Abstract
AcrAB is a constitutively expressed, major multidrug efflux system of Escherichia coli. We have purified the cytoplasmic membrane component, AcrB, to near homogeneity, and reconstituted the protein into proteoliposomes. In the presence of ΔpH (outside acid), the protein catalyzed the extrusion of fluorescent phospholipids, which were then trapped by protein-free acceptor vesicles. Known substrates of AcrAB, such as bile acids, erythromycin, and cloxacillin, inhibited this activity. Addition of various drugs to AcrB-containing proteoliposomes, in the presence of ΔpH (inside acid) resulted in proton efflux, suggesting that AcrB is a proton antiporter. Interestingly, fluorescent lipid extrusion was accelerated strongly by the periplasmic protein AcrA in the presence of Mg2+, and at pH 5.0 AcrA alone produced a slow mixing of lipids of different vesicles, without causing the mixing of intravesicular material. These results suggest that AcrA brings two membranes together, and under certain conditions may even cause the fusion of at least the outer leaflets of the membranes, contributing to the ability of the AcrAB–TolC system to pump drugs out directly into the medium.
Emergence of microbial resistance has shadowed the introduction of every new therapeutic agent. Of particular threat is the presence of a mechanism that, upon the exposure of a microbe to a single agent, causes it to develop cross-resistance to many other unrelated drugs. This phenotype often results from the overexpression of an energy-dependent, multidrug efflux system. In various Gram-negative pathogens these multidrug transporters produce intrinsic resistance to many agents (1–3).
The outer membrane of Gram-negative bacteria is an effective permeability barrier, acting together with multidrug pumps to increase resistance (1). Such synergism becomes possible because of the unique construction of many Gram-negative multidrug transporters: these pumps appear to translocate drug molecules across both the cytoplasmic, inner, membrane and the outer membrane. Because drugs have to come in slowly through the outer membrane barrier, such direct efflux is extremely efficient in producing resistance: computer simulation suggests that in Escherichia coli, direct efflux of tetracycline by the complex of this type is about 100 times more effective in producing resistance than the simple efflux into the periplasm (4). Genetic studies suggest that each of these transporters consists of three components (1). The efflux pump located in the inner membrane presumably forms a complex with a periplasmic protein, belonging to the MFP (membrane fusion protein) family (5), as well as an outer membrane channel, to enable direct drug efflux into the medium (1, 6). The AcrAB–TolC system of E. coli is a typical example of these complexes (1).
In the acrAB locus of E. coli, acrB encodes a 1048-residue protein with 12 putative transmembrane α-helices. The protein belongs to the RND (resistance–nodulation–cell division) transporter family (7), which appears to catalyze efflux at the expense of protonmotive force. The AcrAB system transports drug molecules directly into the medium, bypassing the periplasm (4). An MFP, the periplasmic lipoprotein AcrA, is required for the function of AcrB in intact cells (8). AcrA has an unusual, elongated form, with a predicted length of 17 nm (9), in agreement with the idea that AcrA links or fuses inner and outer membranes. The third component of the system, the outer membrane channel, is most likely TolC (10).
AcrAB–TolC is the major contributor to the intrinsic resistance of E. coli to solvents (11), dyes, detergents, and lipophilic antibiotics such as novobiocin, erythromycin, fusidic acid (8), and cloxacillin (12). When overexpressed, the system generates significant levels of resistance to many commonly used antibiotics, such as tetracycline and chloramphenicol (13). Drug uptake experiments in whole cells (9, 14) suggested that AcrB is a protonmotive force-dependent drug efflux transporter. In this study we describe purification and reconstitution of the AcrAB system.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification.
AcrA here means the unlipidated, yet fully functional AcrA protein, which was purified as described (9). It was stored in 20 mM Tris⋅HCl, pH 8.0/0.2 M NaCl/1 mM EDTA on ice.
The synthetic peptide corresponding to residues 1032–1045 of AcrB was obtained from Zymed Laboratories in a form coupled to keyhole limpet hemocyanin. This conjugate was used to generate rabbit polyclonal antibodies. When inner membrane proteins of E. coli were analyzed by Western blotting with this antibody, only one band with the apparent molecular mass of 110 kDa (expected: 113 kDa) was seen in the acrAB+ parent strain AG102, but not in its ΔacrAB derivative AG102A (13).
