Abstract
Stably transfected PC12 cell lines expressing similar amounts of chimeric receptors composed of the extracellular domain of the human platelet-derived growth factor (PDGF)β receptor and the transmembrane and intracellular domains of the fibroblast growth factor receptors (FGFRs) 1, 3, and 4 undergo ligand-induced differentiation. The FGFR1 chimera (PFR1) is the most potent of the three, and PFR4 requires more frequent (every 24 hr) addition of ligand to maintain the response. Both PFR1 and -3 also show significant ligand-independent autophosphorylation but PFR4 does not. All of the chimeras activated phospholipase Cγ, Shc, FGFR substrate (FRS)2, and the mitogen-activated protein kinases, ERK1 and 2. PFR4 was moderately weaker in stimulating these effects as well; PFR1 and -3 were comparable. None of the chimeras induced Sos association or were coprecipitated with Shc. Cotransfection of a dominant-negative Shc derivative, with tyrosine at 239, 240, and 317 replaced with phenylalanine, in the PFR-expressing cells was without effect on PDGF-induced neurite outgrowth. The same derivative substantially inhibited the response of these cells to NGF. These results indicate that FGFR1, 3, and 4 (i) are capable of signaling in a similar fashion; (ii) primarily use FRS2 and, perhaps, PLCγ; and (iii) do not utilize Shc. The results also suggest that the principal difference between FGFR1, 3, and 4 is in the strength of the tyrosine kinase activity and that qualitative differences in signaling capacity are likely to be less important.
The production of intracellular signals arising from extracellular stimuli involves several strategies in eukaryotic cells. Plasma membrane-bound receptors with cytoplasmic tyrosine kinase domains represent an important subclass of such generators, particularly for the tissue growth factors, as exemplified by epidermal growth factor, nerve growth factor (NGF), and platelet-derived growth factor (PDGF) (1). In general, these growth factors function by forming active dimers that autophosphorylate selected tyrosine residues in their cytoplasmic sequences, thus enhancing kinase activity and leading to the induction of various downstream pathways via association of effector and/or adaptor molecules with the modified receptor. Both the extracellular ligands and the receptors are characterized by homologous structures, and there is often extensive crossover in ligand binding and subsequent receptor activation.
The fibroblast growth factor (FGF) family is composed of 10 well defined members (and several additional ones tentatively identified from sequence studies) with four genes encoding tyrosine kinase-containing receptors (2). The latter genes give rise to several major and minor receptor types (because of alternative splicing); some alterations in the extracellular domain can markedly change ligand selectivity (3, 4). There are also inactive forms that may play regulatory roles. Thus, given the variability in ligand structure, ligand–receptor affinity, and receptor structure, there is an enormous number of potential combinations that could produce a substantial spectrum of intracellular signals. It is perhaps not surprising that this extensive growth factor family is involved in an impressive array of biological responses and physiological functions, whose complexity matches the structural diversity of the ligands and receptors (5, 6).
As a clearer understanding of the major pathways induced by receptor tyrosine kinases has emerged, a number of studies examining the responses of the FGF receptors (FGFRs), both individually and comparatively, have been reported (7–11). In the main, these have been designed to probe ligand contributions and the nature and strength of the signals produced, i.e., the pathways activated. Under basically similar conditions, it was found that FGFR4 produced a weaker signal than either FGFR1 or 2, particularly with respect to responses involving phospholipase Cγ (PLCγ), Shc, and p89/90 [FGF receptor substrate (FRS)2 or 80H-K] (7–9). In addition, at least one signaling entity, an 85-kDa serine/threonine kinase, was found associated uniquely with FGFR4 (12). Although not compared specifically with the others, most studies with FGFR3 are consistent with the view that it produces signals similar to FGFR1 (11).
Previous studies on FGF signaling have been carried out by using both transient and stable transfectants of FGFR in cells with and without (or nearly so) endogenous receptors (7–11). Because these experiments used full-length receptors, the activating ligands were all from the FGF family; these have variable affinities and utilize heparin (or related compounds) as cofactors in forming (stabilizing) the activated receptor. These studies are also complicated by the presence of endogenous receptors and the foreign environment of the host cell.
To circumvent these issues, we have investigated the signaling capacity of three of the FGFRs in a well studied paradigm, the rat PC12 cell, that normally expresses these receptors, by using chimeric constructs that contain a PDGF extracellular domain substituted for the corresponding FGF ligand-binding domain (13, 14). These molecules are readily expressed and activated in stably transfected PC12 cells and do not interact with the endogenous FGFRs (which remain functionally silent). This approach provides a uniform stimulus that is not affected by heparin (or related substances). In this report, we compare the signaling properties of FGFR1, 3, and 4, which have not been previously examined as a group. Although the results agree with the earlier reports that FGFR4 appears to have a weaker kinase than the other members of the family, all three were found to be much more similar in their responses than these earlier observations had suggested.
