Abstract
A specific interaction between the α subunit of RNA polymerase and an A+T-rich upstream sequence (UP element) stimulates transcription at some promoters in Escherichia coli. We found that RNA polymerase formed a heparin-resistant nonproductive initiation complex at the malT promoter which has an A+T-rich upstream sequence that begins 9 bp upstream of the −35 region. Substitution of other sequences for the A+T-rich sequence eliminated both the formation of heparin-resistant complexes and α binding to the malT promoter. A 5-bp deletion between the A+T-rich sequence and the −35 region increased promoter activity. The UP element derived from the rrnB P1 promoter stimulated transcription of the malT core promoter when placed 4 bp upstream from the malT −35 region, but insertion of an additional 4 bp between the rrnB P1 UP element and the −35 element eliminated transcription activity without eliminating heparin-resistant complex formation. Similar UP element effects were observed in hybrids with the lac core promoter, even though the region around the transcription start site was melted in both productive and nonproductive complexes. We conclude that UP elements can mediate the formation of both productive and nonproductive open complexes, depending on their location with respect to the core promoter.
Promoter recognition and open complex formation are central events in transcription initiation by RNA polymerase (RNAP) holoenzyme. In Escherichia coli, the RNAP σ subunit plays an essential role in these processes. The σ70 makes sequence-specific contacts to two hexameric elements located approximately 10 and 35 bp upstream of the transcription start site (1). The similarity of a promoter sequence to the −10 and −35 consensus hexamers (TATAAT and TTGACA, respectively) and to the consensus spacing between them (17 bp) is a major determinant of the strength of a promoter (2, 3). In addition, it was found recently (4, 5) that an A+T-rich sequence located upstream of the −35 hexamer (UP element) acts as a third RNAP recognition element in certain promoters.
The best characterized UP element is that of the rrnB P1 promoter, which increases promoter activity more than 30-fold in vivo (5). The C-terminal domain of the RNAP α subunit (αCTD), which functions as the target for a number of transcription factors (6, 7), binds directly to the UP element and is responsible for the specific interaction of the UP element with RNAP (5, 8–10). In addition, the effect of the UP element on a promoter activity correlates generally with its degree of similarity to the UP element consensus sequence (11). It has been estimated that approximately 3% of E. coli promoter sequences for mRNAs and 19% of promoters for stable RNAs contain a match to the consensus UP element sequence similar in extent to the rrnB P1 UP element (10). However, the functional significance of most of these sequences remains to be studied.
We found previously (12) that RNAP formed a stable nonproductive complex at the malT promoter in the absence of added factors, whereas it formed a productive complex in the presence of the cAMP receptor protein (CRP). The nonproductive complex was resistant to the competitor heparin and made the abortive trinucleotide ApUpU from ApU and UTP, but it did not make a productive transcript in the presence of all four nucleoside triphosphates. The DNA region occupied by RNAP in the nonproductive complex was essentially the same as that in the CRP-dependent productive complex, although there was a significant difference in the DNase I protection pattern of the two complexes, particularly between −20 and −60. We noticed that the malT promoter contains an A+T-rich sequence upstream of the core promoter and that its location relative to the −35 hexamer is 5 bp further upstream of the −35 element than the rrnB P1 UP element. Here, we investigate how the location of an UP element affects the function of the malT promoter and the two hybrid promoters, rrnB P1 UP-malT and rrnB P1 UP-lac. We show that an UP element can mediate the formation of either a productive or a nonproductive complex, depending on its location with respect to the core promoter.
MATERIALS AND METHODS
Media and Growth Conditions.
Cells were grown aerobically at 37°C in Luria–Bertani medium (13). When used, ampicillin was added at 50 μg/ml. Bacterial growth was monitored by determining the optical density at 600 nm.
DNAs and Proteins.