For purification of AcrB, DH5α cells containing the plasmid pUC151A (14) were grown in 8 liters of LB medium containing 100 μg/ml ampicillin. At OD600 of 1.8 the cells were harvested and washed once in ice-cold 20 mM Tris⋅HCl, pH 8.0/0.1 M NaCl (buffer A). Unless noted otherwise, all solutions from this step on contained 1 mM phenylmethylsulfonyl fluoride. The cells were then resuspended in 200 ml of buffer A containing 1 mM MgCl2 and 0.1 mg/ml DNase I. The cells were broken by one pass through a French pressure cell at 12,000 psi (1 psi = 6.89 kPa), and after 10-min incubation on ice, NaEDTA (pH 8.0) was added to 2 mM. The supernatant obtained by centrifugation of the lysate at 7,000 rpm for 20 min was further centrifuged at 40,000 rpm for 60 min in a Ti45 rotor (Beckman). The pellet was resuspended in 100 ml of buffer A. An equal volume of 6 M urea in buffer A (at room temperature) was then slowly added to the ice-cold suspension with stirring. After 10 min, membranes were collected by centrifugation as above and were washed in buffer A. The sediment was resuspended in 20 ml of binding buffer (20 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/5 mM imidazole) followed by the slow addition of an equal volume of 10% (vol/vol) Triton X-100 in binding buffer. After overnight storage at 4°C, the mixture was centrifuged as above, and the supernatant was loaded onto a metal-chelating Sepharose column (HiTrap, Pharmacia) charged with Cu2+, which had been equilibrated with the binding buffer containing 0.2% Triton X-100 (buffer B). The column was washed with buffer B and then eluted with a linear gradient of imidazole (from 5 mM to 200 mM) in buffer B. The fractions containing AcrB, which came off at about 0.1 M imidazole, were identified by immunoblotting with the AcrB-specific antibody. The purity of preparation, as assessed by SDS/PAGE and Coomassie blue staining, was >90% (see below). AcrB was stable for at least 1 week at 4°C.
For reconstitution into proteoliposomes, AcrB was processed as follows. Imidazole was first removed by gel filtration on Sepharose G-25 equilibrated with buffer B. AcrB was then bound again to the Cu2+-column and, after an exhaustive wash with a buffer containing 25 mM Hepes–KOH (pH 7.0), 0.5 M KCl, 5 mM imidazole, 1.2% octyl β-d-glucopyranoside, and 10% glycerol, was eluted with the same buffer containing 0.1 M imidazole. AcrB was used for reconstitution soon after elution, because it was stable only for several hours in this state. (It became aggregated if the concentration exceeded 0.3 mg/ml.)
Protein Assays and Analysis.
Protein was determined with BCA reagent (Pierce) or amido black (15) with bovine serum albumin as standard. SDS/PAGE was performed by standard methods through 5% stacking and 10% separating gels, with samples solubilized at room temperature. Gels were stained with Coomassie blue or silver nitrate. For immunoblotting, proteins were transferred electrophoretically to poly(vinylidene difluoride) (PVDF) membrane (Bio-Rad) in 3-(cyclohexylamino)-1-propanesulfonic acid–NaOH (10 mM, pH 11.0) and methanol (10%), and AcrB was visualized with the anti-AcrB rabbit serum and alkaline-phosphatase-conjugated anti-rabbit antibody (Sigma).
AcrB Reconstitution into Vesicles.
AcrB was reconstituted into proteoliposomes by detergent dilution as described (16), except the following. E. coli polar lipids (Avanti Polar Lipids) were dispersed in reconstitution buffer (25 mM Hepes–KOH, pH7.0/0.1 M KCl/1 mM DTT). AcrB (50 μg) in 1.2% octyl β-d-glucopyranoside was added to 4.5 mg of lipids, and the total volume was adjusted to 0.5 ml with the reconstitution buffer and 20% octyl β-d-glucopyranoside so that the final detergent concentration was 1.1%. As a control, vesicles without AcrB were prepared in the same manner. The mixtures were diluted with 3 ml of precooled reconstitution buffer, and the detergent was removed by dialysis against 1 liter of reconstitution buffer containing 2 g of Bio-Beads SM2 (Bio-Rad) overnight at 4°C.
For the preparation of donor vesicles, 0.25 ml of premixed lipid solution in chloroform (20 mg/ml) containing E. coli phospholipids, 1,2-dipalmitoyl N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)phosphatidylethanolamine (N-NBD-PE), and 1,2-dipalmitoyl N-(lissamine rhodamine B sulfonyl)phosphatidylethanolamine (N-Rh-PE) in a 99:0.5:0.5 molar ratio, was dried down in a glass tube under nitrogen, and the tubes were kept under reduced pressure for at least 2 hr. The lipid was rehydrated in the reconstitution buffer at room temperature and then vesicles were prepared as described above. Unlabeled acceptor vesicles were prepared similarly without adding fluorescent phosphatidylethanolamine derivatives. The size of reconstituted vesicles was measured by dynamic light scattering as described previously (9). All experiments with vesicles were carried out in 25 mM Hepes–KOH, pH 7.0/0.1 M KCl (assay buffer), except sometimes the pH was adjusted to a different value, and in some cases 5 mM MgCl2 was added. For pH ranges of 4.5 to 6, 2-(N-morpholino)ethanesulfonic acid buffer was substituted for Hepes.
Fluorescence Measurements.
The fluorescent probes were obtained from Molecular Probes. Fluorescence measurements were carried out with a Shimadzu RF-5301 spectrofluorimeter under continuous stirring in a thermostated cuvette holder at 30°C. In all experiments vesicles (up to 20 μl) were added to 2 ml of buffer. Excitation and emission wavelengths used for each fluorescent probe were, respectively, pyranine, 455 and 509 nm; N-NBD-PE, 460 and 538 nm; and 1-aminonaphthalene-3,6,8-trisulfonate (ANTS), 360 and 540 nm.