MATERIALS AND METHODS
Construction of Chimeras and Cells.
A general scheme for preparing the cDNA of the FGFR chimeras (PFR1, PFR3, and PFR4) used in these studies, as first reported for a chimera of PDGF receptor (PDGFR) and TrkA (15), has been described (refs. 13 and 14; S.R., B. S. Khatra, J. T. Kurokawa, and R.A.B., unpublished data). Briefly, the 3′ end of the extracellular domain of human PDGFRβ (hPDGFRβ)(16) was fused to the engineered intracellular and transmembrane domains of rat FGFR1 (rFGFR1), hFGFR3, and rFGFR4, respectively, by using a MseI restriction site. The FGFR1 and FGFR4 clones were obtained from a PC12 cell library (S.R., unpublished data); the FGFR3 clone was obtained from human cDNA (17). The MseI site, which occurs normally in PDGFR cDNA, was introduced by using PCR at the 5′ end of the intracellular domain of each FGFR. Introduction of the EcoRI restriction site at the 3′ end of each FGFR and elimination of internal MseI and EcoRI sites were achieved by using PCR and/or single-strand DNA mutagenesis with appropriate oligonucleotides. In no case did these change the amino acid sequence from that of the parent molecules. Clonal PC12 cell lines expressing the chimeras were obtained after retroviral infection and G418 selection (14). Single cell lines expressing similar amounts of PFR1, -3, and -4 were chosen and used throughout the experiments.
Cell Culture and Neurite Outgrowth Assay.
PFR1-, PFR3-, and PFR4-expressing clonal cells were grown in DMEM (Irvine Scientific) containing 5% plasma-derived horse serum, 2.5% plasma-derived fetal calf serum (Cocalico Biologicals, Reamstown, PA) and 1% Pen-strep solution (GIBCO/BRL) (complete medium) in tissue culture flasks (Costar) at 37°C in a 5% CO2 humidified atmosphere. For neurite outgrowth assays, cells were plated in collagen-coated six-well plates (Falcon) and scored as described (13).
Immunoprecipitation and Immunoblotting Analysis.
PC12 cell lines expressing PFR1, -3, and -4 were grown in complete medium in either 100- or 150-mm2 collagen-coated tissue culture dishes until 60–70% confluent, starved in DMEM containing 0.2% plasma-derived horse serum, and treated with PDGF-BB (Austral Biological) (30 ng/ml) at 37°C for various lengths of time. Lysis, immunoprecipitation, and immunoblot analyses were carried out as described (13). For coprecipitation experiments, a Nonidet P-40 lysis buffer was used (18). The following antibodies were used: mouse monoclonal anti-hPDGFRβ for immunoprecipitations (Genzyme) and for immunoblotting (Austral Biological); rabbit polyclonal anti-PLCγ and anti-Sos (Santa Cruz Biotechnology); rabbit polyclonal anti-Shc and mouse monoclonal anti-Grb2 (Transduction Laboratories, Lexington, KY) and rabbit anti-phospho-mitogen-activated protein kinase (Promega). For coprecipitation studies to detect FRS2 (p89/90) and FRS2 binding proteins, P13Suc agarose (Oncogene Science) was used.
Immunocomplex Kinase Assay.
Lysates from PC12 cell lines, untreated or stimulated with PDGF (30 ng/ml) at 37°C for 5, 30, and 120 min, were incubated with 10 μl of agarose-conjugated polyclonal anti-Erk1 (Santa Cruz Biotechnology) for 3 hr at 4°C. After washing the agarose beads once in lysis buffer and once in kinase buffer, kinase reactions were performed in the presence of 2 mg/ml myelin basic protein (as substrate), 50 μM ATP, and 5 μCi (1 Ci = 37 GBq) [γ-32P]ATP essentially as described (19).
Expression of Shc and Immunofluorescence Analysis.
The glutathione S-transferase (GST)–Shc or GST–Shc Y239/240/317F constructs have been described (19). PC12 cells were plated at low density onto collagen-coated glass coverslips, grown for 24 hr in complete medium, and transfected with 4 μg/ml DNA and Lipofectin reagent (GIBCO/BRL) according to the manufacturer’s recommendations. After 36 hr in complete medium, the cells were treated with DMEM containing 1% plasma-derived horse serum and growth factors (NGF, 100 ng/ml; PDGF, 30 ng/ml). After 24 hr, they were processed for the immunofluorescence detection of the GST antigen as described (19).