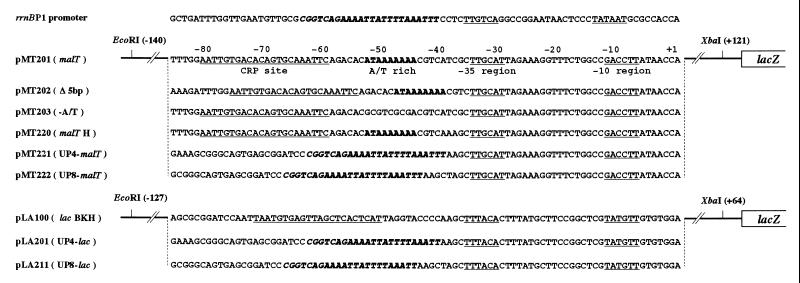
The promoters used in this study are shown in Fig. 1. The HindIII end of the 247-bp EcoRI–HindIII fragment containing the wild-type malT promoter of pMT100 (12) was filled in and cloned between the EcoRI and SmaI sites of pUC19. The 261-bp EcoRI–XbaI fragment from the resulting plasmid was cloned into pLA100 to construct pMT201. pMT202 (Δ5 bp), pMT203 (−A/T), and pMT220 (malT H) were constructed from pMT201 by polymerase chain reaction mutagenesis. The 180-bp PvuII–HindIII fragment containing the lac promoter derived from pUC19 was cloned between the SmaI and HindIII sites of pMS437C (14). The HindIII site of the resulting plasmid was filled in and an XbaI linker was inserted to construct pLA02. The BamHI (−78), KpnI (−45), and HindIII (−37) sites were introduced into the lac promoter region of pLA02 by polymerase chain reaction mutagenesis to form pLA100. To construct pLA200, the BamHI and HindIII region of pLA100 was replaced by a 30-bp synthetic oligonucleotide containing the rrnB P1 UP element. To form pMT221 (UP4–malT), the EcoRI–HindIII region of pMT220 was replaced by the EcoRI–HindIII fragment derived from pLA200. To form pLA201 (UP4-lac), the BamHI and HindIII region of pLA100 was replaced by a 29-bp synthetic oligonucleotide containing the rrnB P1 UP element. To construct pMT222 (UP8–malT) and pLA211 (UP8–lac), pMT221 and pLA201, respectively, were digested with HindIII and filled in. CRP and RNAP were purified as described previously (12). The α subunit of RNAP was purified, according to the method described (15), from cells harboring pGEMAX185.
Figure 1.
Promoters used in this study. The −35 and −10 hexamers, and the CRP binding sites are underlined. The transcription start site is numbered as +1. The upstream A+T-rich sequence of the malT promoter appears in boldface. The UP element derived from the rrnB P1 appears in boldface italic.
Electrophoretic Mobility-Shift Assay (EMSA).
Reaction mixtures (30 μl) containing a 5 nM DNA fragment and 30 nM RNAP were incubated in transcription buffer (20 mM Tris⋅HCl, pH 7.9/100 mM NaCl/3 mM MgCl2/0.1 mM EDTA/0.1 mM DTT/5% glycerol/50 μM cAMP/50 μg/ml BSA) and assayed as described (12), except that the heparin challenge was performed by adding 3 μl of 50 μg/ml of heparin for 3 min. At the times indicated, 3 μl of NTP solution (1 mM ATP, UTP, or 1 mM each of the four NTPs) was added to the mixture before the heparin treatment. For α-subunit binding assays, α (0–0.4 μM) was incubated with a 5 nM DNA fragment for 40 min at 22°C in 20-μl reaction mixtures containing 50 mM Tris⋅HCl (pH 7.9), 50 mM NaCl, 3 mM DTT, and 5% glycerol; the complexes were analyzed by native gel electrophoresis at room temperature. The gel contained 6% polyacrylamide, 7.5% glycerol, and 45 mM Tris⋅borate (pH 8.3)/1 mM EDTA (TBE). The running buffer contained TBE and 2% glycerol.
In Vitro Transcription.
Complexes were formed as described above in 30 μl of transcription buffer, and transcription was started by adding 3 μl of a substrate solution containing 0.05 mM [α-32P]UTP (5 μCi) and 0.5 mM each of ATP, GTP, and CTP. After 15 min of incubation, the reaction was terminated by adding 60 μl of phenol and 30 μl of stop buffer (0.6 M sodium acetate, pH 5.5/20 mM EDTA/200 μg/ml tRNA). The products were precipitated with ethanol and fractionated by electrophoresis on 8% polyacrylamide gels containing 8 M urea. Abortive initiation assays were performed at 37°C in 30-μl reaction mixtures containing transcription buffer, 5 nM UP4-lac or UP8-lac hybrid promoter DNA fragment, and 30 nM RNAP. The reaction was started by adding 3 μl of a solution containing 1 mM ATP and 0.1 mM [α-32P]UTP (10 μCi). After 15 min of incubation, products were precipitated with ethanol and analyzed on 20% polyacrylamide/8 M urea gels.
Permanganate Footprinting.
