Abstract
A key question in our understanding of the cis-regulation of gene expression during embryonic development has been the molecular mechanism that directs enhancers to specific promoters within a gene complex. Promoter competition and insulators are thought to play a role in regulating these interactions. In the bithorax complex of Drosophila, the IAB5 enhancer is located 55 kb 3′ of the Abdominal-B (Abd-B) promoter and 48 kb 5′ of the abdominal-A (abd-A) promoter. Although roughly equidistant from the two promoters, IAB5 specifically interacts only with the Abdominal-B promoter, even though the enhancer and promoter are separated by at least two insulators. Here we demonstrate that a 255 bp element, located 40 bp 5′ of the Abd-B transcriptional start site, has a novel cis-regulatory activity as it is able to tether IAB5 to the Abd-B promoter in transgenic embryos. The tethering element is sufficient to direct IAB5 to an ectopic promoter in competition assays. Deletion of the promoter-tethering element results in the redirection of enhancer-driven gene expression on transgenes. Taken together, these results provide evidence that specific long-range enhancer-promoter interactions in the bithorax complex are regulated by a tethering element 5′ of the Abd-B promoter. We discuss a bioinformatic analysis of the tethering element across different Drosophila species and a possible molecular mechanism by which this element functions. We also examine existing evidence that this novel class of cis-regulatory elements might regulate enhancer-promoter specificity at other gene complexes.
Keywords: cis-regulation, Enhancer, Drosophila, Bithorax, Abdominal-B, Promoter
INTRODUCTION
A long-standing issue in our comprehension of gene expression has been the mechanism by which enhancers are recruited to specific promoters over long distances. Insulators, tissue-specific silencer elements and promoter competition are three possible mechanisms by which enhancers are directed to specific promoters (for summary see Fig. 1A). Insulator DNAs have been identified at a number of genetic loci and act to disrupt the interaction of a shared enhancer with a promoter (Fig. 1A, promoter A) when positioned between them (Geyer, 1997; West et al., 2002). Tissue-specific silencer elements act to repress transcription in a sub-set of cells where the promoter might otherwise be activated by an enhancer (Fig. 1A, promoter B) (Drewell et al., 2000). Neither of these mechanisms prevents the enhancer from interacting with other promoters. Promoter competition is responsible for the preferred interaction of an enhancer with a specific promoter when linked to two or more promoters. In this way the distal ‘strong’ promoter (Fig. 1A, promoter D) competitively recruits the enhancer in preference to the proximal ‘weak’ promoter (Fig. 1A, promoter C). This is thought to occur via selective recognition of distinct core promoter elements by the shared enhancer (Butler and Kadonaga, 2002). However, at the bithorax complex (BX-C) in Drosophila none of these mechanisms can explain enhancer-promoter specificity. We present evidence of a fourth mechanism, promoter tethering, capable of regulating enhancer-promoter specificity.
Fig. 1. Molecular organization of cis-regulatory sequences at the Drosophila bithorax complex.
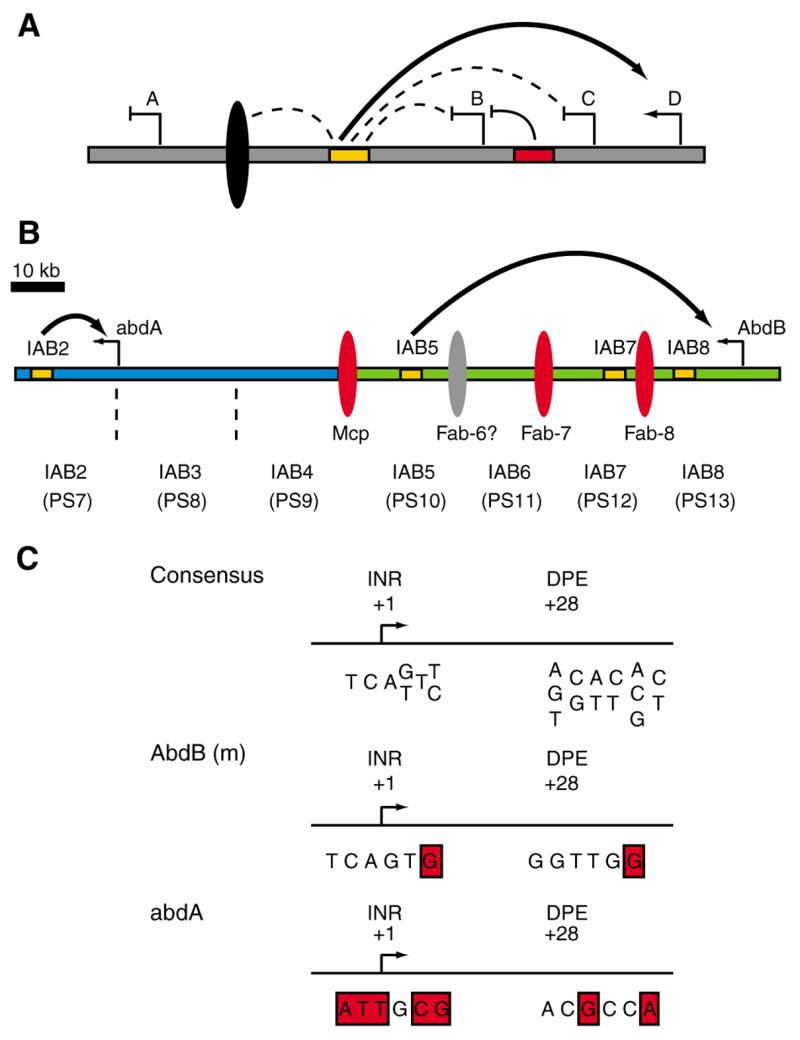
(A) Regulation of promoter-enhancer interactions. The diagram depicts a common enhancer (orange rectangle) at a model gene complex. The enhancer is able to interact with any of the promoters A, B, C or D. An insulator DNA (black ellipse) located between the enhancer and gene A prevents the enhancer from interacting with gene A but leaves it free to activate other promoters. A silencer element (red rectangle) represses gene B in cell types where it would otherwise be activated by the enhancer. Promoter competition regulates the interaction of the enhancer with gene C and gene D. The enhancer can activate either gene, but prefers the core promoter region of gene D. (B) Summary of the Abd-B-abd-A region of the BX-C. The abd-A and Abd-B transcription start sites are indicated by leftward arrows. The intergenic region is ~100 kb in length. The iab regions that control expression of the two homeotic genes are indicated: IAB2–IAB8 (shown with respect to the corresponding embryonic parasegment). IAB2, IAB3 and IAB4 (shown in blue) regulate expression of abd-A. IAB5, IAB6, IAB7 and IAB8 (shown in green) direct Abd-B expression. The insulator DNAs that separate the different iab regions are marked by red ellipses. Characterized enhancers within the iab regions are shown as orange rectangles. The IAB5 enhancer is located 55 kb 3′ of the Abd-B promoter and 48 kb 5′ of the abd-A promoter, but only interacts with Abd-B over the intervening insulator sequences. The IAB2 enhancer is located 18 kb 3′ of the abd-A promoter and directs expression specifically from abd-A. (C) Core promoter sequences at abd-A and Abd-B. The consensus sites for initiator (INR) and downstream promoter elements (DPE) in Drosophila are shown (Butler and Kadonaga, 2002). The sequences from the Abd-B and abd-A promoters do not match these consensus sites (mismatches shown in red). In addition, neither of the homeotic promoters contains a recognizable TATA box, suggesting that the core promoter elements at Abd-B(m) and abd-A are not distinctive. Abd-B(m), morphogenetic transcript.