RESULTS
Morphological Responses of PFR1, -3, and -4.
After retroviral transfection and selection, stable lines of PC12 cells bearing each FGFR chimera were established, either clonally or as pools of clones. The clonal lines showed variable levels of receptor expression (14); the pooled lines routinely displayed moderate to high expression levels. The experiments reported herein were all performed with lines expressing high, but comparable, levels of receptor chimera as determined independently by 125I-PDGF binding (data not shown).
As shown in Fig. 1, all three FGFR chimeras stimulated the production of neurites on addition of PDGF. In cells that were mock-transfected or transfected with kinase-inactive chimera, there was no response at any concentration of ligand, in keeping with the absence of PDGFR in these cells (18). At the highest concentration of ligand tested (30 ng/ml) and longest time point (96 hr), maximal differentiation was achieved in all cases. However, at shorter intervals and lesser concentrations of growth factor, significant differences were observed. In cells bearing PFR1, 10 ng/ml PDGF produced 30–40% differentiation, even after only 4 hr of stimulation. In contrast, PFR3 and -4 did not show significant responses at this dose until 24 hr. The more potent capacity of PFR1 to induce differentiation is reflected in the modest but significant levels (≈10%) of neurite-bearing cells at 1 ng/ml PDGF. Neither PFR3 nor PFR4 showed measurable morphological responses at either 1 ng/ml PDGF or in unstimulated controls (data not shown). Importantly, PFR4 required the addition of more ligand after 24 hr to maintain these responses, whereas neither PFR1- nor PFR3-expressing cells did. Thus, PFR1 appears to be generally the strongest inducer of PC12 differentiation and PFR4 the weakest, with PFR3 occupying an intermediate position. Although these findings are at best semiquantitative, they are quite similar to the mitogenic potencies of these receptors (in their native form) expressed in L6 myoblasts (7–9).
Figure 1.
Time- and dose-dependent induction of neurites in PC12 cells expressing chimeric receptors: PFR1 (A); PFR3, (B); and PFR4 (C). Cells, plated as described in Materials and Methods, were treated with various concentrations of PDGF (1, 10, and 30 ng/ml) for 4, 8, 24, 48, and 96 hr, as indicated, and scored for the presence of neurites. After 48 and 72 hr (also 24 hr for PFR4), the medium was changed, and fresh PDGF was added to the cells. In the case of PFR1, the 24-hr time point could not be included because most of the cells expressing this chimera lifted in the presence of PDGF. However, they subsequently reattached to the cell culture plates and presented neurites comparable to the other cell lines at the longer time points. Values are the average of duplicate determinations from a representative experiment.
Ligand Activation of PFR1, -3, and -4.
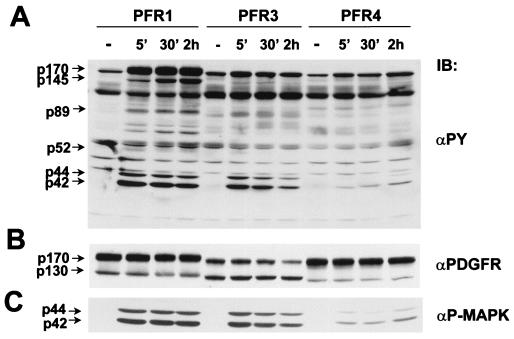
Receptor activation, as measured by autophosphorylation of each chimera 10 min after the addition of PDGF (30 ng/ml), is shown in Fig. 2. The level of tyrosine phosphorylation in immunoprecipitates, by using anti-PDGFR, clearly indicates a more robust modification of PFR1 than either PFR3 or -4. Some ligand-independent phosphorylation of both PFR1 and -3 (Fig. 2A and more clearly in Fig. 2B), but not PFR4, is evident despite the fact that, as judged by the PDGFR blots, shown in Fig. 2A Lower, and by 125I-PDGF binding experiments (data not shown), there is a comparable amount of PFR4 expressed. This ligand-independent phosphorylation is usually attributed to the high density of receptors in the stably transfected cell and, in this case, emphasizes the weak kinase activity of PFR4. Native or mock-transfected cells or cells expressing kinase-negative mutants showed no detectable responses with any of the immune reagents (data not shown). These experiments also clearly show the expression of the mature (p170) and the immature (p130) forms of the receptor chimeras. Both forms are routinely observed in virtually all tyrosine kinase receptor expression experiments. The p130 form has been suggested to correspond to an incompletely glycosylated intermediate and is normally not activated (phosphorylated) (18).
Figure 2.