Fifty microliter reactions were incubated for 30 min at 37°C in a transcription buffer containing 1 nM 32P-end-labeled DNA fragment and 110 nM RNAP. At the times indicated, 100 nM CRP was added before RNAP. The mixtures were treated with 5 μl of 50 μg/ml heparin for 3 min at 37°C. Two millimolar KMnO4 was added for 1 min at 25°C, and the reaction was then quenched with 2-mercaptoethanol. After the addition of 20 μl of 1.5 M sodium acetate/20 mM EDTA/100 μg/ml tRNA, the mixture was precipitated with ethanol. DNA was cleaved by incubation in 1 M piperidine at 90°C for 30 min, and products were analyzed on a 6.5% polyacrylamide/8 M urea sequencing gel.
β-Galactosidase Assay.
KI70 (Δlac) and OK6201 (Δlac crp−) cells (16) harboring the promoter lacZ fusion plasmids were grown to an OD600 of 0.8. The β-galactosidase activity was determined as described (13).
S1 Nuclease Assay.
Cells harboring the promoter-lacZ fusion plasmids were grown to an OD600 of 0.5 and RNA was prepared as described (17). EcoRI–XbaI fragments of pMT201 and pLA100, 32P-labeled at the XbaI 5′ end were used as DNA probes for the malT and lac transcripts, respectively. RNA was hybridized with the DNA probes and treated with 100 units of S1 nuclease at 37°C for 15 min, and the reaction products were analyzed on 8% polyacrylamide/8 M urea gels.
RESULTS
The A+T-Rich Sequence Is Responsible for the Formation of Nonproductive Complex at the malT Promoter.
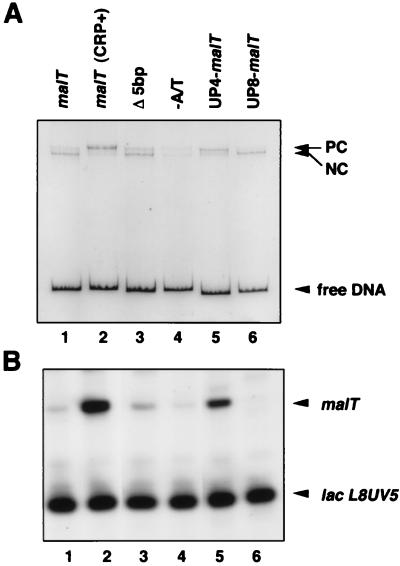
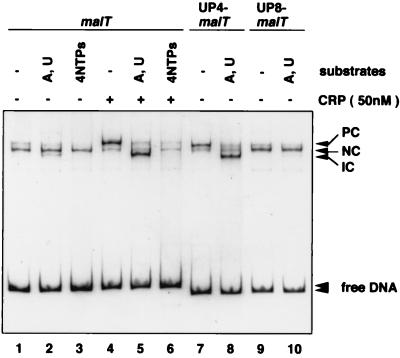
In the absence of CRP, the malT promoter forms, with RNAP, a heparin-resistant binary complex that is unable to make run-off transcripts (12). The malT promoter has an A+T-rich sequence upstream of the −35 hexamer (Fig. 1). To examine the potential role of this A+T-rich sequence in the formation of the nonproductive complex, we constructed several variants of the malT promoter (Fig. 1) and analyzed RNAP-promoter complexes by EMSA. The nonproductive complex exhibits slightly more increased mobility than the productive complex formed after incubation of the malT promoter with both RNAP and CRP, and after being challenged with heparin (Fig. 2) (12). When the A+T-rich sequence was replaced by a G+C-rich sequence, formation of the nonproductive complex was markedly reduced (Fig. 2A, lanes 1 and 4). The reduction in the amount of nonproductive complex was even more significant in the lac-malT hybrid promoter when the malT A+T-rich sequence was replaced by the upstream region from the lac promoter (data not shown). We conclude that the A+T-rich sequence is required for the formation of the nonproductive heparin-resistant complex. The small amount of productive complex formed in the absence of CRP–cAMP was not affected by elimination of the A+T-rich sequence (lane 4).
Figure 2.
Effect of the UP element on the malT core promoter in vitro. (A) EMSA for RNAP binding. The EcoRI–XbaI fragments containing the malT promoter derivatives were incubated with RNAP, treated with heparin, and analyzed by EMSA. In lane 2, CRP (50 nM) was added before RNAP. PC and NC represent the productive and nonproductive complexes, respectively. (B) In vitro transcription assay. The EcoRI–XbaI fragments containing the malT promoter derivatives were the templates, and the 205-bp EcoRI fragment containing the lacL8UV5 promoter (39) was an internal control.
Effect of the Location of UP Elements on RNAP–malT Promoter Interaction.