The BX-C contains over 300 kb of genomic DNA but codes for only three homeotic transcription factors which pattern the thorax and abdomen: Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B) (Lewis, 1978; Martin et al., 1995; Sanchez-Herrero et al., 1985). In the case of abd-A and Abd-B, the cis-regulatory DNA required for accurate spatial and temporal expression during embryonic development lies in an organized array of genetically defined domains: infraabdominal (iab) regions, iab2 to iab8 (Fig. 1B) (Akbari et al., 2006; Celniker et al., 1990; Maeda and Karch, 2006; Sanchez-Herrero, 1991). Each iab region is thought to contain an enhancer capable of directing expression in the corresponding abdominal parasegment (Hogga et al., 2001; Karch et al., 1985; Mihaly et al., 2006). For example, the IAB5 enhancer is located 55 kb 3′ of the Abd-B promoter and 48 kb 5′ of the abd-A promoter (Fig. 1B) but preferentially directs expression only of Abd-B in presumptive abdominal segment 5 (parasegment 10) (Busturia and Bienz, 1993; Ohtsuki et al., 1998). By contrast, the IAB2 enhancer is located 18 kb 3′ of the abd-A transcription start site (Fig. 1B) and interacts only with the abd-A promoter, directing expression in presumptive abdominal segment 2 (parasegment 7) (Shimell et al., 2000). Disruption of IAB5-Abd-B or IAB2-abd-A interactions disrupts normal embryonic development and results in homeotic segment transformations (Busturia and Bienz, 1993; Karch et al., 1985; Shimell et al., 2000).
A critical question is how the IAB5 enhancer is selectively recruited only to the Abd-B promoter. IAB5 has been shown to have a preference for TATA-containing promoters on synthetic transgenes with exogenous promoters (Ohtsuki et al., 1998). However, at the endogenous locus neither of the Abd-B or abd-A promoters contains a TATA box. We also examined whether the Abd-B or abd-A promoters contained the other known core promoter elements: initiator (Inr) or downstream promoter element (DPE) (for a review, see Butler and Kadonaga, 2002). Neither promoter has 100% matching sequence for the weakly defined consensus sites for these elements (Fig. 1C). A weakly defined consensus site allows for more variation in the nucleotides present at any particular position within the site. Any variation from a weakly defined site is indicative that the site is not present. It is, therefore, unlikely that a promoter competition model accounts for the specific enhancer-promoter interactions in the BX-C. In order for long-range activation of Abd-B expression to occur, IAB5 has to bypass at least two known insulator sequences, Fab-7 and Fab-8, that have enhancer-blocking activity (Fig. 1B) (Barges et al., 2000; Hagstrom et al., 1996; Zhou et al., 1999; Zhou et al., 1996). One characterized mechanism for IAB5 to bypass these insulator elements involves a novel class of regulatory elements known as the promoter targeting sequences (PTS). Two PTS elements have been characterized; PTS-7, which is located adjacent to Fab-8, and PTS-6, which is located adjacent to Fab-7 (Chen et al., 2005). The PTS elements are thought to function as anti-insulators, facilitating the correct promoter-enhancer interactions. In addition, earlier studies suggested that genomic regions 5′ of the Abd-B promoter may be capable of recruiting enhancers in trans (Sipos et al., 1998). In this study we examined whether this 5′ upstream region was able to direct IAB5 to Abd-B in cis. In vivo analysis of transgenes identified a 255 bp DNA element that facilitates IAB5-Abd-B interactions. This regulatory DNA is located 40 bp 5′ of the Abd-B transcription start site and permits IAB5 to activate a distal Abd-B-CAT reporter gene in preference to a more proximal abd-A-lacZ reporter. This element is also sufficient to direct IAB5 to an ectopic promoter in competition assays. Deletion of this element results in the redirection of enhancer-promoter interactions on transgenes. We suggest that the 255 bp cis-regulatory sequence is a promoter-tethering element (PTE) capable of selectively recruiting enhancers from the iab regions, including the IAB5 enhancer, to the Abd-B promoter at the endogenous locus.
MATERIALS AND METHODS
Preparation of genomic regions
Genomic regions from the bithorax complex of Drosophila melanogaster were PCR amplified using conventional methods and cloned into either a modified pBluescript or pGemTEasy vectors. The Abd-B promoter region for the class A transcript (Martin et al., 1995) was isolated from genomic DNA as a 1522 bp PCR fragment (48,905–50,427 in EMBL:DM31961), which includes 294 bp of 5′ flanking sequence and 1228 bp of 3′ sequence. The abd-A promoter region was isolated as a 957 bp PCR fragment (152,853–153,810) which includes 538 bp of 5′ flanking sequence and 419 bp of 3′ sequence. The IAB5 enhancer (Ohtsuki et al., 1998) was isolated as a 1026 bp PCR fragment (104,011–105,037). The IAB2 enhancer (Shimell et al., 2000) was isolated as a 1969 bp PCR fragment (171,019–172,988). The PTE was isolated as a 255 bp PCR fragment that extends from −294 bp to −40 bp relative to the Abd-B transcription start site. The Fab-8 insulator element was isolated as a 743 bp PCR fragment (63,627–64,370) and specifically designed to exclude the adjacent PTS7 element. The Abd-BΔPTE region was isolated as a 1268 bp PCR fragment (49,159–50,427).