Comparison of the induced tyrosine kinase activity of PFR1, -3, and -4. PC12 cells expressing the chimeric receptors were incubated at 37°C for 10 min without (−) or with (+) PDGF (30 ng/ml). Lysates (2 mg) were immunoprecipitated with anti-PDGFR and immunoblotted with anti-phosphotyrosine (αPY). (A) Upper, high-molecular-weight region of the anti-phosphotyrosine blot; Lower, the same blot stripped and reprobed with monoclonal anti-PDGFR. The arrows indicate the two forms (mature and incompletely glycosylated) of the chimeric receptors. (B) Upper, longer exposure of the same (full) antiphosphotyrosine blot shown in A. Tyrosine-phosphorylated proteins of 145, 89, and 43 kDa are indicated by arrows; Lower, the same blot stripped and reprobed with anti-PLCγ. Molecular mass standards (kDa) are shown.
Longer exposure of a larger portion of the same gel shown in Fig. 2A reveals some coprecipitated proteins at molecular mass values of 145, 89, and 43 kDa with ligand-stimulated PFR1 (Fig. 2B). The 145-kDa band, seen with all three receptors, was identified as PLCγ as judged by using immunoblotting (Fig. 2B Lower), and this is consistent with previous reports that all three of these receptors can activate this signaling entity in other cell types (albeit variably) (7–11). The activation of PLCγ, as well as other signaling entities, particularly by PFR3 and -4, is more clearly identified by direct immunoprecipitation (see below).
The early kinetics of PDGF-induced activation of each chimera is shown in Fig. 3. The downstream activation of the mitogen-activated protein kinases, ERK1 and -2, by all three receptors (in a ligand-dependent fashion) is shown in Fig. 3A and, more specifically, in Fig. 3C. In keeping with the other responses, activation by PFR1 is most pronounced, but it occurs in all three cases, albeit with different kinetics of activation. Similar results were obtained by using a kinase assay on anti-ERK immunoprecipitates of PFR1, -3, and -4 cell lysates with myelin basic protein as substrate (data not shown). The -fold increase of activation obtained for each lysate, after normalizing the basal level of ERK activity, ranged from 40 (PFR1), 30 (PFR3), and 7 (PFR4) after 5 min of stimulation with PDGF to 50 (PFR1), 10 (PFR3), and 16 (PFR4) after 2 hr of stimulation. This is in keeping with the well established link between ERK 1/ERK2 activation and the differentiation in PC12 cells (15, 20, 21).
Figure 3.
Time course of induced protein tyrosine phosphorylation of PC12 cell lines expressing PFR1, -3, and -4. Cells were untreated (−) or incubated at 37°C with PDGF (30 ng/ml) as indicated. (A) Total cell extracts (50 μg) were immunoblotted with an anti-phosphotyrosine (αPY). Molecular mass standards (kDa) are shown. (B) The same blot was stripped and the top part reprobed with anti-PDGFR. The two forms of the chimeras are indicated. (C) The bottom part of the gel in A was reprobed with anti-active MAPK. p44ERK-1 and p42ERK-2 are indicated.
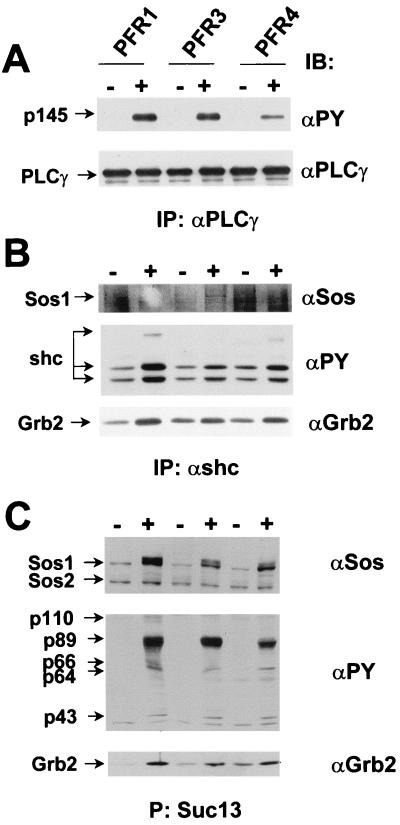
The activation of other signaling entities that have been attributed to the FGFR family, i.e., PLCγ, Shc, Grb2, and p89/90 (FRS2), by each chimera, can be seen in Fig. 3, and more specifically in Fig. 4. As described above, each chimera activates PLCγ, as judged by tyrosine phosphorylation, when stimulated with PDGF (Fig. 4A). Similar to the responses seen in transfected L6 cells (7–9), the activation by PFR4 is less robust. However, activation by PFR1 and -3 are basically indistinguishable. Anti-Shc immunoprecipitates (Fig. 4B) show significant levels of ligand-dependent phosphorylation and some increase in Grb2 binding, but little, if any, association of Sos, the linker protein to Ras (and the downstream activation of ERK1/ERK2). In contrast, by using recombinant p13Suc bound to agarose, which recognizes p89/90 (Fig. 4C), lysates from cells expressing PFR1, -3, and -4 collected 5 min after the addition of PDGF showed strong ligand-dependent activation of p89/90, with associated Grb2 and Sos as well as other unidentified proteins of molecular mass 110, 66, 64 and 43 kDa. Interestingly, the levels of activation of all of these entities were similar for each chimera.