Transport of Fluorescent Phospholipids Between Vesicles.
AcrB-dependent extrusion of fluorescent phospholipids was measured by quenching of NBD fluorescence, caused by resonance energy transfer to N-Rh-PE (17). Typically, 5 μl of donor AcrB-vesicles containing these two fluorescent phospholipids (40 nmol of total phospholipid, 0.36 μg of AcrB) were mixed with 10 μl of unlabeled acceptor vesicles (650 nmol of phospholipid). The reaction was initiated at 30°C by addition of MgCl2, and the increase of NBD fluorescence was followed. After termination of the reaction by addition of EDTA (final concentration 10 mM), the N-NBD-PE fluorescence was completely dequenched by solubilizing vesicles with the detergent dodecyl maltoside (final concentration 0.5%). AcrA-induced transfer of fluorescent lipids was followed in a similar way, using AcrB-free donor vesicles.
Detection of Transmembrane H+ Flux.
The method of Clement and Gould (18) was used. A hydrophilic fluorescent pH probe, pyranine, was entrapped in the intravesicular space by rehydrating lipids in solutions containing 1 mM probe, and the suspensions were then frozen in a dry ice/ethanol bath and thawed at room temperature twice. Between cycles liposomes were briefly bath-sonicated (19). Untrapped probe was removed by gel filtration (P-40 column, Pharmacia). Our vesicles bound little pyranine on their external surface, and less than 5% of the fluorescence changes arose from the externally bound probe. A standard curve was developed by calibrating the fluorescent response of the entrapped pyranine to changes in the external pH in the presence of nigericin (0.1 μM) (20). The fluorescence response of pyranine was linear in the region from pH 6.8 to pH 8.0, as found previously (18).
Content Mixing.
ANTS solution was entrapped in one vesicle population and N,N′-p-xylenebis(pyridinium bromide) (DPX) solution in the other. Fusion between the two populations causes quenching of ANTS fluorescence by DPX, through the mixing of aqueous contents of the two vesicle species (21). Simple release of liposome contents does not decrease fluorescence, because quenching by DPX is highly concentration dependent. ANTS and DPX were encapsulated as described for pyranine. These vesicles were mixed in a 1:1 ratio in the assay buffer.
AcrA Binding to Vesicles.
This binding was measured by coflotation of AcrA and vesicles in a Nycodenz (Sigma) step gradient. Incubation mixtures (0.3 ml) containing AcrA (24 μg) and fluorescent vesicles (650 nmol) in the assay buffer were prepared and MgCl2 was added, when appropriate, to 5 mM. After 10 min at 37°C, 0.3 ml of 80% (wt/vol) Nycodenz dissolved in the same buffer was added. The mixtures were loaded into 11 × 34 mm ultraclear centrifuge tubes and overlaid with 1 ml of 30% (wt/vol) Nycodenz in assay buffer followed by 0.1 ml of the same buffer. The samples were then centrifuged in a TLS55 rotor (Beckman) at 45,000 rpm for 3 hr at 4°C. The vesicles were harvested from the 0/30% Nycodenz interface. Protein concentration was determined by amido black assay and normalized to the phospholipid concentration as determined by total phosphorus assay (22).
In the fluorescence energy transfer assay, N-NBD-PE was mixed with phospholipids at a molar ratio of 1:99. NBD-labeled vesicles were added into assay buffer to 43 μM, and fluorescence was recorded with excitation at 290 nm (INBD0). Then AcrA was added and mixture was allowed to equilibrate at 30°C for 5 min. Emission from NBD via energy transfer from tryptophan was recorded (INBD). The extent of the energy transfer was calculated as (INBD − INBD0)/INBD0.
RESULTS AND DISCUSSION
Purification and Reconstitution of AcrB.
Antibody detection showed that AcrB, produced by E. coli DH5α/pUC151A, was present only in the membrane fraction, even after washing with 3 M urea (not shown). For purification of AcrB we used the histidine cluster in the C-terminal part of AcrB (Experimental Procedures). This feature promoted strong adsorption of AcrB onto a copper affinity column. AcrB was eluted with 0.1 M imidazole, and was more than 90% pure (Fig. 1A). We used this affinity-purified AcrB, although the remaining low molecular weight contaminants could be removed by hydroxyapatite chromatography (not shown).
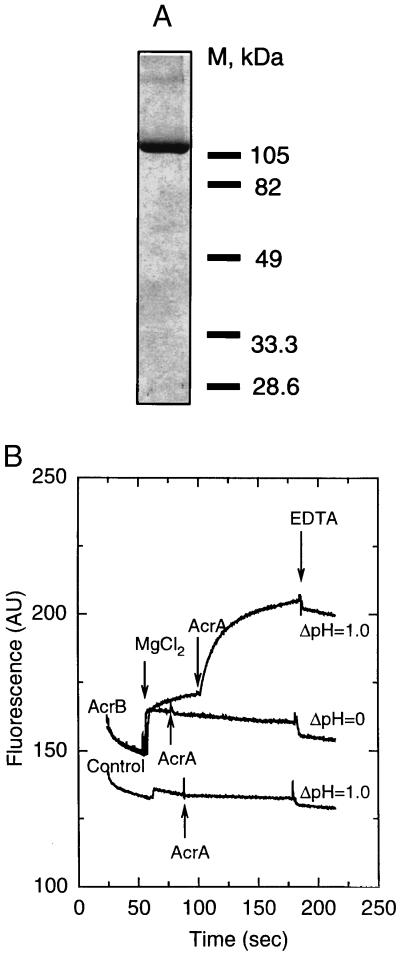
Figure 1.
