Abstract
We have tested the role of the polar loop of subunit c of the Escherichia coli ATP synthase in stabilizing the hairpin structure of this protein. The structure of the c32–52 peptide corresponding to the cytoplasmic region of subunit c bound to the dodecylphosphocholine micelles was solved by high-resolution NMR. The region comprising residues 41–47 forms a well-ordered structure rather similar to the conformation of the polar loop region in the solution structure of the full-length subunit c and is flanked by short α-helical segments. This result suggests that the rigidity of the polar loop significantly contributes to the stability of the hairpin formed by the two helices of subunit c. This experimental system may be useful for NMR studies of interactions between subunit c and subunits γ and ɛ, which together form the rotor of the ATP synthase.
Keywords: ATP synthase, protein NMR, peptide structure, membrane protein
The FoF1 ATP synthase is a multisubunit protein complex, which catalyzes ATP synthesis using the driving force of transmembrane gradient of protons in mitochondria, chloroplasts, and most microorganisms, or sodium ions in some bacteria (Senior 1988). In Escherichia coli and other bacteria the membrane domain of ATP synthase (F0) consists of three types of subunits in a ratio of a:b:c = 1:2:10–12 (Schneider and Altendorf 1986; Fillingame et al. 2000; Jiang et al. 2001), and contains the transmembrane ion channel. The F1-domain protruding into the cytoplasm consists of two major subunits, α and β, arranged in a hexamer, and three minor subunits present in a ratio of (αβ)3γδɛ.
The flow of protons through the F0-channel induces rotation of the cylindrical oligomer built of the c subunits. This rotation is transmitted inside the core of the F1 complex through the shaft built of the elongated subunit γ. Cyclical conformational changes in the three substrate binding centers, which are located on the β subunits, caused by the subunit γ rotation constitute the structural basis of ATP formation in F1 (Cross 2000; Yoshida et al. 2001).
Molecular steps of enzymatic catalysis in mitochondrial F1 have been illuminated by a series of high-resolution X-ray structures capturing the protein at different stages of the catalytic cycle (Abrahams et al. 1994; Braig et al. 2000; Menz et al. 2001). In contrast, structure of the F0 complex is still largely unknown, with the exception of the rotor module built of 10–12 copies of the c subunit and the membrane anchor of the subunit b dimer (Dmitriev et al. 1999a). Model structures of the subunit c oligomer in E. coli were calculated from the solution structure of the subunit c monomer and extensive intersubunit cross-linking data (Dmitriev et al. 1999b; Rastogi and Girvin 1999). Recently, a high-resolution structure of the subunit c oligomer from Ilyobacter tartaricus was solved by X-ray crystallography (Meier et al. 2005).
In an organic solvent–water mixture, subunit c monomer folds into a hairpin consisting of two long α-helices connected by a short well-ordered loop (Girvin et al. 1998). The number of atomic contacts between the helices is relatively small, which limits the number of possible interactions stabilizing the hairpin structure in the solution and in the native enzyme within the membrane. Stacking interactions between aromatic rings of Y10, Y73, and F76 located close to the periplasmic ends of the helices were proposed to stabilize the hairpin structure (Girvin et al. 1998). However, such interactions are absent in the structure of subunit c at pH 8 (Rastogi and Girvin 1999), as well as in the structures of the subunit c monomer from thermophilic bacterium PS3 in organic solvent (Nakano et al. 2006) and of the subunit c oligomer from Ilyobacter tartaricus (Meier et al. 2005).
The loop connecting the two helices of subunit c is well-ordered, and its rigid conformation could conceivably stabilize the folding of the helices in the hairpin structure. The sequence of the loop region includes a highly conserved sequence motif RQP(E/D) corresponding to R41–D44 in E. coli. Mutations in the R41–D44 region result in uncoupling of ATP hydrolysis from proton transport (Fillingame 1997; Mosher et al. 1985), presumably by disrupting functionally important interactions between the F1 and F0 complexes. Close contacts between the polar loop region of the subunit c and the subunits γ (Watts et al. 1995, 1996) and ɛ (Zhang and Fillingame 1995; Hermolin et al. 1999) were directly demonstrated by Cys–Cys cross-linking analysis in cell membranes. Thus, the polar loop region of subunit c is directly involved in maintaining stability of the rotor assembly in the ATP synthase. The available high-resolution structures of the E. coli subunit c were solved in a mixture of chloroform, methanol, and water. In the native enzyme in cell membranes, the polar loop region is believed to be located at the interface between the lipid bilayer and the cytoplasm. Therefore, detergent micelles in water may provide a closer approximation to the native environment than organic solvent.