Figure 4.
Tyrosine phosphorylation of the SH2-containing proteins PLCγ, Shc and p89/90 (FRS2) and associated proteins following activation of PFR1, -3, and -4 by PDGF. (A) Tyrosine phosphorylation of immunoprecipitated PLCγ before (−) and after (+) stimulation at 37°C for 10 min with PDGF (30 ng/ml). Upper, immunoblot with anti-phosphotyrosine (αPY); Lower, immunoblot, after stripping and reprobing, with an anti-PLCγ. (B) Tyrosine phosphorylation of immunoprecipitated Shc and its association with Sos and Grb2 after stimulation of PC12 cells at 37°C for 5 min with PDGF (30 ng/ml). Top, immunoblot with anti-Sos; Middle, immunoblot with anti-phosphotyrosine; Bottom, immunoblot with anti-Grb2. (C) Tyrosine phosphorylation of p89/90 (FRS2) and its association with Sos and Grb2 after stimulation of PC12 cells at 37°C for 5 min with PDGF (30 ng/ml). Lysates (PFR1 and PFR3, 2 mg, and PFR4, 3 mg) were treated with recombinant p13Suc agarose to precipitate the proteins. Top, immunoblot with anti-Sos; Middle, immunoblot with anti-phosphotyrosine; Bottom, immunoblot with anti-Grb2. The arrows indicate the tyrosine-phosphorylated polypeptides of 110, 66, 64, and 43 kDa, which coprecipitate with the FRS2-phosphorylated protein.
Role of Shc in FGFR-Induced PC12 Cell Differentiation.
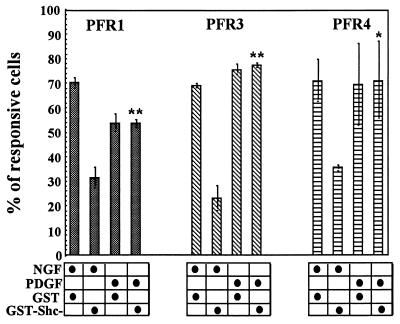
As shown in Fig. 4 and as reported by others (10, 11, 13, 14, 22), various FGFRs do cause phosphorylation of Shc, but evidence of direct interaction with any FGFR is lacking. The absence of Sos association with Shc–Grb2 complexes (Fig. 4B) also suggests that Shc activation by FGFRs may not directly activate ERK1/ERK2 phosphorylation/translocation. However, Shc clearly is specifically bound to and activated by TrkA, which leads directly to NGF-induced PC12 differentiation (15, 21). To test the role of Shc activation by the FGFR chimeras in PC12 cells, a dominant-negative Shc derivative (expressed as a GST–fusion protein), in which Tyr-239, -240, and -317 were mutated to phenylalanine (19), was transiently transfected into cells expressing the chimeras. As shown in Fig. 5, after 24 hr of stimulation by PDGF (to activate the chimera), the cells containing the Shc derivative were indistinguishable from those transfected with the control. However, in all cases, the Shc dominant-negative mutant decreased the differentiative response to NGF in the same cells by ≈50%. Thus, unlike TrkA, none of the three FGFR tested appear to require Shc for this response.
Figure 5.
Effect of the expression of GST–Shc Y239/240/317F fusion protein on NGF- and PDGF-induced neurite outgrowth in PC12 cells expressing PFR chimeric receptors. Clonal cell lines were transfected with DNA coding for GST or GST–Shc Y239/240/317F (SHC). After 36 hr, PC12 cells were treated with NGF (100 ng/ml) or PDGF (30 ng/ml) for 24 hr and processed for immunofluorescence detection of the GST antigen. Each histogram represents the mean of the percentage of immunolabeled cells responsive to NGF or PDGF obtained in three independent experiments. Vertical bars indicate the standard error. ∗, P < 0.05; ∗∗, P < 0.01.