(A) SDS/PAGE of affinity-purified AcrB, stained with Coomassie blue. (B) Intermembrane transport of fluorescent phospholipids by reconstituted AcrB. Donor vesicles containing AcrB pump (AcrB) and protein-free control vesicles (control) were mixed with a large excess of acceptor vesicles, and NBD fluorescence was monitored at 30°C (AU, arbitrary units). The ΔpH was generated by dilution of the vesicles (internal pH, 7.0) into buffer of pH 6.0. The transport reaction was initiated by addition of 5 mM MgCl2, followed by addition of AcrA to 15 μg/ml. The transport was terminated by addition of EDTA (10 mM).
To reconstitute AcrB into the vesicles, Triton X-100 was exchanged with octyl β-d-glucopyranoside. Phospholipids were mixed with AcrB, and octyl β-d-glucopyranoside was diluted out. The recovery of phospholipids in vesicles was 50–90%. About 90% of the added AcrB typically comigrated with the assembled lipid vesicles (not shown). The mean hydrodynamic radii of both AcrB-containing and AcrB-free vesicles were 50 ± 14 nm. The molar ratio of lipid to protein in vesicles was between 5,000 and 10,000. Assuming a cross section of 65 Å2 per phospholipid molecule, we estimate that the average proteoliposome contained 5–10 copies of AcrB.
Intermembrane Transport of Fluorescent Phospholipid Derivatives by Reconstituted AcrAB System.
The major mechanism for the intrinsic resistance of Gram-negative bacteria to many antibacterial agents appears to be the unique efflux systems, such as the AcrAB–TolC complex, that expel drugs directly into the medium, so that they cannot easily reenter the cells because of the effective outer membrane barrier. The sequence of the inner membrane protein AcrB suggested that it is an efflux pump driven by the protonmotive force (7, 14). However, our attempts to demonstrate directly the accumulation of radioactive drugs by everted membrane vesicles were mostly unsuccessful, presumably because the substrates of AcrB are lipophilic molecules that readily diffuse back across the membrane into the medium. In fact, such an assay was successful so far only with membrane-impermeant taurocholate (23). Nor was it possible to show the extrusion of lipophilic fluorescent dyes from within the bilayer, probably because they reentered spontaneously into the bilayer.
The lipophilicity of the substrates suggested that AcrB recognizes and expels its substrates from within the phospholipid bilayer (1), similar to the mechanism suggested for the mammalian Mdr pump (24). Encouraged by the knowledge that the mouse Mdr2 pump recognizes a phospholipid as substrate (25), we examined whether AcrB extruded fluorescent derivatives of phosphatidylethanolamine, N-NBD-PE and N-Rh-PE, which were initially incorporated at high concentrations into the donor (AcrB-containing) vesicles so that the NBD fluorescence was quenched due to energy transfer to rhodamine. Active extrusion of these fluorescent phospholipids should decrease their concentrations in the bilayer and produce a decreased quenching, or increased fluorescence of NBD. To prevent the reentry of expelled lipids into the bilayer of the original vesicles, we added, as a trap, an excess of “acceptor” vesicles that contained neither AcrB nor fluorescent lipids.
As the driving force for transport, a pH gradient was generated by dilution of the donor and acceptor vesicles, made in a pH 7.0 buffer, into a medium of pH 6.0. When Mg2+ was added, there was a small, rapid increase in fluorescence (Fig. 1B). This increase occurred in the absence of ΔpH and was largely reversible by EDTA (Fig. 1B); it was probably caused by the binding of Mg2+. Importantly, only with the AcrB-containing proteoliposomes was there a sustained increase in NBD fluorescence, which was not seen in the absence of the pH gradient (Fig. 1B, see AcrB, ΔpH = 1.0 curve, between the addition of MgCl2 and AcrA), a result suggesting the ΔpH-driven, AcrB-catalyzed, expulsion of phospholipid derivatives from the donor vesicles. This increase did not occur in the absence of acceptor vesicles (not shown).
To confirm that the transport was dependent on AcrB pump, the donor vesicles were treated with 0.1 μg/ml trypsin. The treatment decreased (to 50% after 15-min digestion; data not shown) the slow, AcrB-dependent fluorescence increase, which presumably reflects lipid transport.
Interestingly, when AcrA, which appears to act by connecting two membranes (9), was present together with Mg2+, the AcrB-dependent transport activity was greatly stimulated (Fig. 1B; Fig. 2 Inset). AcrA, however, did not induce transport in AcrB-free vesicles at pH 6.0 (Fig. 1B).