UP elements that have been characterized (to date) in detail facilitate formation of productive open complexes (refs. 4 and 5; see also ref. 30). The malT A+T-rich sequence exhibits a moderate similarity to the rrnB P1 (5) and consensus UP elements (10). We addressed the question of why the A+T-rich sequence leads to nonproductive complex formation at the malT promoter. We noticed that the right-most end of the A+T-rich malT, with respect to the −35 hexamer, is 5 bp further upstream than that of the UP element in rrnB P1. Interestingly, analysis by EMSA revealed that the deletion of 5 bp between the A+T-rich sequence and the −35 region of the malT promoter moderately increased (about 2-fold) the amount of the upper band, corresponding to the productive complex (Fig. 2A, lanes 1 and 3). The Δ5-bp mutant promoter also exhibited a 2.5-fold increase in promoter activity (Fig. 2B, lane 3).
We next examined the effect of the location of the rrnB P1 UP on malT core promoter activity. We constructed two hybrid promoters (UP4–malT and UP8–malT) in which the rrnB P1 UP element was placed 4 and 8 bp, respectively, upstream of the malT −35 hexamer (Fig. 1). The two rrnB P1 UP–malT hybrid promoters were analyzed by EMSA. RNAP preferentially formed a lower mobility complex with the UP4–malT promoter whereas it formed predominantly a higher mobility complex with the UP8–malT promoter (Fig. 2A, lanes 5 and 6). The UP4–malT promoter was active in transcription in vitro, but little activity was detected with the UP8–malT promoter (Fig. 2B, lanes 5 and 6). These experiments clearly indicate that only a suitably located UP element stimulates the formation of a productive complex at the malT promoter, thereby activating transcription.
The Binding of the α Subunit to the A+T-Rich Sequence in the malT Promoter.
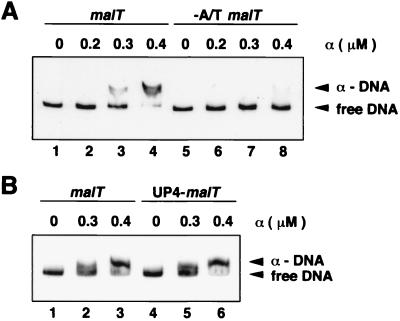
To examine whether the malT A+T-rich sequence interacts with the α subunit, we performed EMSA with purified α subunits and DNA fragments containing the malT promoter variants. α formed a complex with the wild-type malT promoter (Fig. 3A, lanes 1–4), but much less complex was formed at the −A/T malT promoter (lanes 5–8). The affinity of the wild-type malT promoter for α subunits was approximately equivalent to that of the UP4-malT promoter, in which the rrnB P1 UP element was fused to the malT core promoter (Figs. 1 and 3B).
Figure 3.
EMSA for α subunit binding. (A) Comparison of the wild-type and −A/T malT promoters. The 261-bp EcoRI–XbaI fragment derived from pMT201 (lanes 1–4) or from pMT203 (lanes 5–8) was incubated with the indicated concentrations of α subunits. (B) Comparison of the wild-type malT and UP4–malT promoters. The EcoRI–XbaI fragment derived from pMT201 (lanes 1–3) or from pMT221 (lanes 4–6) was incubated with the indicated concentrations of α subunits.
The Transcription Initiation Region Is Melted in Nonproductive Complexes.
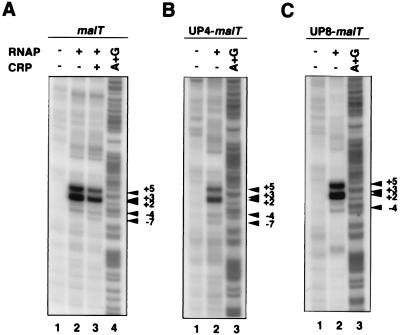
To examine whether the region around the transcription start site was melted, we performed permanganate footprinting. The 32P-labeled DNA fragment carrying the wild-type malT promoter was incubated with CRP–cAMP and/or with RNAP to form the productive or nonproductive complexes. After the addition of heparin, KMnO4 modification was carried out. Permanganate preferentially modifies T residues in single-stranded regions of DNA (18). As shown in Fig. 4A, melting around the transcription start site was detected in both the productive and nonproductive complexes. The two functionally different complexes produced essentially the same melting pattern, although there was a slight difference in band intensity. The same melting pattern was also observed with the UP4-malT (Fig. 4B) and UP8-malT (Fig. 4C) promoters.
Figure 4.