Construction of P-element transgenes
The P-transformation vector used in this study is a modified pCaSpeR and contains divergently transcribed white, CAT and lacZ reporter genes (Ohtsuki et al., 1998). Genomic promoter regions from the Bithorax complex were PCR amplified using conventional methods and cloned as AscI-BamHI fragments in a modified pBluescript at the 5′ end of either the lacZ or CAT reporter genes (Calhoun et al., 2002). The abd-A-lacZ fusion gene was isolated as an AscI-XbaI fragment and used to replace the AscI-XbaI lacZ fragment in the pCaSpeR vector. The Abd-B-CAT fusion gene was isolated as an AscI-NotI fragment and used to replace the AscI-NotI CAT fragment in pCaSpeR. The IAB5 enhancer was isolated as a PstI fragment and cloned into the pCaSpeR vector in the unique PstI site 3′ of lacZ. The IAB2 enhancer was isolated as a NotI fragment and cloned into the unique NotI site 3′ of CAT. The Fab-8 insulator element was isolated as an AscI fragment and cloned into the unique AscI site between the Abd-A-lacZ and Abd-B-CAT fusion genes. The previously described 1.6 kb spacer from bacteriophage λ alone was sub-cloned into the AscI site in pCaSpeR (Calhoun et al., 2002). The PTE element was isolated as an AscI fragment and cloned into the unique AscI site 5′ of eve-lacZ and its orientation was determined by sequencing. The eve-lacZ gene used was as previously described (Ohtsuki et al., 1998). The Abd-BΔPTE promoter was isolated as an AscI-NotI fragment and used to replace the AscI-NotI CAT fragment in pCaSpeR.
P transformation assays and in situ hybridization
Reporter transgenes were introduced into the Drosophila germline using standard methods (Small et al., 1992). Multiple transgenic lines were generated for each construct and at least three independent lines were analyzed by in situ hybridization. Embryos were collected, fixed and hybridized with digoxigenin-labeled lacZ or CAT probe as previously described (Bae et al., 2002).
Bioinformatic analysis
Levels of sequence conservation were calculated using VISTA (Frazer et al., 2004) for the PTE (chr3R:12,760,005–12,760,259), PTE 5′ (chr3R:12,760,259–12,760,513), PTE 3′ (chr3R:12,759,751–12,760,005), BX-C (chr3R: 12,470,945–12,809,178) and iab5-iab8 (chr3R: 12,695,347–12,755,125) regions of the D. melanogaster genome (April 2004) using the following parameters: calc window, 24 bp; cons window, 24 bp; cons identity, 80%. Absolute levels of conservation were obtained by comparing the number of perfectly aligned base pairs to the total length of the promoter-tethering element in D. melanogaster.
RESULTS
Dissecting promoter-enhancer interactions at the BX-C
On transgenic reporter constructs, IAB5 activates expression only from the Abd-B promoter in parasegments 10, 12 and 14 (presumptive abdominal segments 5, 7 and 9) in the developing embryo (Fig. 1B) (Drewell et al., 2002a; Ohtsuki et al., 1998). To identify the cis-regulatory elements responsible for this specificity, an Abd-B promoter was attached to a CAT reporter gene. The Abd-B promoter region is 1.5 kb in length and includes 294 bp of 5′ flanking sequence and approximately 1.2 kb of 3′ sequence. On a reporter transgene the IAB5 enhancer is able to direct strong CAT expression from the Abd-B promoter in the characteristic three posterior abdominal stripes in blastoderm-stage embryos (Fig. 2A). One possible mechanism by which promoter-enhancer specificity is established at the BX-C could be the existence of promoter-proximal elements, which inhibit enhancers from interacting with specific promoters. For example, IAB5 may be prevented from interacting with the abd-A promoter by a negative regulatory element close to the abd-A transcription start site. However, this does not appear to be the case, as IAB5 is able to interact strongly with a 1 kb abd-A promoter-lacZ reporter gene when located close to the abd-A promoter and in the absence of any other competing promoters (Fig. 2B). If the IAB2 enhancer is also added to this transgenic construct, a much broader band of lacZ expression expanding towards the anterior of the blastoderm-stage embryo is seen (Fig. 2C), indicating that IAB2 and IAB5 are both able to drive expression from the abd-A promoter. On transgenic constructs, IAB2 directs expression from the abd-A promoter predominantly in parasegments 7 and 9 (presumptive abdominal segments 2 and 4) and more weakly in parasegments 11 and 13 (segments 6 and 8), as previously described for a Ubx-lacZ reporter gene (Shimell et al., 2000). However, on transgenes the Abd-B promoter is also capable of interacting with the IAB2 enhancer. When the IAB2 and IAB5 enhancers are present on the same construct, bands of CAT expression are detected extending from presumptive abdominal segment 2 towards the posterior of the blastoderm-stage embryo (Fig. 2D). The promoters from the BX-C, therefore, appear to be responsive to both of the enhancers tested, indicating that the promoter regions themselves are not able to inhibit enhancer-promoter interactions.
Fig. 2. Enhancer-promoter interactions at the BX-C.
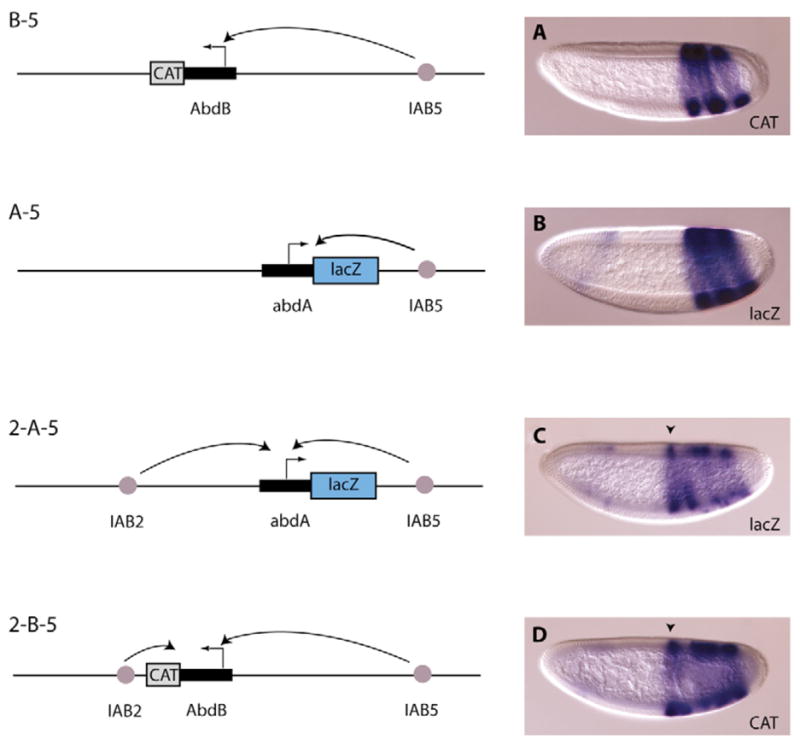
Transgenic embryos were hybridized with digoxigenin-labeled antisense RNA probes (diagrams on left). The embryos shown on the right are at blastoderm stage and orientated with anterior to the left and dorsal up. (A) On the B-5 construct the IAB5 enhancer directs expression of the Abd-B-CAT reporter gene in three characteristic stripes in presumptive abdominal segments 5, 7 and 9. (B) IAB5 is also able to interact with the abd-A promoter in the absence of a competing promoter, as it activates transcription from the abd-A-lacZ reporter gene. Ectopic expression of the lacZ and CAT reporter genes is also detected in the anterior of some transgenic embryos, as previously described (Ohtsuki et al., 1998; Zhou et al., 1999). (C,D) IAB2 and IAB5 will simultaneously activate expression from the abd-A or Abd-B promoter, indicated by a broad band of expression extending from presumptive abdominal segment 2 (indicated by black arrowhead) towards the posterior in 2-A-5 (C) and 2-B-5 (D) transgenic embryos. These expression patterns indicate that, in the absence of competing promoters, both homeotic gene promoters are responsive to interaction with the enhancers from the BX-C.