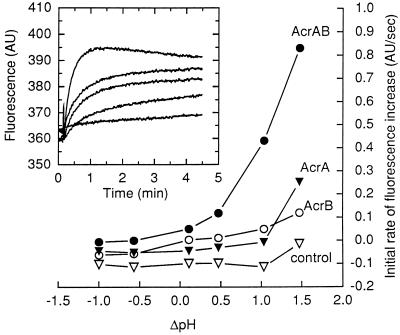
Figure 2.
Initial rates of the intermembrane transport of fluorescent phospholipids as a function of ΔpH. The magnitude of ΔpH across the membrane was adjusted by a 200-fold dilution of the donor vesicles (internal pH, 7.0) into buffers of different pH values. All reaction mixtures contained 5 mM MgCl2. AcrA, when added, was at 25 μg/ml. (Inset) Time course of NBD fluorescence dequenching by AcrB-containing vesicles in the presence of AcrA, which was present at 90 (top curve), 60, 30, 15, or 0 μg/ml (bottom curve). Intravesicular pH was 7.0, and the pH of the assay buffer was 6.0. MgCl2 was present at 5 mM.
The dependence of AcrB transport activity on ΔpH was systematically studied by altering the external pH with vesicles of internal pH 7.0 (Fig. 2). Increase of ΔpH (exterior acid) resulted in the stimulation of the activity. [External pH of 7.0 caused a similar transport in vesicles with an internal pH of 8.0, indicating that the effect was not caused simply by the acidic external pH alone (not shown).] When ΔpH was abolished or reversed, the presence of AcrB and/or AcrA had little effect on the intermembrane phospholipid transfer. Orientation of reconstituted AcrB in the proteoliposomes is not known. However, imposition of outside-acid ΔpH presumably caused only those AcrB proteins inserted in the natural orientation to function—i.e., to extrude the substrates into the medium and onto the acceptor vesicles.
At ΔpH of 1.5, AcrA alone caused a slow transport of fluorescent lipids (Fig. 2). The role of AcrA in lipid movement is examined further in the last section of this paper.
Known Substrates of AcrAB Efflux Pump Inhibit the Extrusion of Fluorescent Phospholipids.
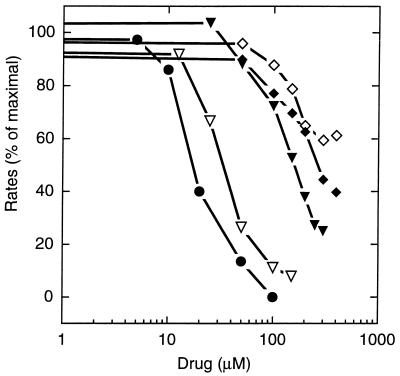
We have used NBD- and rhodamine-labeled phospholipids because of their energy transfer characteristics. However, these fluorescent lipids will not readily cross the outer membrane, and we therefore cannot tell whether they are good substrates of the AcrAB efflux system by examining intact cells. For this reason, we tested whether known substrates of this pump, including lipophilic antibiotics (8) and bile salts (23), competed against the extrusion of the fluorescent phospholipids in our system. Significant inhibition was observed in most cases (Fig. 3). The inhibitory effect did not depend on the charge or pKa of the agent (Table 1). Of the compounds tested the most potent were the bile salts (Fig. 3; Table 1). Because the IC50 values of these compounds were sufficiently low, nonspecific effects, such as the binding of Mg2+, can be excluded as their mode of action. Among antibiotics, erythromycin and cloxacillin inhibited lipid transport, but chloramphenicol did not. However, chloramphenicol caused proton flux (see below), and the result here may be due to suboptimal experimental conditions.
Figure 3.
Transport of fluorescent phospholipid by AcrB is inhibited by various drugs. The initial rates of the intermembrane lipid transfer were measured in the presence of erythromycin (⋄), cloxacillin (♦), glycochenodeoxycholate (▾), glycocholate (▿), and taurocholate (●). Each experimental point is the average of duplicates. The experiments were performed as in the “AcrB, ΔpH = 1.0” curve of Fig. 1B, except that the vesicles were first equilibrated with the drug. Initial rates were measured after the addition of AcrA to 25 μg/ml.
Table 1.
Activity of various substrates of the AcrAB efflux system
| Agent | AcrAB-mediated resistance* | Charge | pKa | IC50 for fluorescent lipid extrusion,† μM | t1/2 for ΔpH dissipation in AcrB vesicles,‡ min | t1/2 for transmembrane diffusion,§ sec |
|---|---|---|---|---|---|---|
| Novobiocin | 32 | − | 4.3 | ND¶ | 2.3 | <2 |
| Fusidic acid | 32 | − | 5.4 | ND¶ | ND | <2 |
| Cloxacillin | 256 | − | 2.7 | 115 | 1.2 | 8.5 |
| Erythromycin | 32 | + | 8.8 | 110 | 2.6 | 6 |
| Chloramphenicol | 8 | No charge | —‖ | 1.9 | ND | |
| Glycochenodeoxycholate | ND | − | 4.2 | 50 | ND | 23 |
| Glycocholate | ND | − | 4.0 | 105 | 1.5 | >1,200 |
| Taurocholate | >16 | − | 1.9 | 15 | 1.0 | >1,200 |
Figures represent the ratio between the minimal inhibitory concentration of acrAb+ parent strain and that of the ΔacrAB mutant strain (7, 8, 13, 14). ND, not determined.