DISCUSSION
PC12 cells respond to external stimuli in a variety of ways, reflecting the chemical signals induced and the concomitant alterations in cell physiology and structure. The benchmark change is the reversible differentiation that is characterized by the cessation of cell division and the outgrowth of neurites produced by factors such as NGF and FGF (23). Although the full spectrum of responses that these agents elicit, and therefore the complete description of the molecular mechanisms involved, has not been determined, it is clear that certain pathways contribute significantly to this ligand-generated differentiation. These emanate from the receptor tyrosine kinases specific for each factor, and studies with chimeric receptors have demonstrated that the ligands themselves (as internalized agents) as well as other entities affecting ligand binding, such as heparinoids, are not required (13, 14, 18). Thus, the specificity of receptor responses can be traced directly to the qualitative and quantitative nature of the events catalyzed by the activated tyrosine kinase domains.
The differentiation of PC12 cells by FGF is also complicated by the expression of multiple kinase-containing receptors, which are variably activated by members of the FGF family (3, 5). FGFR1 and -4 are the dominant species in these cells, but some FGFR3 is also present (14). There is apparently little or no FGFR2 (S.R., unpublished observations). To eliminate these complications and thus allow a more accurate assessment of the signaling capacity of individual receptors, chimeras bearing the extracellular domain of hPDGFRβ and the transmembrane/intracellular domains of the corresponding FGFR were constructed and stably transfected into PC12 cells (13, 14, 18). These experiments demonstrated that FGFR1 and, contrary to an earlier report (24), FGFR3, could produce differentiation without involvement of any endogenous receptors. As described herein, similar experiments now establish that FGFR4 is also capable of inducing this response, although it is not as effective. In fact, repeated addition of ligand (at more frequent intervals than the other PFRs) to maintain the differentiated state is required. Nonetheless, the ability of PFR4 to induce PC12 cell differentiation does demonstrate a greater similarity to the other FGFRs than previously supposed, including the apparent activation of pathways not detected in earlier work (7–9) (see below).
A common feature of tyrosine kinase-containing receptors is their ability to activate multiple pathways, presumably simultaneously. The FGFR family has been linked to three such pathways, two of which are potentially overlapping. PLCγ can be activated by direct interaction with the FGFR1 receptor at Tyr-766 (residue numbering from human FGFR1) after its phosphorylation (25), and this site, as a consensus sequence for the SH2 domain of PLCγ, is conserved in the FGFR family (3). The other two pathways, which use different adaptor structures, i.e., Shc and FRS2 [also denoted SNT (or SLP) and 80K-H (26–29)], both lead to stabilized Ras complexes that in turn initiate a kinase cascade that activates ERK1 and/or -2. Although there are other sites of autophosphorylation besides Tyr-766, none of these are required for the activation of either Shc or FRS2 by FGFR1 (and presumably the other FGFRs) (22).
Previous studies on the activation of these three pathways by FGFRs, done both individually and comparatively, are summarized in Table 1. The studies listed were mainly performed in L6 (or derivative) cells, which have few, if any, endogenous FGFRs (7–11). The semiquantitative values (0–3+) assigned in making this compilation show relative levels of response between receptor types in a given study and are not comparable between reports. Presented in this fashion, a few important features stand out. FGFR1 was consistently found to robustly activate all three pathways; FGFR4 generally did not. Of the three signaling mechanisms, PLCγ was the best for FGFR4, but it was always found to be consistently weaker than with either of the other receptors. Where measured, neither Shc nor FRS2 could be detected. However, ERK1/ERK2, the mitogen-activated protein kinases that are ultimately activated by these adapters, were found to be partially phosphorylated. Finally, other as-yet-unidentified proteins were found to be specifically associated with FGFR1, -3, and -4 (7, 8, 11).
Table 1.
Comparison of ligand-induced FGFR responses
| Receptor/Effector | Cells
|
|||||
|---|---|---|---|---|---|---|
| L6/3T3 | L6/BaF3 | L6E9 | L6 | L6 | PC12 | |
| FGFR1 | ||||||
| PLCγ | +++ | +++ | +++ | ND | +++ | |
| Shc | +++ | +++ | ND | +++ | ++ | |
| SNT | ND | ND | +++ | +++ | ++ | |
| ERK1/2 | +++ | +++ | +++ | ND | +++ | |
| Other | +++† | |||||
| FGFR3 | ||||||
| PLCγ | +++ | +++ | ||||
| Shc | +++ | ++ | ||||
| SNT | +++ | +++ | ||||
| ERK1/2 | +++ | ++ | ||||
| Other | +++‡ | |||||
| FGFR4 | ||||||
| PLCγ | + | + | + | ++ | ||
| Shc | − | − | ND | ++ | ||
| SNT | ND | ND | − | ++ | ||
| ERK1/2 | + | − | + | + | ||
| Other | +++* | |||||
| Ref. | 7 | 8 | 9 | 10 | 11 | This study |
ND, not determined.