It is a formal possibility that promoter-enhancer specificity at the BX-C is established as a result of the endogenous enhancers interacting with promoters in an orientation-dependent manner. This is unlikely to be a general rule as the enhancers in the IAB3 and IAB4 regions must activate the abd-A promoter in the 3′ direction, while IAB5 and IAB2 both interact with their respective target promoters in the 5′ direction (see Fig. 1B). To test whether the enhancer activities are directional we also reversed the orientation of IAB2 and IAB5 relative to their target promoters on the transgenic constructs. This failed to disrupt enhancer-driven expression of the reporter genes (data not shown). Therefore, the enhancers from the BX-C appear to be orientation independent and promiscuous, able to activate any promoter. In addition, the homeotic gene promoters do not appear to harbor inhibitory regulatory elements capable of repressing activation from either enhancer. As a result, when only a single promoter is present, both IAB2 and IAB5 will drive expression strongly from either abd-A-lacZ or Abd-B-CAT.
The Abd-B promoter contains a tethering element
In order to determine if the Abd-B and abd-A promoters are able to specifically recruit either the IAB5 or IAB2 enhancers in competition with another promoter, we introduced the two promoters in tandem on transgenic constructs. On the B-A-5 construct, the IAB5 enhancer is placed 3′ of the abd-A-lacZ gene. In this stringent test, IAB5 fails to activate the proximal abd-A-lacZ gene (Fig. 3C) but directs strong expression of the more distal Abd-B-CAT gene in both blastoderm-stage (Fig. 3A) and germband-elongation-stage embryos (Fig. 3B). These staining patterns mimic the endogenous IAB5-driven activation pattern in parasegments 10, 12 and 14 and indicate that the 1.5 kb Abd-B promoter region is sufficient to selectively recruit the IAB5 enhancer. To test whether the abd-A promoter has a similar tethering activity for an endogenous enhancer, IAB2 was placed 3′ of the Abd-B-CAT gene on the 2-B-A-5 construct. However, unlike IAB5, IAB2 is not directed to its endogenous target promoter but is instead recruited to the proximal Abd-B promoter. This was shown by broad stripes of CAT staining in transgenic embryos in parasegments 7, 9, 11 and 13 driven from IAB2 and 10, 12 and 14 driven from IAB5 (Fig. 3D,E), while no abd-A-lacZ expression was observed (Fig. 3F). This result indicates that the abd-A promoter does not contain a regulatory element sufficient to tether the IAB2 enhancer. It is possible that at the endogenous locus there is no requirement for this activity, as the abd-A promoter is relatively close to IAB2 and therefore may not have to compete for the enhancer. As discussed earlier, in contrast to IAB2, IAB5 is roughly equidistant from both homeotic promoters in the BX-C but is selectively recruited only to Abd-B over 55 kb of intervening DNA sequence.
Fig. 3. Regulatory specificity determines cis-interactions at the BX-C.
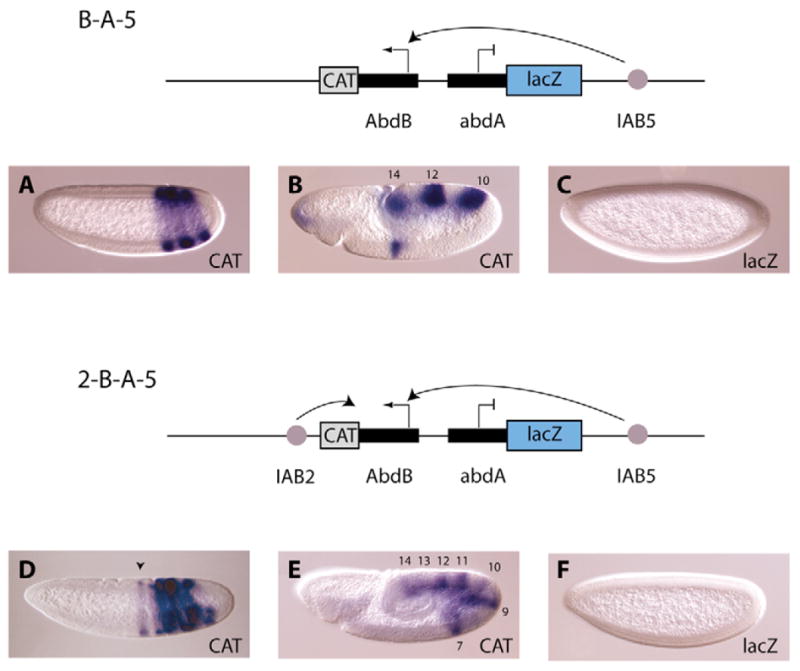
On the B-A-5 construct (top diagram), the IAB5 enhancer was placed 3′ of the proximal abd-A-lacZ reporter gene. The distal CAT gene is under the control of the Abd-B promoter. In this configuration the IAB5 enhancer selects Abd-B over abd-A, so that the Abd-B-CAT reporter exhibits a three-stripe IAB5 expression pattern in parasegments 10, 12 and 14; this is readily detected in blastoderm-stage embryos (A) and germband-elongation-stage embryos (B), whereas abd-A-lacZ is silent (C). By contrast, the IAB2 enhancer on the 2-B-A-5 construct (lower diagram) does not interact with its normal endogenous target (abd-A), but directs expression from the Abd-B-CAT gene in parasegments 7 (black arrow), 9, 11 and 13. In blastoderm-stage (D) and germ band elongation-stage embryos (E), a composite IAB2-IAB5-driven expression pattern is detected. The inactivity of the lacZ reporter on this construct (F) suggests that IAB2 only interacts with the most proximal promoter.