From Fig. 3.
Not determined because stock solutions at high concentrations could not be prepared at pH 6.0.
No activity detected in the range tested.
From Fig. 4.
Measured as described in text, by adding drugs (up to 0.2 mM) to AcrB-free vesicles at pH 7.0 and ΔpH = 0.
These results, showing inhibition by known AcrAB substrates, thus strongly suggest that the extrusion of fluorescent phospholipids reflects the activity of AcrB pump. It is not known whether AcrB pumps out unmodified natural phospholipids. However, a mammalian multidrug transporter Mrp1 uses, as substrates, those phospholipids labeled with fluorescent fatty acids, but not natural phospholipids (26). In any case, the observed extrusion of phospholipid derivatives is consistent with the idea that AcrB captures its substrates from within the bilayer (1). Similarly, the Mdr pump of mammalian cells (24) and LmrA transporter from Lactococcus lactis (27) were shown to bind their substrates within the bilayer.
Drug-Induced Proton Efflux by Reconstituted AcrB.
We tested whether drug transport by the reconstituted AcrB was accompanied by proton flux, by following the intravesicular pH with the water-soluble fluorescent pH probe pyranine, which does not cross the vesicle membrane (18, 20). When protein-free vesicles with entrapped pyranine were pre-equilibrated in the pH 7.0 buffer and then were diluted into the pH 6.0 buffer, the pyranine fluorescence (therefore intravesicular pH) decreased slowly with an apparent half-time of more than 5 min, through the slow, spontaneous leakage of H+ (not shown). Upon addition of nigericin, which makes membrane permeable to H+, the pH gradient collapsed instantaneously (not shown). When erythromycin (0.2–0.8 mM) was added to the vesicles, the entry of the uncharged erythromycin base resulted in a small, immediate increase of internal pH, which was complete within seconds. After this phase, the membrane showed the same degree of leakiness as found in the absence of the drug (not shown). Erythromycin (0.8 mM) did not affect fluorescence when added directly to a solution of pyranine. Similar results were obtained for several other known substrates of the AcrB pump, with lipophilic acids producing a small acidification of intravesicular space (Table 1, last column). Proton leak was not visibly enhanced by the presence of agents shown in Table 1 (up to 0.4 mM).
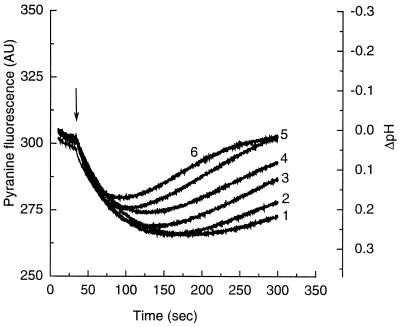
Because many charged drugs produced initial changes in intravesicular pH as described above, we chose to let the drugs come to an equilibrium with vesicles and then start the reaction by generating ΔpH. Thus valinomycin was added to K+-containing vesicles in an isoosmotic NaCl-containing buffer. This generated a transmembrane electrical potential, which then was rapidly converted to an interior-acid ΔpH (Fig. 4) in the presence of 0.1 M Cl− (28). In our system, ΔpH generated was stable for at least 4 min and only then began to dissipate with a half-life of 5 min. When known substrates of AcrAB pump (8, 12, 14, 23) were present, ΔpH was dissipated rapidly from the beginning, because of the proton efflux presumably accompanying the inward transport of drugs (Fig. 4; Table 1). The rate of dissipation was also higher at higher drug concentration, as shown by using erythromycin (not shown). Bile salts, which appeared to have the highest affinity to the AcrB transporter (Fig. 3), also produced the most rapid flux of protons. The drug-stimulated proton efflux depended on the presence of AcrB, since the presence of drugs did not increase proton flux in the AcrB-free control vesicles (not shown). Further, the drug-dependent proton efflux was not observed when ΔpH was absent, that is, when valinomycin was not added or when it was added to vesicles resuspended in a K+-containing buffer (not shown).
Figure 4.
Drug-induced proton efflux in AcrB-containing vesicles. AcrB-containing vesicles reconstituted in a buffer containing 0.1 M KCl were diluted into the same buffer containing 0.1 M NaCl instead, and drugs were added at 0.2 mM. Upon the subsequent addition of valinomycin (arrow), the fluorescence of the pyranine decreased rapidly, indicating the generation of interior-acid ΔpH. The generated ΔpH was dissipated only slowly in the absence of drugs (curve 1). The rate of decay, and hence the proton efflux, increased in the presence of various drugs: erythromycin (curve 2), chloramphenicol (curve 3), cloxacillin (curve 4), sodium glycocholate (curve 5), and sodium taurocholate (curve 6). Curves were displaced slightly in the vertical direction to minimize the effect of the initial entry of drugs (see text).