, p85; †, p80; ‡, p66.
The results reported herein by using PC12 cells (Table 1) show some similarities and some differences with the various L6 cell data. In keeping with those reports, the FGFR1 chimera (PFR1) was the most active, requiring the least amount of ligand to show significant PC12 cell differentiation and giving the highest levels of receptor autophosphorylation. It strongly activated PLCγ and FRS2 and induced significant levels of Shc phosphorylation. PFR3 was also highly responsive but was not as potent in producing differentiation (at lower ligand concentrations) and in activating ERK1/ERK2. However, its ability to activate FRS2 and PLCγ was about equal to PFR1.
PFR4 showed the greatest differences with the L6 cell data. The activation of all three signaling pathways was comparable to the other two chimeras, although the levels of ERK1/ERK2 activation were reduced, indicating that its signaling specificity is not greatly different than PFR1 and -3. In fact, its decreased potency seems rather to be because of a weaker kinase, reflected in the greatly decreased amount of receptor autophosphorylation and the absence of ligand-independent phosphorylation, which is consistently observed in stable transfectants where receptor is expressed in high amounts and can readily be seen with PFR1 and PFR3 (Fig. 1).
Although all of the PFR chimeras tested stimulated the phosphorylation of Shc (and the association of Grb2), the absence of Sos in the precipitates again raises the question of the contribution of this apparent activation to FGFR signaling. Previous experiments (10, 13, 14, 29) have consistently failed to show Shc association with FGFRs, suggesting that the interaction is either too transient to detect or that the activation arises through the intermediate agency of other adapters/effectors. The identification of FRS2 as a strong signaling device for all three FGFRs tested in these experiments also potentially eliminates any need for this effector. That it may not signal at all in FGF-stimulated cells is suggested by the effect of the dominant-negative Shc construct in which the three tyrosine residues that provide sites of phosphorylation were converted to phenylalanine (19). This derivative did not impair the neurite outgrowth activity of any of the PFRs but significantly decreased this response when NGF, known to use Shc as a mediator (15, 20, 21), was used as stimulus. This is particularly significant because NGF (via TrkA) also uses FRS2 (SNT) as mediator (21) and, in fact, the residual activity may result, in part, from its activity. Clearly, FRS2 cannot fully substitute for Shc in the NGF response. Although these experiments alone do not rule out that Shc is an adaptor for FGFRs, taken with the other observations, it strongly suggests that this is not a productive activation leading to signals that contribute to PC12 cell differentiation (and possibly not to other functions as well). In fact, the activation of Shc may be secondary to the receptor activation and may reflect other Shc functions/activities, such as translocation from the cytoplasm to the cytoskeleton, where phosphorylation can occur (30).
Three proteins of unknown function have been shown to be associated with FGFR1, -3, and -4 (Table 1). These entities were either not observed or were seen at very low levels in the receptor immunoprecipitations or the whole-cell lysates blotted with phosphotyrosine antibodies. In the absence of specific antibodies, it is uncertain whether these entities are even present (and associated with the corresponding FGFRs) in PC12 cells.
This study describes the comparison of FGFR1, -3, and -4 signaling in a single paradigm. In the absence of ligand effects, eliminated by the chimeric constructs, the intracellular signaling properties of these receptors show a greater similarity than has been previously observed. This emphasizes the importance of cellular context when evaluating signaling properties and the fact that the same receptors may behave quite differently from cell type to cell type.
Acknowledgments
This work was supported by U.S. Public Health Service Research Grant AG09735 and program project Grant HD22657. E.D.F. was supported by U.S. Public Health Service Research Training Grant GM-07311.