Potassium permanganate footprinting of malT promoter derivatives. The 261-bp EcoRI–XbaI fragments derived from pMT201 (A), the 236-bp EcoRI–XbaI fragment derived from pMT221 (B), and the 240-bp EcoRI–XbaI fragment derived from pMT222 (C) were used for KMnO4 footprinting. The arrowheads indicate the permanganate-sensitive sites. The numbers represent positions relative to the transcription start site.
Effect of Ribonucleotides on Transcription Complexes.
We next characterized the effects of ribonucleotides on wild-type malT open complexes with EMSA. The addition of the initiating nucleotides (ATP and UTP) should allow a productive complex to proceed to +8. In the presence of CRP, most of these initiated complexes had a mobility greater than that of the complexes formed in the absence of NTPs (Fig. 5, lanes 4 and 5). When all 4 NTPs were added in the presence of CRP, the amount of heparin-resistant complex was dramatically reduced, indicating that most of the RNAP ran off the template (lane 6). On the other hand, the addition of NTPs had essentially no effect on the complex formed in the absence of CRP (lanes 2 and 3). Similar experiments were done with complexes formed by the UP4-malT and UP8-malT promoters. Again a productive UP4-malT complex (lane 7) was converted to an initiated complex by the addition of ATP and UTP (lane 8), whereas a nonproductive UP8-malT complex (lane 9) remained unaffected (lanes 10).
Figure 5.
Effect of ribonucleotides on RNAP-promoter complexes. The EcoRI–XbaI fragments containing the malT promoter derivatives were incubated with or without CRP (50 nM) in the presence of cAMP (50 μM) and then with RNAP. The mixtures were incubated with the indicated NTPs (1 mM each) for 15 min before treatment with heparin. PC, NC, and IC represent the productive, nonproductive, and initiated complexes, respectively.
Effect of UP Element Location on malT Transcription in Vivo.
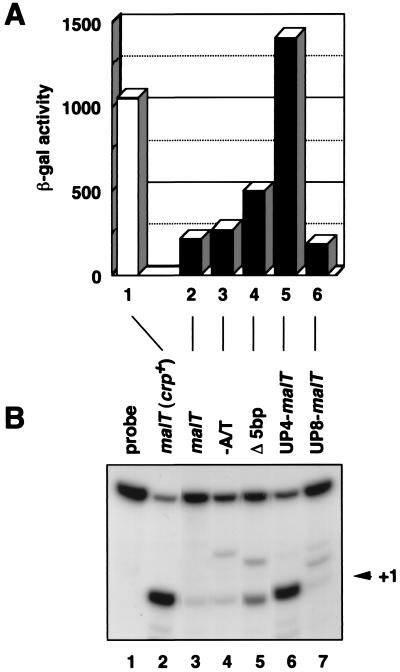
To study the effect of an UP element on transcription from the malT promoter in vivo, β-galactosidase activities were measured in Δlac crp− cells carrying the promoter-lacZ fusion plasmids (Fig. 6A). The β-galactosidase activity in Δlac crp+ cells carrying the malT promoter-lacZ fusion (pMT201) was about 5-fold higher than that in crp− cells (lanes 1 and 2). Elimination of the malT A+T-rich sequences caused little change in transcription in the absence of activation by CRP (compare lanes 2 and 3). On the other hand, the 5-bp deletion between the malT A+T-rich sequences and the −35 region increased basal transcription 2.5-fold (lanes 2 and 4). Furthermore, the rrnB P1 UP element at its normal position with respect to the −35 element markedly stimulated the malT core promoter (UP4-malT; lane 5), but not when fused 4 bp further upstream (UP8-malT; lane 6). Quantitative S1 nuclease assays confirmed that differences in β-galactosidase activities reflected differences in promoter strength (Fig. 6B).
Figure 6.
Effect of the location of the UP element on promoter activity in vivo. (A) β-Galactosidase activity of the malT-lacZ fusions. The β-galactosidase activity of KI70 (Δlac) (lane 1) and OK6201 (Δlac crp−) (lanes 2–6) cells harboring the malT–lacZ fusion plasmids were measured. Each bar is expressed in Miller units (13). The value is an average obtained from three independent experiments. (B) S1 assay. Total RNAs (100 μg) prepared from KI70 (Δlac) (lane 2) and OK6201 (Δlac crp−) (lanes 3–7) cells harboring the indicated plasmids were subjected to S1 nuclease assay.
Effect of an UP Element on the lac Core Promoter.