Anti-insulator activity in the Abd-B promoter
In order to activate Abd-B expression at the endogenous gene complex, IAB5 has to bypass at least two known insulator sequences, Fab-7 and Fab-8, that have enhancer-blocking activity (Fig. 1B) (Hagstrom et al., 1996; Zhou et al., 1999; Zhou et al., 1996). These insulator elements have previously been shown to disrupt promoter-enhancer interactions when placed between a promoter and enhancer on transgenes (Barges et al., 2000; Zhou et al., 1999; Zhou et al., 1996; Zhou and Levine, 1999). To further characterize the IAB5-Abd-B interaction, we therefore needed to test the interaction in the presence of an insulator from the BX-C. The 2-B-Fab-8-A-5 construct was created in which the previously characterized Fab-8 insulator element (Zhou and Levine, 1999) was placed between the homeotic promoter elements. The Fab-8 insulator sequence was specifically designed to exclude the adjacent PTS7 element. The addition of the Fab-8 insulator sequence did not disrupt the IAB5-Abd-B interaction, as strong CAT expression was detected in posterior stripes in blastoderm-stage embryos (Fig. 4A) whereas no abd-A-lacZ expression was detected (Fig. 4B). To confirm that the CAT expression in these embryos was not solely an IAB2-driven pattern, we generated transgenic lines carrying the B-Fab-8-A-5 transgene, from which the IAB2 enhancer was removed. In these embryos the IAB5 enhancer was recruited to the Abd-B promoter, as strong IAB5-driven expression was detected for the Abd-B-CAT gene (Fig. 4C) and no abd-A-lacZ expression was observed (Fig. 4D). To ensure that altering the enhancer-promoter spacing would not modulate the IAB5-Abd-B interaction, we created the 2-B-1.6λ-A-5 construct. To generate this construct, a 1.6 kb lambda DNA fragment (Calhoun et al., 2002) was inserted between the promoter regions on the 2-B-A-5 construct (Fig. 3). The addition of the spacer had no effect on the IAB5-Abd-B interaction, as strong CAT expression in a composite IAB2-IAB5-driven pattern was observed (Fig. 4E) whereas no lacZ expression was detected (Fig. 4F). These results indicate that the Abd-B promoter may contain a tethering activity that allows the IAB5 enhancer to drive expression of Abd-B-CAT across the Fab-8 insulator. This anti-insulator activity is important in the context of the endogenous BX-C, as IAB5 must bypass at least two insulators to interact with the Abd-B promoter.
Fig. 4. Anti-insulator activity in the Abd-B promoter.
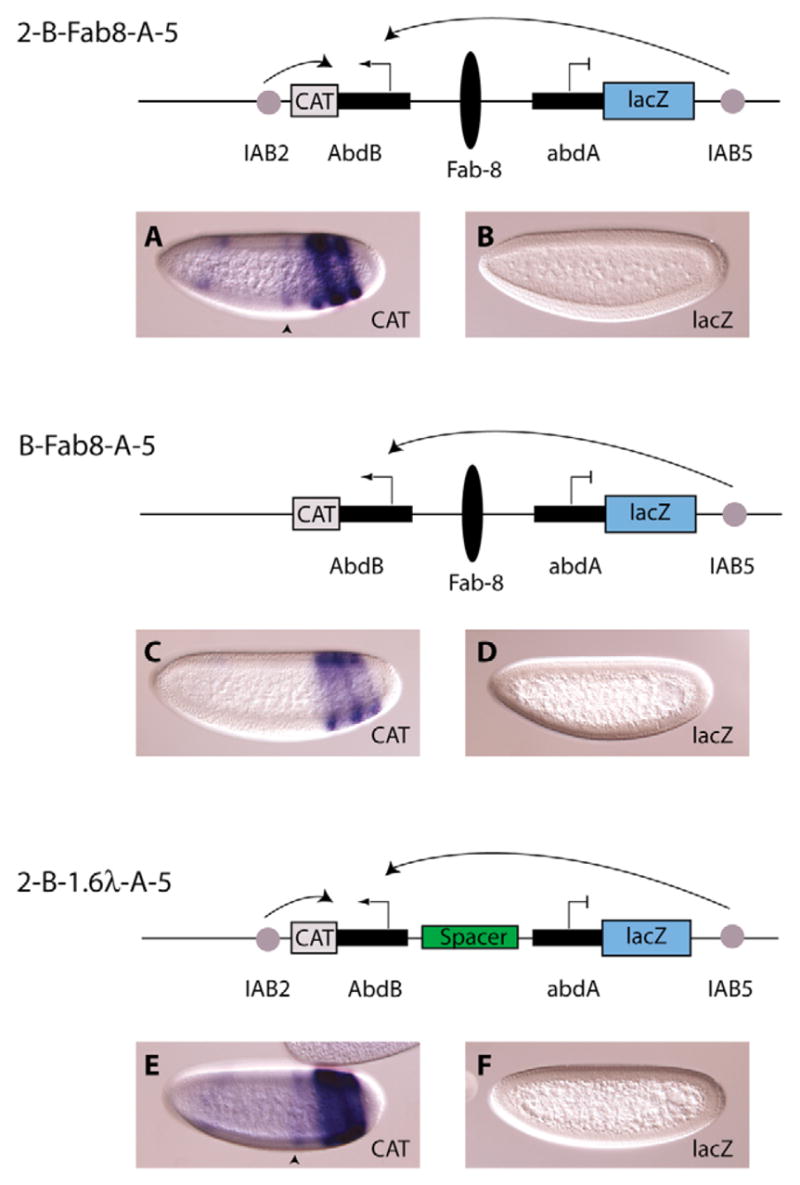
On the 2-B-Fab-8-A-5 construct (top diagram), the Fab-8 insulator was inserted between the two homeotic promoters. In this configuration both the IAB2 and IAB5 enhancers activate the Abd-B promoter, as CAT is expressed in the broad stripes, extending from parasegment 7 (black arrowhead) towards the posterior of the embryo (A), whereas no lacZ expression can be detected (B). To verify that IAB5 is activating expression from the Abd-B promoter, the IAB2 enhancer was removed to generate the B-Fab-8-A-5 construct (middle diagram). Transgenic embryos confirmed that IAB5 is indeed recruited the Abd-B promoter as CAT expression can be seen in a distinctive IAB5 pattern (C), whereas lacZ is not activated (D). To examine whether spacing had an effect on these interactions, a 1.6 kb λ-spacer DNA was placed between the two homeotic promoters on the 2-B-1.6λ-A-5 construct (bottom diagram). The addition of this λ-spacer did not modulate the activation of expression from the Abd-B promoter by the IAB2 and IAB5 enhancers (E) and no lacZ expression is detected (F).
Identification of the promoter-tethering element
In an effort to identify the region in the Abd-B promoter responsible for the tethering of IAB5 to the Abd-B promoter, a 255 bp DNA sequence from −40 to −294 bp 5′ of the Abd-B transcription start site was deleted from the Abd-B promoter. Although no consensus core elements are present at the Abd-B promoter, this region was selected to exclude the possibility of including any potential core promoter sequences (Butler and Kadonaga, 2002) close to the transcriptional start site but leaving the promoter region capable of activating transcription. This 255 bp sequence also overlaps with the 5′ region previously shown to be functionally important in regulating promoter-enhancer communication at the BX-C in trans (Sipos et al., 1998).
