J. Infect. Dis. 167: 203–207. [DOI] [PubMed] [Google Scholar]
- Munoz V. and Serrano, L. 1994. Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1: 399–409. [DOI] [PubMed] [Google Scholar]
- Nair D.T., Singh, K., Siddiqui, Z., Nayak, B.P., Rao, K.V., and Salunke, D.M. 2002. Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response. J. Immunol. 168: 2371–2382. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Kent, S.B., Strick, N., and Parker, K. 1986. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell 46: 429–436. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Seto, B., and Strick, N. 1989. Antibodies to synthetic peptides from the preS1 region of the hepatitis B virus (HBV) envelope (env) protein are virus-neutralizing and protective. Vaccine 7: 234–236. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Strick, N., and Sproul, P. 1992. Search for hepatitis B virus cell receptors reveals binding sites for interleukin 6 on the virus envelope protein. J. Exp. Med. 175: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran N., Geiger, B., and Shaul, Y. 2001. HBV infection of cell culture: Evidence for multivalent and cooperative attachment. EMBO J. 20: 4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.A., Dubanchet, S., and Capel, F. 1989. A monoclonal antibody specific for the hepatocyte receptor binding site on hepatitis B virus. Mol. Immunol. 26: 531–537. [DOI] [PubMed] [Google Scholar]
- Poland G.A. and Jacobson, R.M. 2004. Clinical practice: Prevention of hepatitis B with the hepatitis B vaccine. N. Engl. J. Med. 351: 2832–2838. [DOI] [PubMed] [Google Scholar]
- Pontisso P., Ruvoletto, M.G., Gerlich, W.H., Heermann, K.H., Bardini, R., and Alberti, A. 1989. Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology 173: 522–530. [DOI] [PubMed] [Google Scholar]
- Pontisso P., Ruvoletto, M.G., Tiribelli, C., Gerlich, W.H., Ruol, A., and Alberti, A. 1992. The preS1 domain of hepatitis B virus and IgA cross-react in their binding to the hepatocyte surface. J. Gen. Virol. 73: 2041–2045. [DOI] [PubMed] [Google Scholar]
- Ramelot T.A., Gentile, L.N., and Nicholson, L.K. 2000. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry 39: 2714–2725. [DOI] [PubMed] [Google Scholar]
- Ryu C.J., Cho, D.Y., Gripon, P., Kim, H.S., Guguen-Guillouzo, C., and Hong, H.J. 2000. An 80-kilodalton protein that binds to the pre-S1 domain of hepatitis B virus. J. Virol. 74: 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saag M.S. 2006. Emtricitabine, a new antiretroviral agent with activity against HIV and hepatitis B virus. Clin. Infect. Dis. 42: 126–131. [DOI] [PubMed] [Google Scholar]
- Sayers E.W., Gerstner, R.B., Draper, D.E., and Torchia, D.A. 2000. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry 39: 13602–13613. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S., Kroon, G.J.A., Foss, T.R., Chung, J., Wright, P.E., and Dyson, H.J. 2001. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 123: 2970–2978. [DOI] [PubMed] [Google Scholar]
- Shin D., Kim, S.I., Kim, M., and Park, M. 2006. Efficient inhibition of hepatitis B virus replication by small interfering RNAs targeted to the viral X gene in mice. Virus Res. 119: 146–153. [DOI] [PubMed] [Google Scholar]
- Sominskaya I., Pushko, P., Dreilina, D., Kozlovskaya, T., and Pumpen, P. 1992. Determination of the minimal length of preS1 epitope recognized by a monoclonal antibody which inhibits attachment of hepatitis B virus to hepatocytes. Med. Microbiol. Immunol. (Berl.) 181: 215–226. [DOI] [PubMed] [Google Scholar]
- Stephenne J. 1990. Development and production aspects of a recombinant yeast-derived hepatitis B vaccine. Vaccine 8: S69–S73. doi: 10.1016/0264-410X(90)90221-7. [DOI] [PubMed] [Google Scholar]
- Stibbe W. and Gerlich, W.H. 1983. Structural relationships between minor and major proteins of hepatitis B surface antigen. J. Virol. 46: 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. and Sykes, B.D. 1994. Chemical shifts as a tool for structure determination. Methods Enzymol. 239: 363–392. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Bigam, C.G., Yao, J., Abildgaard, F., Dyson, H.J., Oldfield, E., Markley, J.L., and Sykes, B.D. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6: 135–140. [DOI] [PubMed] [Google Scholar]
- Wright T.L. 2006. Introduction to chronic hepatitis B infection. Am. J. Gastroenterol. 101: S1–S6. doi: 10.1111/j.1572-0241.2006.00469.x. [DOI] [PubMed] [Google Scholar]
- Zitzewitz J.A., Ibarra-Molero, B., Fishel, D.R., Terry, K.L., and Matthews, C.R. 2000. Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system. J. Mol. Biol. 296: 1105–1116. [DOI] [PubMed] [Google Scholar]