If the short range interactions within the connecting polar loop segment of subunit c are sufficient to maintain a rigid structure, which in turn would stabilize the folding of the two helices into a hairpin, then the isolated loop segment should possess a well-ordered structure similar to its conformation in the full-length subunit c. The NMR structures of soluble protein fragments corresponding to the loops connecting the transmembrane helices in bacteriorhodopsin and lactose permease have suggested an important role of short-range interactions in the well-structured connecting segments in determining the global fold of polytopic membrane proteins (Katragadda et al. 2001; Bennett et al. 2004). To clarify the role of the polar loop in stabilizing the structure of subunit c and to determine its conformation in an aqueous detergent, we have solved the structure of a subunit c fragment corresponding to residues 32–52 in the presence of dodecylphosphocholine.
Results and Discussion
The c32–52 peptide corresponds to the polar loop connecting the two long α-helices of subunit c (residues 41–47), and the adjacent several turns of helices I and II (Fig. 1A). As expected from the high content of polar amino acids in this region of subunit c, the c32–52 peptide was found to be easily soluble in water. To facilitate NMR assignments, the peptide was 15N-labeled on glycine, alanine, and leucine residues. The Fmoc derivatives of these amino acids are commercially available and relatively inexpensive, allowing routine incorporation of the corresponding 15N-labeled amino acids in the synthetic peptides for structure determination.
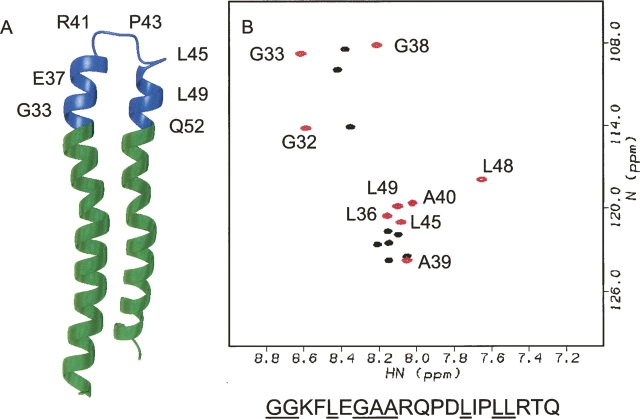
Figure 1.
(A) NMR structure of subunit c (1C0V) with the region corresponding to the c32–52 peptide shown in blue. (B) 1H,15N-HSQC spectra of c32–52 peptide recorded in the presence of 100 mM DPC (red) and without any detergent added (black). Sequential assignments made in the presence of DPC are shown. The residue numbers correspond to the full-length subunit c. Amino acid sequence of the c32–52 peptide is shown below with 15N-labeled residues underlined.
Chemical shift dispersion of the c32–52 sample in water was poor (Fig. 1B) and appeared to indicate a disordered state lacking secondary structure. In the native ATP synthase complex in the cell membrane residues 32–52 are predicted to extend from the lipid polar head group region into the cytoplasm. To simulate this environment, dodecylphosphocholine was added to the sample. Addition of dodecylphosphocholine to the peptide solution dramatically improved chemical shift dispersion and caused large chemical shift changes of several residues (Fig. 1B). These changes strongly indicate binding of the peptide to DPC micelles. A single set of signals was observed for all the 15N-labeled amino acid residues in the peptide in the presence of 100 mM DPC.
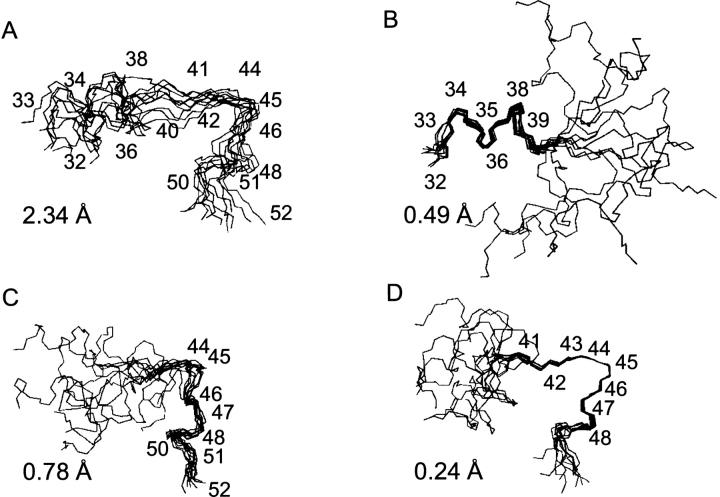
We have calculated a structure of the c32–52 peptide bound to DPC micelles (Supplemental Table 1). The peptide is folded, and comprises three distinct structural regions (Fig. 2A–D). Residues 33–39 form a short α-helix hinged to a rigid loop including residues 41–47 followed by another turn of an α-helix (residues 48–51). This structure strongly resembles the corresponding region of the full-length subunit c structure determined in the chloroform–methanol–water mixture (Girvin et al. 1998). The RMSD for the backbone atoms of residues 41–47 between the ensembles of the 10 best structures of the c32–52 and the full-length subunit c was 1.1 Å (Fig. 3A).