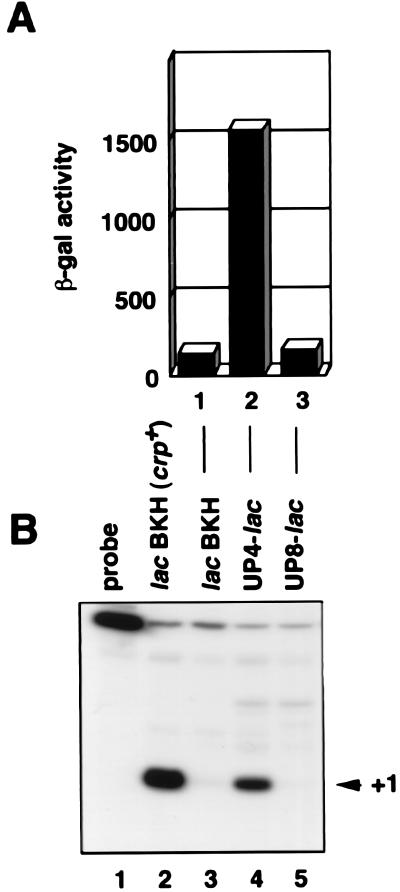
To examine the effect of an UP element on the lac core promoter, rrnB P1 UP element-lac hybrid promoters were constructed, the promoters were fused to lacZ (Fig. 1), and promoter activities were assessed by measuring the β-galactosidase activity of the Δlac cells harboring the fusion plasmids (Fig. 7A). The UP element markedly enhanced lac core promoter activity when placed 4 bp upstream of the −35 region as reported (4), whereas it had no effect when placed 8 bp upstream. S1 assays confirmed that the UP element affected transcription from lac P1 in the UP4-lac promoter construct (Fig. 7B).
Figure 7.
Characterization of UP-lac hybrids promoters in vivo. (A) Effect of the location of UP elements on promoter activity. The β-galactosidase activity of OK6201 cells harboring the indicated promoter-lacZ fusion plasmids were determined. Bars are expressed in Miller units (13). The value is an average obtained from two independent experiments. (B) Analysis of the transcription start site. Total RNAs (100 μg) prepared from KI70 (Δlac) (lane 2) and OK6201 (Δlac crp−) (lanes 3–5) cells harboring the indicated plasmids were subjected to S1 nuclease assay.
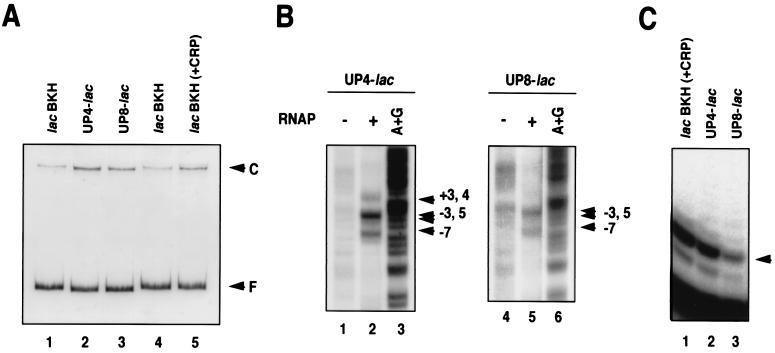
We also used EMSA to test whether the rrnB P1 UP element could stimulate the formation of a heparin-resistant complex at the transcriptionally inactive UP8-lac promoter. As shown in Fig. 8A, the formation of heparin-resistant complexes was significantly increased at both the UP4-lac and UP8-lac promoters compared with the wild-type lac promoter. The heparin-resistant complex formed with the wild-type lac promoter in the absence of CRP-cAMP was at lac P2 (19, 20). The −35 region of P2 no longer existed in UP4-lac and UP8-lac promoters, suggesting that RNAP occupied P1 in these hybrid promoters. This was confirmed by DNase I footprinting (data not shown). In addition, permanganate footprinting revealed that a region around the P1 start point was melted in both hybrid promoter complexes, although DNA melting at the UP8-lac promoter was rather weak (Fig. 8B). We also performed abortive initiation assays on the UP-lac hybrid promoters complexes and the initiating nucleotides ATP and UTP. As shown in Fig. 8C, the pppApApUpU lac P1 product (19) was clearly produced from both promoters. Thus, the heparin-resistant complex formed at the UP8-lac promoter had the ability to produce abortive RNAs, but not run-off transcripts. We conclude from these results that the UP element increases RNAP complex formation at lac P1; whether these complexes are productive or nonproductive, however, depends on the location of the UP element, as in the case of the malT promoter.