When comparing CAT reporter gene expression in transgenic embryos containing the full-length Abd-B promoter (Fig. 5A) to those in which the 255 bp sequence is deleted (Fig. 5C), it appears that this deletion abrogates recruitment of the IAB5 enhancer to the Abd-B promoter. In fact, deletion of the 255 bp sequence in the BΔPTE-A-5 construct resulted in re-direction of IAB5-driven expression from the Abd-B promoter (Fig. 5C) to the abd-A promoter directing lacZ expression (Fig. 5D) in transgenic embryos. We therefore named the 5′ 255 bp sequence the promoter-tethering element (PTE). In a few transgenic lines carrying the BΔPTE-A-5 construct very weak IAB5-driven expression of the BΔPTE-CAT reporter gene was observed (data not shown), probably because of position effects. This experiment indicates that the Abd-B promoter is functional in this configuration, but only at certain integration points in the genome. It is possible that the lack of expression of the CAT reporter gene on these constructs could simply be a result of a non-functional Abd-B promoter, caused by the truncation of the promoter. To verify that the Abd-BΔPTE promoter was still functional in a non-competitive situation, IAB2 was inserted 1 kb 3′ of the Abd-BΔPTE promoter to generate the 2-BΔPTE-A-5 construct. This resulted in IAB2-driven activation of the Abd-BΔPTE promoter (Fig. 5G), indicating that this promoter is still functional, whereas IAB5 was solely recruited to the abd-A promoter. (Fig. 5H). The level of expression detected from the truncated Abd-BΔPTE promoter was comparable to other transgenic lines analyzed in this study, suggesting the promoter is fully active. In addition, these embryos clearly demonstrate the different expression patterns driven by the two IAB enhancers (Fig. 5G,H). The deletion of the PTE, therefore, confirms that this cis-regulatory sequence is necessary for the recruitment of IAB5 to the Abd-B promoter on transgenes.
Fig. 5. Identification of the promoter-tethering element.
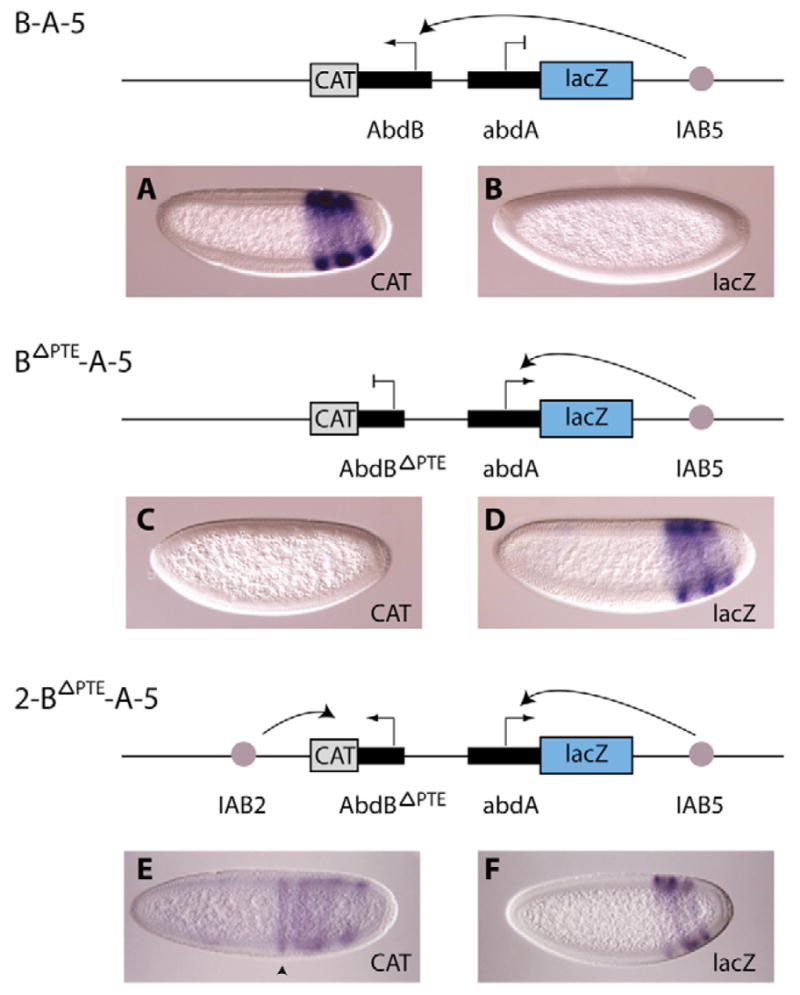
On the B-A-5 construct (top diagram), the IAB5 enhancer specifically bypasses the abd-A promoter (B) to activate expression from the Abd-B promoter (A). When the 255 bp region extending from −40 to −294 bp relative to the transcription start site is removed from the Abd-B promoter (BΔPTE-A-5 construct; middle diagram), the IAB5 enhancer is now re-directed to the abd-A promoter, as no CAT expression is detected (C) and strong lacZ expression is detected (D) in the majority of embryos. Extremely weak CAT expression in an IAB5-directed pattern was detected in a few transgenic embryos (data not shown). The integrity of the Abd-BΔPTE promoter was confirmed by insertion of the IAB2 enhancer 3′ of the Abd-BΔPTE-CAT reporter gene on the 2-BΔPTE-A-5 construct (bottom diagram). This resulted in IAB2 activation of the Abd-BΔPTE promoter indicated by an IAB2-driven CAT expression pattern in parasegments 7 (black arrowhead), 9, 11 and 13 in transgenic embryos (E), whereas lacZ is activated in an IAB5-driven expression pattern in the more posterior parasegments 10, 12 and 14 (F). Embryos exhibiting weaker staining are shown to clearly demonstrate the different expression patterns driven by the two IAB enhancers, although overall the expression in the 2-BΔPTE-A-5 embryos was comparable to other transgenic lines analyzed in this study.