Figure 2.
Best fit superpositions of the 10 lowest energy c32–52 structures calculated for the backbone atoms of residues 32–52 (A), 32–39 (B), 45–52 (C), and 41–47 (D). The corresponding RMSD values are shown in each panel.
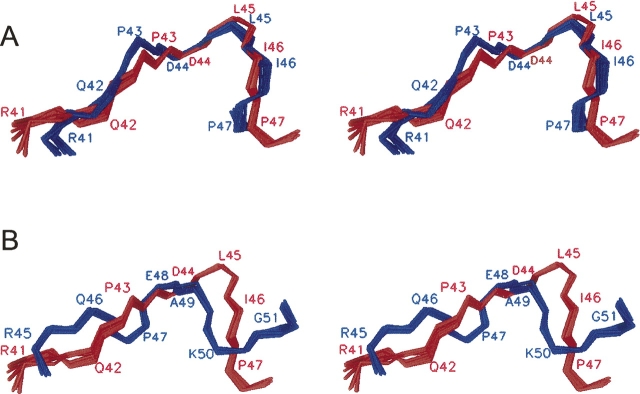
Figure 3.
Stereoview of the superimposed polar loop regions (residues 41–47) in the c32–52 peptide and in the full-length subunit c structures. (A) The c32–52 structure (red) and the NMR structure of the E. coli subunit c (1C0V, blue). The RMSD for backbone atoms is 1.1 Å. (B) The c32–52 structure (red) and the X-ray crystal structure of the subunit c from Ilyobacter tartaricus (1YCE, blue). The RMSD for backbone atoms is 2.4 Å.
The rigidity of the loop region in the subunit c and the c32–52 peptide is most likely determined by steric constraints introduced by two proline residues (P43 and P47) and the bulky side chains of the other residues in the sequence R41–QPDLI–P47 combined with spatial restrictions caused by peptide binding to the detergent micelle. Proximity of the Q42 side-chain amide to the backbone carbonyl of I46 in the c32–52 structure suggests the possibility of an H-bond formation, but the Q42 side-chain conformation is not defined well enough to detect such an H-bond with certainty.
In the c32–52 peptide, the short α-helical segments flanking the polar loop region do not fold together. The connection of the N-terminal α-helix to the polar loop appears to be very flexible. An amphipathic nature of this short helix combined with the hydrophilic profile of the adjacent loop region suggests that the c32–52 peptide is bound to the surface, rather than inserted inside the micelle. The C-terminal α-helical segment displays less flexibility probably due to the presence of P47.
The structure of the monomeric subunit c of the E. coli ATP synthase in chloroform–methanol–water mixtures solved by NMR resembles a hairpin of two long α-helices connected by a short well-structured loop. This architecture is also observed in the high-resolution X-ray structure of the subunit c oligomer from Ilyobacter tartaricus (Meier et al. 2005), even though the relative orientation of the two helices and the conformation of polar loop are somewhat different. A remarkable feature of the NMR solution structure is that the helices fold together even though the number of interhelical contacts that may stabilize the hairpin structure is relatively small.
A rigid connecting loop could limit the conformational space available to the two helices, and thus the other weak interactions could stabilize the hairpin structure of the subunit c monomer in the solution and of the oligomeric complex in the membrane. The structure of the loop region in the c32–52 peptide is indeed very well defined and quite similar to the corresponding region in the NMR structure of the full-length subunit c (Fig. 3A). Importantly, this structural similarity is not significantly affected by the difference in the solvent systems used for the c32–52 peptide and the full-length subunit c, respectively. This observation indicates that the short-range interactions, which determine the conformation of the polar loop, are strong enough to outweigh the difference in the energy contribution from peptide–solvent interactions, and further validates the use of organic solvent–water mixtures in structural studies of membrane proteins. In contrast, polar loop conformation in the c32–52 peptide and in the NMR structure of the E. coli subunit c differs significantly from the conformation observed in the crystal structure of the Ilyobacter tartaricus subunit c oligomer (Fig. 3B). Since the choice of the solvent does not significantly affect polar loop structure, this difference may be accounted for by structural rearrangement taking place when the individual c subunits form an oligomeric complex.