ABBREVIATIONS
- NGF
nerve growth factor
- PDGF
platelet-derived growth factor
- PDGFR
PDGF receptor
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- GST
glutathione S-transferase
- PLCγ
phospholipase Cγ
- FRS2
FGF receptor substrate 2
References
- 1.Schlessinger J, Ullrich A. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 2.McKeehan W L, Wang F, Kan M. Prog Nucleic Acid Res Mol Biol. 1998;50:136–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 3.Jaye M, Schlessinger J, Dionne C A. Biochem Biophys Acta. 1992;1135:185–199. doi: 10.1016/0167-4889(92)90136-y. [DOI] [PubMed] [Google Scholar]
- 4.Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D. J Mol Evol. 1997;44:43–56. doi: 10.1007/pl00006120. [DOI] [PubMed] [Google Scholar]
- 5.Johnson D E, Williams L T. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 6.Slavin J. Cell Biol Int. 1995;19:431–444. doi: 10.1006/cbir.1995.1087. [DOI] [PubMed] [Google Scholar]
- 7.Vainikka S, Joukov V, Wennström S, Bergman M, Pelicci P G, Alitalo K. J Biol Chem. 1994;269:18320–18326. [PubMed] [Google Scholar]
- 8.Wang J-K, Gao G, Goldfarb M. Mol Cell Biol. 1994;14:181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaoul E, Reich-Slotky R, Berman B, Ron D. Oncogene. 1995;10:1553–1561. [PubMed] [Google Scholar]
- 10.Klint P, Kanda S, Claesson-Welsh L. J Biol Chem. 1995;270:23337–23344. doi: 10.1074/jbc.270.40.23337. [DOI] [PubMed] [Google Scholar]
- 11.Kanai M, Goke M, Tsunekawa S, Podolsky D K. J Biol Chem. 1997;272:6621–6628. doi: 10.1074/jbc.272.10.6621. [DOI] [PubMed] [Google Scholar]
- 12.Vainikka S, Joukov V, Klint P, Alitalo K. J Biol Chem. 1996;271:1270–1273. doi: 10.1074/jbc.271.3.1270. [DOI] [PubMed] [Google Scholar]
- 13.Thompson L M, Raffioni S, Wasmuth J J, Bradshaw R A. Mol Cell Biol. 1997;17:4169–4177. doi: 10.1128/mcb.17.7.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foehr E D, Raffioni S, Fujii R, Bradshaw R A. Immunol Cell Biol. 1998;76:406–413. doi: 10.1046/j.1440-1711.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 15.Obermeier A, Bradshaw R A, Seedorf K, Choidas A, Schlessinger J, Ullrich A. EMBO J. 1994;13:1585–1590. doi: 10.1002/j.1460-2075.1994.tb06421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claesson-Welsh L, Eriksson A, Moren A, Severinsson L, Ek B, Ostman A, Betsholtz C, Heldin C-H. Mol Cell Biol. 1988;8:3476–3486. doi: 10.1128/mcb.8.8.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson L M, Plummer S, Schalling M, Altherr M R, Gusella F, Housman D E, Wasmuth J J. Genomics. 1991;11:1133–1142. doi: 10.1016/0888-7543(91)90041-c. [DOI] [PubMed] [Google Scholar]
- 18.Raffioni S, Zhu Y-Z, Bradshaw R A, Thompson L M. J Biol Chem. 1998;273:35250–35259. doi: 10.1074/jbc.273.52.35250. [DOI] [PubMed] [Google Scholar]
- 19.Thomas D, Bradshaw R A. J Biol Chem. 1997;272:22293–22299. doi: 10.1074/jbc.272.35.22293. [DOI] [PubMed] [Google Scholar]
- 20.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 21.Stephens R M, Loeb D M, Copeland T D, Pawson T, Greene L A, Kaplan D R. Neuron. 1994;12:691–705. doi: 10.1016/0896-6273(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi M, Dikic I, Sorokin A, Burgess W H, Jaye M, Schlessinger J. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene L A, Tischler A S. Adv Cell Neurobiol. 1982;3:373–414. [Google Scholar]
- 24.Lin H-Y, Xu J, Ornitz D M, Halegoua S, Hayman M J. J Neurosci. 1996;16:4579–4587. doi: 10.1523/JNEUROSCI.16-15-04579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammadi M, Honegger A M, Rotin D, Fischer R, Bellot F, Li W, Dionne C A, Jaye M, Rubinstein M, Schlessinger J. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabin S J, Cleghon V, Kaplan D R. Mol Cell Biol. 1993;4:2203–2213. doi: 10.1128/mcb.13.4.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J K, Xu H, Li H C, Goldfarb M. Oncogene. 1996;13:721–729. [PubMed] [Google Scholar]
- 28.Ong S H, Goh K C, Lim Y P, Low B C, Klint P, Claesson-Welsh L, Cao X, Tan Y H, Guy G R. Biochem Biophys Res Commun. 1996;225:1021–1026. doi: 10.1006/bbrc.1996.1288. [DOI] [PubMed] [Google Scholar]
- 29.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 30.Thomas D, Patterson S D, Bradshaw R A. J Biol Chem. 1995;270:28924–31. doi: 10.1074/jbc.270.48.28924. [DOI] [PubMed] [Google Scholar]