PTE can regulate ectopic enhancer-promoter interactions
To further analyze the functional activity of the PTE in Drosophila embryos, the W-5-EZ construct was created in which the IAB5 enhancer was positioned between the mini-white reporter gene and an even-skipped-lacZ fusion reporter gene. As previously described, in this configuration the IAB5 enhancer has a strong preference for the TATA-box-containing even-skipped (eve) promoter in transgenic blastoderm-stage embryos and only weakly activates white (Fig. 6A,B) (Ohtsuki et al., 1998). Insertion of the 255 bp PTE adjacent to the eve-lacZ gene on the W-5-PTE-EZ transgenic construct resulted in redirection of the IAB5 enhancer-driven expression solely to the eve promoter as lacZ was expressed in the three characteristic posterior abdominal stripes (Fig. 6D), whereas no white expression was detected (Fig. 6C). The expression patterns from these constructs demonstrate that the PTE from 5′ of Abd-B will regulate ectopic promoter-enhancer interactions. The regulatory switch induced by the juxtapositioning of the PTE with the eve promoter indicates that the tethering of IAB5 is not promoter-specific. This fits with the regulatory logic required at the endogenous locus as a PTE in a gene complex will only be required to recruit specific enhancers to the promoter at which it is located.
Fig. 6. The PTE is able to regulate expression of an ectopic promoter.
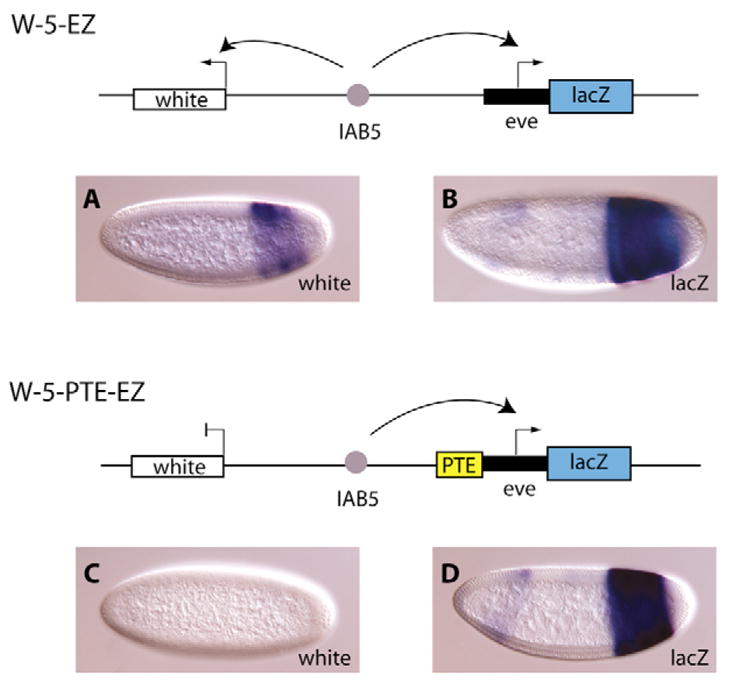
In the W-5-EZ construct (top diagram), the IAB5 enhancer is located between the white and even-skipped (eve) promoters. In this configuration, IAB5 is able to drive expression from white (A) and eve-lacZ (B). Insertion of the 255 bp PTE sequence adjacent to the eve-lacZ promoter in the W-5-PTE-EZ construct (bottom diagram) resulted in strong expression of lacZ (D) and an absence of expression of white (C), indicating that IAB5 is now exclusively activating the eve-lacZ promoter.
Bioinformatic analysis of the PTE sequence in different Drosophila species
Our transgenic studies indicate that the PTE is a functional element in the BX-C. One potential model for the tethering function of this regulatory sequence is that it may contain binding sites for trans factors that directly interact with the enhancers in the BX-C to bring them into close proximity with the Abd-B promoter. A prediction of this model is that the putative binding sites in the PTE should be conserved in different Drosophila species. As a result it should be possible to identify a high level of conservation for short stretches of sequence within the PTE that contain binding sites. To test this we carried out bioinformatic studies across seven different Drosophila species (Fig. 7A). The PTE sequence as a whole does not demonstrate a significantly higher level of conservation in the different species when compared to other sequences from the BX-C, including the IAB5 enhancer with known regulatory activity (Fig. 7B). However, within the PTE there are two short sequences that are more highly conserved: a 24-mer and a 27-mer (Fig. 7B,C). It is therefore possible that these short sequences represent conserved protein binding sites and that the trans factors involved in the functional activity of the PTE interact specifically with these conserved sequences (see Discussion).
Fig. 7. Conservation of the PTE sequence across Drosophila species.
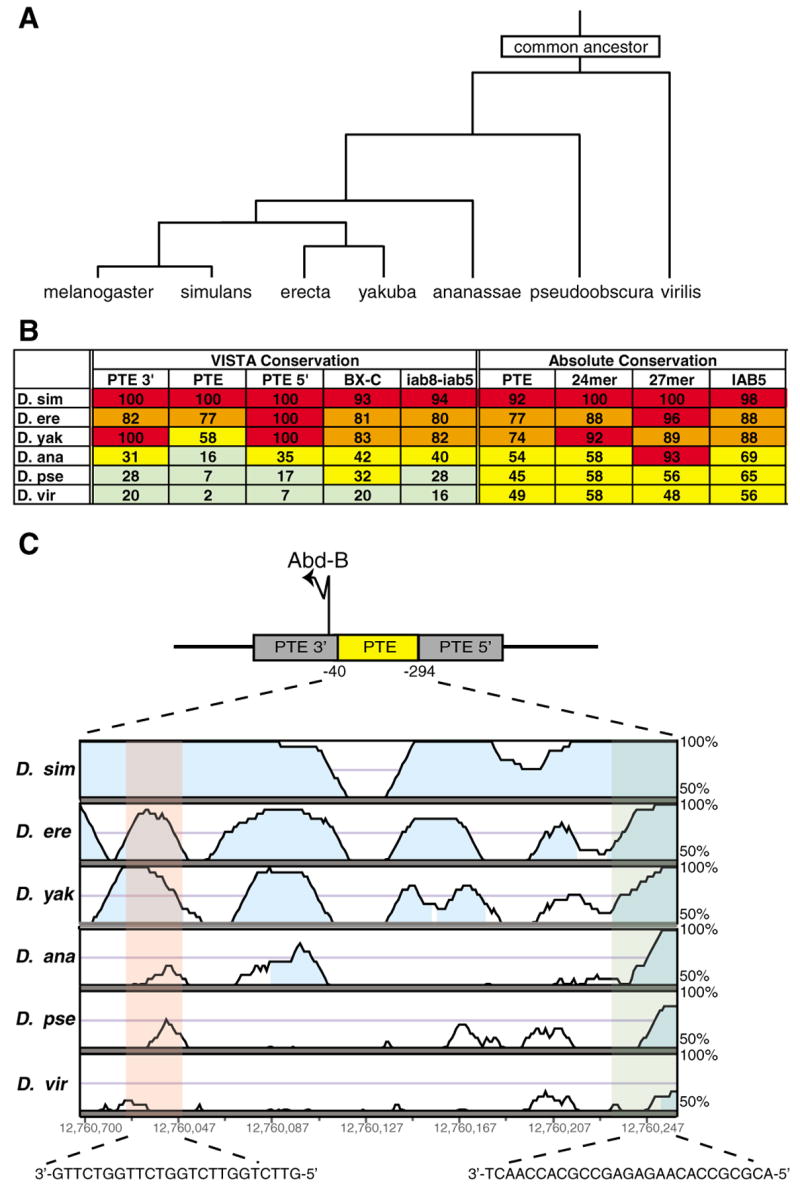
(A) An informal consensus tree (Crosby et al., 2007) illustrating evolutionary relationships among Drosophila species. (B) Sequence conservation levels for the 255 bp PTE and PTE 5′ and PTE 3′ (which represent the 255 bp regions 5′ and 3′ of the PTE with respect to Abd-B). For comparison, the conservation within the entire bithorax complex (BX-C) and the complete chromosomal control regions that direct Abdominal-B gene expression (iab8-iab5) and the IAB5 enhancer are also shown. Level of conservation between D. melanogaster and six other Drosophila species is indicated by color code: >90% red, 70–89% orange, 30–69% yellow, <30% green for different sequences. Conservation values were calculated using VISTA genome browser (see Materials and methods). Absolute conservation is the total percentage of D. melanogaster nucleotides that were conserved in the homologous region identified by VISTA alignment from the other species examined. (C) VISTA plots of the alignments indicate that the level of conservation is variable across the length of the PTE sequence. Overall, the PTE is less highly conserved than neighboring sequences or the non-genic regions in the BX-C, but there are two short, highly conserved sequences within it, a 24-mer (pink) and a 27-mer (green). The height of the peaks in the plots represents the level of conservation over a 24 bp window. Regions at least 24 bp long that are >80% conserved are indicated in cyan.