The interactions between the polar loop region of subunit c and subunits ɛ and γ appear to be essential for maintaining the structural integrity of the ATP synthase rotor. However, no high-resolution structure of the c–γɛ contact region is available. The NMR structure of the subunit ɛ (Wilkens and Capaldi 1998) and the present work provide the basis for mapping the interactions between the c32–52 peptide bound to DPC micelles and the isolated subunit ɛ.
In conclusion, the short-range interactions define a well-ordered structure of the subunit c polar loop. These interactions constitute an important factor stabilizing the hairpin structure of the subunit c. In general, the short structured connecting loops are likely to play an important role in defining the global fold of the polytopic membrane proteins.
Materials and Methods
Peptide synthesis
A 21-residue peptide corresponding to residues 32–52 of the E. coli subunit c (GGKFLEGAARQPDLIPLLRTQ) was synthesized at the University of Wisconsin Biotechnology Center on an Applied Biosystems Synergy 432A instrument using Fmoc (N-(9-fluorenyl)-methoxycarbonyl) chemistry. The alanine, glycine, and leucine precursors were 15N-labeled (Cambridge Isotope Laboratories, Inc.). The N terminus was acetylated and the C-terminal carboxyl was amidated. The peptide was purified from the crude synthesis mixture by reverse-phase high-pressure liquid chromatography. Identity of the purified peptide was confirmed by amino acid analysis and electrospray mass spectrometry. The final product was judged to be >98% pure based on analytical high-pressure liquid chromatography.
NMR spectroscopy and structure calculation
Samples for NMR were 2 mM peptide in 10 mM sodium phosphate, pH 6.3 prepared in D2O, or a 9:1 mixture of H2O and D2O. The samples used for structure determination also contained 100 mM uniformly deuterated dodecylphosphocholine (Cambridge Isotope Laboratories, Inc.). Details of NMR experiments are provided in the Supplemental material. The structure was calculated from 208 NOE-derived inter- and intraresidue distance restraints, and further refined by the addition of eight hydrogen bond constraints involving backbone amide protons of residues 36–40 and 50–52, where the NOE pattern was characteristic of an α-helix. Distance calibration and structure calculation by simulated annealing were performed with the DYANA software package (Guntert et al. 1997). The MOLMOL program (Koradi et al. 1996) was used for visual analysis of the structure and for preparing molecular graphics figures.
Acknowledgments
We are grateful to Dr. Gary Case for synthesizing the c32–52 peptide. This study was supported by United States Public Health Service Grant GM23105 to R.H.F. and the University of Saskatchewan start-up funds to O.Y.D. Data were collected at the National Magnetic Resonance Facility at Madison, supported by funding from NIH, the University of Wisconsin, the National Science Foundation, and the Department of Agriculture.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Oleg Dmitriev, University of Saskatchewan, 107 Wiggins Road, Saskatoon, Saskatchewan S7N 5E5, Canada; e-mail: Oleg.Dmitriev@usask.ca; fax: (306) 966-4390.
Abbreviation: DPC, dodecylphosphocholine.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072776307.