The AcrB-containing proteoliposomes thus showed proton efflux when a substrate was added to the external medium in the presence of interior-acid ΔpH. These observations suggest that AcrB acts as a proton antiporter. Interestingly, positively charged, negatively charged, and uncharged substrates caused similar efflux of protons. If AcrB pumps out its substrate from the cytosol, this will create different changes in membrane potential depending on the charge on the substrate. Such an unexpected outcome may perhaps be avoided if AcrB extrudes those substrates that partition into the bilayer from outside, before they cross into the cytoplasm (1). This hypothesis remains to be tested.
Role of AcrA in the Intermembrane Probe Transfer by AcrAB Pump.
The elongated shape of AcrA suggests that it connects the inner and outer membranes in intact cells (9). This concept suggests that AcrA stimulates probe transport (Fig. 2 Inset) by interacting with vesicles and bringing them together. We therefore investigated the potential interaction of AcrA with phospholipid bilayers.
AcrB-containing and control vesicles were incubated with AcrA, and the mixture was analyzed by flotation in a Nycodenz step gradient. The binding of AcrA to vesicles did not require AcrB, and was around 0.4 μg/μmol of lipid at pH 5, 6, and 7 in the absence of Mg2+. Addition of Mg2+ increased AcrA binding, and at pH 6.0, 2.5 μg of AcrA became associated with 1 μmol of lipids.
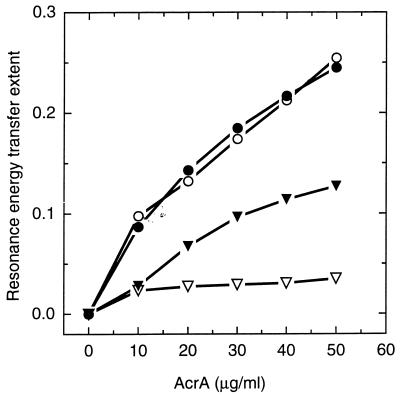
We confirmed this conclusion by fluorescence energy transfer between the unique tryptophan residue of AcrA and N-NBD-PE in the lipid bilayer. Trp-346 is located close to the conserved C-terminal hydrophobic domain of AcrA. At pH 5.0, we found an efficient energy transfer which was nearly proportional to the AcrA concentration up to 50 μg/ml (Fig. 5), indicating that the Trp-346 became located close to the bilayer surface. The extent of energy transfer decreased with increasing pH. However, in the presence of 5 mM Mg2+ a significant energy transfer was observed even at pH 6.0 (Fig. 5). AcrA thus appears to become associated with lipid bilayers in a manner dependent on pH and Mg2+.
Figure 5.
Binding of AcrA to the vesicles detected by the resonance energy transfer from Trp-346 of AcrA to N-NBD-PE in the vesicles. AcrA at indicated concentrations was incubated with 86 nmol of NBD-labeled vesicles for 5 min at pH 6.0 (▿, ▾) or 5.0 (○, ●) and the extent of resonance energy transfer was determined. Empty and filled-in symbols show incubation in the absence and presence of 5 mM Mg2+.
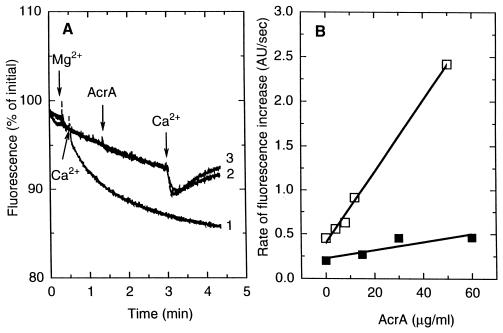
We examined whether the binding of AcrA to vesicles results in membrane fusion. Such fusion events are usually followed by the intermixing of aqueous contents of the vesicles. Therefore ANTS was entrapped in one liposome population and DPX in another. Intermixing of contents will cause quenching of ANTS fluorescence by DPX. The reliability of this assay was confirmed by demonstrating the fast content mixing upon addition of Ca2+ at pH 7.0 (both internal and external)(Fig. 6A). In contrast, the addition of 5 mM Mg2+ followed by AcrA (15 μg/ml) did not produce any quenching of ANTS fluorescence even at acidic pH values (Fig. 6A).
Figure 6.
AcrA may induce hemifusion of membranes under certain conditions. (A) AcrA does not produce mixing of vesicle contents. ANTS fluorescence was measured in the 1:1 mixture of ANTS-containing vesicles and DPX-containing vesicles. Ca2+ (5 mM) immediately induced fusion at pH 7.0 (curve 1), but 5 mM MgCl2 plus 25 μg/ml AcrA did not, even at pH 6.0 (curve 2) and 5.0 (curve 3). (B) AcrA-catalyzed intermixing of lipids of different vesicles at external pH of 5.0 (□) and 6.0 (■). AcrB-free donor vesicles prepared in the pH 7.0 buffer were mixed with acceptor vesicles in the presence of 5 mM MgCl2. Fluorescent lipid transfer was initiated by the addition of AcrA.