Figure 8.
Characterization of UP-lac hybrid promoters in vitro. (A) EMSA. The indicated lac promoter derivatives were incubated with RNAP. Lane 5 shows the CRP-dependent P1 complex of the wild-type lac promoter formed in the presence of CRP and cAMP. Lanes 1 and 4 correspond to the P2 complex of the wild-type lac promoter. (B) Potassium permanganate footprinting of UP4- and UP8-lac promoters. The arrowheads indicate the permanganate-sensitive sites. The numbers represent positions relative to the transcription start site of P1. (C) Abortive initiation assay. The products of the abortive initiation reaction were analyzed by electrophoresis on a 20% polyacrylamide gel containing 8 M urea. Lane 1 represents the products derived from the wild-type lac promoter in the presence of CRP and cAMP. The arrowhead indicates the major product, which presumably corresponds to the pppApApUpU product synthesized at the lac P1 promoter.
DISCUSSION
The rrnB P1 UP element was identified as a third RNAP recognition element because it increased promoter strength both in vivo and in vitro (5). A major finding in the present study is that UP elements can lead to the formation of transcriptionally inactive open complexes when placed a half-turn upstream of the position where they stimulate transcription. In other words, the location of an UP element with respect to the core promoter is crucial in determining the nature of transcription complexes. The helical phase-dependent effect of the UP element on promoter function is analogous to that of upstream A-tracts (21–23) and CRP (24, 25).
It has been proposed that a UP element can contain two separable sections that can function independently and may be recognized by two flexible αCTD monomers (4, 10, 26). We observed that the rrnB P1 UP element could stimulate malT transcription when separated 14 bp from the −35 hexamer, although the level of activation was reduced compared with the UP4-malT promoter (data not shown). It is known that insertion of 5 bp at −46 or deletion of 3 bp from −38 to −40 results in loss of UP element function at the rrnB P1 promoter (27). It would be interesting to test whether RNAP forms nonproductive complexes at these mutant rrnB P1 promoters.
It was reported previously that RNAP could form two open complexes (openupper and openlower), differing in electrophoretic mobility and in ability to escape abortive cycling at the lacUV5 promoter, with the openupper being more proficient in escape (28, 29). The relative amounts of the two complexes varied with temperature, the openlower being favored at 16°C, and the major openupper at 37°C. In contrast, no temperature dependence of the ratios of the two complexes was observed in our system (data not shown).
How does an UP element lead to the formation of nonproductive complexes? It is known that upstream A-tracts, which were shown recently (30) to interact with α subunits, can inhibit transcription initiation at synthetic promoters (31). More recently, it was reported that UP elements can inhibit promoter clearance (32). The inhibition of promoter clearance by a transcription factor through overstabilization of RNAP binding has also been reported (33, 34). The RNAP–promoter interaction, however, is not enhanced in the nonproductive complexes at the malT promoter compared with the productive complexes (unpublished data). We prefer a model where the α subunit–UP interaction leads to a transcription complex with an inactive conformation. A similar model was proposed for the repression of the gal P1 promoter by GalR or LacI binding to an upstream operator (35). In addition, it is interesting to note that RNAP carrying a specific mutation in the β subunit forms a stable inactive open complex at a normal promoter (36).
What is the functional significance of the malT UP element and the nonproductive complex in the regulation of malT expression? We showed previously that the role of CRP in malT transcription is to lead to the formation of a productive open complex (12). We found that the malT nonproductive complex is converted to a productive complex by CRP and that CRP-dependent activation is markedly reduced if the promoter lacks its UP element (unpublished results). In other words, the major effect of CRP is not on the recruitment of RNAP to the promoter. One interpretation of our results is that the malT UP element recruits RNAP to the promoter, and that CRP predominantly acts at a post-recruitment step to make the complex productive. We cannot exclude the possibility, however, that CRP also helps the UP element recruit RNAP. The nonproductive complex could be a simple intermediate in the formation of the productive complex; or it could be the product of a branched pathway at an early stage in transcription initiation, a complex that must be disrupted before a productive complex can be formed (37).
One of the major roles of CRP is apparently to increase initial binding of RNAP through CRP–α subunit interaction at promoters where the CRP binding site is located, −61.5 (7, 38). We speculate that recruitment of RNAP by CRP may be insufficient to fully activate a promoter when the CRP site is located farther upstream (e.g., at −70.5, the site of malT). In this case, an UP element may be needed to facilitate recruitment. Further studies are needed to understand how CRP and UP elements cooperate to activate transcription.