References
- Abrahams J.P., Leslie, A.G., Lutter, R., and Walker, J.E. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628. [DOI] [PubMed] [Google Scholar]
- Bennett M., Yeagle, J.A., Maciejewski, M., Ocampo, J., and Yeagle, P.L. 2004. Stability of loops in the structure of lactose permease. Biochemistry 43: 12829–12837. [DOI] [PubMed] [Google Scholar]
- Braig K., Menz, R.I., Montgomery, M.G., Leslie, A.G., and Walker, J.E. 2000. Structure of bovine mitochondrial F1-ATPase inhibited by Mg(2+) ADP and aluminium fluoride. Structure 8: 567–573. [DOI] [PubMed] [Google Scholar]
- Cross R.L. 2000. The rotary binding change mechanism of ATP synthases. Biochim. Biophys. Acta 1458: 270–275. [DOI] [PubMed] [Google Scholar]
- Dmitriev O., Jones, P.C., Jiang, W., and Fillingame, R.H. 1999a. Structure of the membrane domain of subunit b of the Escherichia coli F0F1 ATP synthase. J. Biol. Chem. 274: 15598–15604. [DOI] [PubMed] [Google Scholar]
- Dmitriev O.Y., Jones, P.C., and Fillingame, R.H. 1999b. Structure of the subunit c oligomer in the F1F0 ATP synthase: Model derived from solution structure of the monomer and cross-linking in the native enzyme. Proc. Natl. Acad. Sci. 96: 7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame R.H. 1997. Coupling H+ transport and ATP synthesis in F1F0-ATP synthases: Glimpses of interacting parts in a dynamic molecular machine. J. Exp. Biol. 200: 217–224. [DOI] [PubMed] [Google Scholar]
- Fillingame R.H., Jiang, W., and Dmitriev, O.Y. 2000. Coupling H(+) transport to rotary catalysis in F-type ATP synthases: Structure and organization of the transmembrane rotary motor. J. Exp. Biol. 203: 9–17. [DOI] [PubMed] [Google Scholar]
- Girvin M.E., Rastogi, V.K., Abildgaard, F., Markley, J.L., and Fillingame, R.H. 1998. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry 37: 8817–8824. [DOI] [PubMed] [Google Scholar]
- Guntert P., Mumenthaler, C., and Wuthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Hermolin J., Dmitriev, O.Y., Zhang, Y., and Fillingame, R.H. 1999. Defining the domain of binding of F1 subunit ɛ with the polar loop of F0 subunit c in the Escherichia coli ATP synthase. J. Biol. Chem. 274: 17011–17016. [DOI] [PubMed] [Google Scholar]
- Jiang W., Hermolin, J., and Fillingame, R.H. 2001. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. 98: 4966–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katragadda M., Alderfer, J.L., and Yeagle, P.L. 2001. Assembly of a polytopic membrane protein structure from the solution structures of overlapping peptide fragments of bacteriorhodopsin. Biophys. J. 81: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Meier T., Polzer, P., Diederichs, K., Welte, W., and Dimroth, P. 2005. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus . Science 308: 659–662. [DOI] [PubMed] [Google Scholar]
- Menz R.I., Walker, J.E., and Leslie, A.G. 2001. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell 106: 331–341. [DOI] [PubMed] [Google Scholar]
- Mosher M.E., White, L.K., Hermolin, J., and Fillingame, R.H. 1985. H+-ATPase of Escherichia coli. An uncE mutation impairing coupling between F1 and F0 but not F0-mediated H+ translocation. J. Biol. Chem. 260: 4807–4814. [PubMed] [Google Scholar]
- Nakano T., Ikegami, T., Suzuki, T., Yoshida, M., and Akutsu, H. 2006. A new solution structure of ATP synthase subunit c from thermophilic Bacillus PS3, suggesting a local conformational change for H+-translocation. J. Mol. Biol. 358: 132–144. [DOI] [PubMed] [Google Scholar]
- Rastogi V.K. and Girvin, M.E. 1999. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature 402: 263–268. [DOI] [PubMed] [Google Scholar]
- Schneider E. and Altendorf, K. 1986. Proton-conducting portion (F0) from Escherichia coli ATP synthase: Preparation, dissociation into subunits, and reconstitution of an active complex. Methods Enzymol. 126: 569–578. [DOI] [PubMed] [Google Scholar]
- Senior A.E. 1988. ATP synthesis by oxidative phosphorylation. Physiol. Rev. 68: 177–231. [DOI] [PubMed] [Google Scholar]
- Watts S.D., Zhang, Y., Fillingame, R.H., and Capaldi, R.A. 1995. The γ subunit in the Escherichia coli ATP synthase complex (ECF1F0) extends through the stalk and contacts the c subunits of the F0 part. FEBS Lett. 368: 235–238. [DOI] [PubMed] [Google Scholar]
- Watts S.D., Tang, C., and Capaldi, R.A. 1996. The stalk region of the Escherichia coli ATP synthase. Tyrosine 205 of the γ subunit is in the interface between the F1 and F0 parts and can interact with both the ɛ and c oligomer. J. Biol. Chem. 271: 28341–28347. [DOI] [PubMed] [Google Scholar]
- Wilkens S. and Capaldi, R.A. 1998. Solution structure of the ɛ subunit of the F1-ATPase from Escherichia coli and interactions of this subunit with β subunits in the complex. J. Biol. Chem. 273: 26645–26651. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki, E., and Hisabori, T. 2001. ATP synthase—A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2: 669–677. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Fillingame, R.H. 1995. Subunits coupling H+ transport and ATP synthesis in the Escherichia coli ATP synthase. Cys–Cys cross-linking of F1 subunit ɛ to the polar loop of F0 subunit c . J. Biol. Chem. 270: 24609–24614. [PubMed] [Google Scholar]