Interestingly, when the dilution of fluorescent lipid concentration in the donor vesicle membrane was examined by mixing the donor vesicles containing N-NBD-PE and N-Rh-PE (but not AcrB) with acceptor vesicles composed of only phospholipids, AcrA and Mg2+ produced a slow but steady transfer of lipids at pH 5.0, but not at pH 6.0, where the experiment of Fig. 1 was performed (Fig. 6B). This effect was not seen when another lipid-binding protein, bovine serum albumin, was used instead of AcrA (not shown). Thus at pH 5.0, AcrA may cause the fusion of bilayers without the intermixing of aqueous contents, a phenomenon sometimes referred to as hemifusion (29), although it may alternatively act only by bringing the two bilayers to proximity.
These observations are provocative in relation to the mechanism of action of this protein in intact cells. The Donnan equilibrium concentrates protons and Mg2+ in the periplasmic space (30), and indeed AcrA showed a strong stimulation of AcrB activity at acidic pH and in 5 mM Mg2+ (Fig. 2). Most interestingly, under these conditions AcrA promoted the close association of two membranes, and in addition possibly a hemifusion event. The structural features of AcrA, such as its highly asymmetric shape, the presence of coiled-coil domain, and two hydrophobic regions (9), are reminiscent of viral membrane fusion proteins. AcrA could thus act as a true membrane fusion protein under conditions found in intact cells, perhaps creating the adhesion sites between the inner and outer membranes.
Acknowledgments
This study was supported by grants from the U.S. Public Health Service (AI-09644) and from Microcide Pharmaceuticals, Inc.
ABBREVIATIONS
- N-NBD-PE
1,2-dipalmitoyl N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)phosphatidylethanolamine
- N-Rh-PE
1,2-dipalmitoyl N-(lissamine rhodamine B sulfonyl)phosphatidylethanolamine
- ANTS
1-aminonaphthalene-3,6,8-trisulfonate
- DPX
N,N′-p-xylenebis(pyridinium bromide)
References
- 1.Nikaido H. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X-Z, Livermore D M, Nikaido H. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma D, Cook D N, Hearst J E, Nikaido H. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 4.Thanassi D G, Suh G S B, Nikaido H. J Bacteriol. 1995;177:998–1007. doi: 10.1128/jb.177.4.998-1007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinh T, Paulsen I T, Saier M H., Jr J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis K. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 7.Saier M H, Jr, Tam R, Reizer A, Reizer J. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 9.Zgurskaya H, Nikaido H. J Mol Biol. 1999;235:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 10.Fralick J A. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White D G, Goldman J D, Demple B, Levy S B. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikaido H, Basina M, Nguyen V, Rosenberg E Y. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okusu H, Ma D, Nikaido H. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaffner W, Weissmann C. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 16.Hall J A, Davidson A L, Nikaido H. Methods Enzymol. 1998;292:20–29. doi: 10.1016/s0076-6879(98)92004-3. [DOI] [PubMed] [Google Scholar]
- 17.Struck D K, Hoekstra D, Pagano R E. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 18.Clement N R, Gould J M. Biochemistry. 1981;20:1534–1544. doi: 10.1021/bi00509a019. [DOI] [PubMed] [Google Scholar]
- 19.Mayer L D, Hope M J, Cullis P R, Janoff A S. Biochim Biophys Acta. 1985;817:193–196. doi: 10.1016/0005-2736(85)90084-7. [DOI] [PubMed] [Google Scholar]
- 20.Kamp F, Hamilton J A. Proc Natl Acad Sci USA. 1992;89:11367–11370. doi: 10.1073/pnas.89.23.11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellens H, Bentz J, Szoka F C. Biochemistry. 1984;23:1532–1538. doi: 10.1021/bi00302a029. [DOI] [PubMed] [Google Scholar]
- 22.Ames B N. Methods Enzymol. 1973;8:115–118. [Google Scholar]
- 23.Thanassi D G, Cheng L W, Nikaido H. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homolya L, Hollo Z, Germann U A, Pastan I, Gottesman M M, Sarkadi B. J Biol Chem. 1993;268:21493–21496. [PubMed] [Google Scholar]
- 25.Ruetz S, Gros P. Cell. 1994;77:1071–1081. doi: 10.1016/0092-8674(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 26.Dekkers D W C, Comfurius P, Schroit A J, Bevers E M, Zwaal R F A. Biochemistry. 1998;37:14833–14837. doi: 10.1021/bi981011w. [DOI] [PubMed] [Google Scholar]
- 27.Bolhuis H, van Veen H W, Poolman B, Driessen A J M, Konings W N. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 28.Perlin D S, Kasano K, Brooker R J, Slayman C W. J Biol Chem. 1984;259:7884–7892. [PubMed] [Google Scholar]
- 29.Kemble G W, Danieli T, White J U. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 30.Sen K, Hellman J, Nikaido H. J Biol Chem. 1988;263:1182–1187. [PubMed] [Google Scholar]