Acknowledgments
We thank Dr. Rick Gourse (University of Wisconsin) for thoughtful comments on the manuscript and the referees for helpful criticism and comments. We also thank Drs. Akira Ishihama and Katsuhiko Murakami (National Institute of Genetics) for the gift of pGEMAX185 and their technical comments on the purification of α subunits. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan.
ABBREVIATIONS
- RNAP
RNA polymerase
- αCTD
C-terminal domain of the RNAP α subunit
- CRP
cAMP receptor protein
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 2.McClure W. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 3.Mulligan M E, Hawley D K, Entriken R, McClure W R. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 5.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 6.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby S, Ebright R H. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 8.Murakami K, Fujita N, Ishihama A. EMBO J. 1996;15:4358–4367. [PMC free article] [PubMed] [Google Scholar]
- 9.Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 10.Estrem S T, Gaal T, Ross W, Gourse R L. Proc Natl Acad Sci USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross W, Aiyar S E, Salomon J, Gourse R L. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tagami H, Aiba H. EMBO J. 1998;17:1759–1767. doi: 10.1093/emboj/17.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 14.Kawamukai M, Kishimoto J, Utsumi R, Himeno M, Komano T, Aiba H. J Bacteriol. 1985;164:872–877. doi: 10.1128/jb.164.2.872-877.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igarashi K, Ishihama A. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 16.Morita T, Shigesada K, Kimizuka F, Aiba H. Nucleic Acids Res. 1988;16:7315–7332. doi: 10.1093/nar/16.15.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiba H, Adhya S, de Crombrugghe B. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 18.Borowiec J A, Gralla J D. J Mol Biol. 1987;195:89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- 19.Malan T P, McClure W R. Cell. 1984;39:173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- 20.Straney D C, Straney S B, Crothers D M. J Mol Biol. 1989;206:41–57. doi: 10.1016/0022-2836(89)90522-6. [DOI] [PubMed] [Google Scholar]
- 21.Bracco L, Kotlarz D, Kolb A, Diekmann S, Buc H. EMBO J. 1989;8:4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gartenberg M R, Crothers D M. J Mol Biol. 1991;219:217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- 23.McAllister C F, Achberger E C. J Biol Chem. 1989;264:10451–10456. [PubMed] [Google Scholar]
- 24.Gaston K, Bell A, Kolb A, Buc H, Busby S. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 25.Ushida C, Aiba H. Nucleic Acids Res. 1990;18:6325–6330. doi: 10.1093/nar/18.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newlands J T, Josaitis C A, Ross W, Gourse R L. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josaitis C A, Gaal T, Ross W, Gourse R L. Biochim Biophys Acta. 1990;1050:307–311. doi: 10.1016/0167-4781(90)90186-6. [DOI] [PubMed] [Google Scholar]
- 28.Straney D C, Crothers D M. Cell. 1985;43:449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- 29.Straney D C, Crothers D M. J Mol Biol. 1987;193:279–292. doi: 10.1016/0022-2836(87)90219-1. [DOI] [PubMed] [Google Scholar]
- 30.Aiyar S E, Gourse R L, Ross W. Proc Natl Acad Sci USA. 1998;95:14652–14657. doi: 10.1073/pnas.95.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellinger T, Behnke D, Knaus R, Bujard H, Gralla J D. J Mol Biol. 1994;239:466–475. doi: 10.1006/jmbi.1994.1389. [DOI] [PubMed] [Google Scholar]
- 32.Strainic M G, Jr, Sullivan J J, Velevis A, deHaseth P L. Biochemistry. 1998;37:18074–18080. doi: 10.1021/bi9813431. [DOI] [PubMed] [Google Scholar]
- 33.Monsalve M, Calles B, Mencia M, Salas M, Rojo F. Mol Cells. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- 34.Monsalve M, Calles B, Mencia M, Rojo F, Salas M. J Mol Biol. 1998;283:559–569. doi: 10.1006/jmbi.1998.2084. [DOI] [PubMed] [Google Scholar]
- 35.Choy H E, Park S W, Aki T, Parrack P, Fujita N, Ishihama A, Adhya S. EMBO J. 1995;14:4523–4529. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashlev M, Lee J, Zalenskaya K, Nikiforov V, Goldfarb A. Science. 1990;248:1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- 37.Kubori T, Shimamoto N. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 38.Hochschild A, Dove S L. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 39.Backman K, Ptashne M, Gilbert W. Proc Natl Acad Sci USA. 1976;73:4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]