DISCUSSION
Previous genetic studies demonstrated that genomic regions 5′ of the Abd-B promoter are capable of recruiting the enhancers from the BX-C in trans (Sipos et al., 1998). At the endogenous BX-C, the IAB5 enhancer specifically directs expression from the Abd-B promoter in cis, although it is located slightly closer to the abd-A promoter and is separated from the target promoter by at least two known insulator sequences. The present study provides evidence that a novel promoter-tethering element (PTE), located −40 to −294 bp relative to the Abd-B transcription start site, is responsible for mediating these regulatory interactions and consequently plays a critical role in controlling promoter-enhancer communication at the BX-C.
Functional properties of the PTE
A 1.5 kb Abd-B promoter specifically recruits the IAB5 enhancer on reporter transgenes. The tethering activity contained in the Abd-B promoter region is able to interact with IAB5 over a long distance (>5 kb) and is capable of facilitating the bypass of an intervening promoter (abd-A) from the BX-C (see Figs 3 and 4). Deletion of a 255 bp sequence located in the 5′ region of the Abd-B promoter reveals the existence of a novel cis-element responsible for tethering of the IAB5 enhancer to the promoter. Removal of this PTE sequence from the Abd-B promoter is sufficient to redirect the IAB5 enhancer to the adjacent abd-A promoter on transgenes (see Fig. 5). In addition, fusion of the PTE sequence to an ectopic promoter results in complete recruitment of the IAB5 enhancer to the promoter (Fig. 6).
PTE mediated cis-regulatory interactions in the BX-C
The transgenic experiments presented here demonstrate that the 255 bp DNA sequence located 5′ of the Abd-B promoter contains a promoter-tethering element (PTE) that serves a key regulatory function by specifically recruiting the IAB5 enhancer to the Abd-B promoter. It is conceivable that at the endogenous complex the PTE may be involved in recruiting multiple intergenic enhancers to the Abd-B promoter, such as IAB6 and IAB7. Although this has yet to be tested on transgenes, the available genetic evidence supports this idea, as a relatively small deletion in the Abd-B upstream region is sufficient to disrupt activation by the IAB7 enhancer in trans (Sipos et al., 1998). By contrast, the IAB8 enhancer may not require a tethering element to activate the Abd-B promoter, possibly due to the close proximity of this enhancer to the Abd-B promoter and the fact that there are no intervening insulator elements (see Fig. 1B).
Previous studies demonstrated that a deletion 5′ of the Abd-B promoter region, which included the PTE, resulted in reduced IAB enhancer-Abd-B promoter interactions in trans (Sipos et al., 1998). As larger deletions were made, the interactions between the IAB7 enhancer and the target Abd-B promoter became increasingly weaker. One explanation for this observation could be the existence of additional elements in the extended 5′ promoter sequence which may play a role in the tethering of the IAB enhancers to the Abd-B gene at the endogenous BX-C. Our bioinformatic studies across different Drosophila species support this idea. The neighboring sequence 5′ of the PTE is highly conserved; suggesting that part of this upstream region may also contain sequences that aid in the tethering activity (Fig. 7). In this case, the critical in vivo function of the PTE may be supported by, as yet unidentified, additional cis-regulatory sequences capable of facilitating promoter-enhancer tethering. Transgenic constructs containing a PTE sequence extended to include part of this 5′ region will be important in determining the function of this region.
The PTE may also function in conjunction with a different class of anti-insulator elements at the BX-C, the promoter targeting sequences (PTS), which are known to facilitate promoter-enhancer interactions (Chen et al., 2005). Future transgenic and genetic experimental approaches will help to unravel the combinatorial regulatory activities of these complex cis-elements. However, it is clear that the promoter-enhancer interactions facilitated by the PTE are relatively strong, as neither spacer DNA nor endogenous insulator elements were capable of disrupting these interactions in our transgenic assays. These results provide insight into the regulatory requirements at the endogenous locus. It seems likely that these strong interactions are necessary for the IAB5 enhancer element to bypass the two known insulator elements to activate the Abd-B promoter approximately 55 kb away in cis.
The precise molecular mechanism by which the tethering element functions is not clear. It is possible that common trans factors may bind to both the IAB5 enhancer and PTE and establish protein-protein interactions, although the Abd-B PTE and IAB5 enhancer do not share any extensive sequence homology. There is, however, a precedent for this type of interaction as Sp1 has been shown to form DNA loops between binding sites proximal to promoter sequences and distant binding sites to mediate an increased concentration of activator protein at the promoter (Mastrangelo et al., 1991). Bioinformatic analysis reveals two separate short sequences within the PTE that are highly conserved in different Drosophila species, when compared to the other sequences in the PTE (Fig. 7). It is possible that these sequences harbor binding sites critical for the recruitment of the trans factors involved in the molecular function of the PTE. As previously proposed, it is possible that a mechanism involving the IAB enhancers looping to interact with the PTE and drive expression from the target Abd-B promoter could be facilitated by specific chromatin structures in the BX-C (Akbari et al., 2006; Sipos and Gyurkovics, 2005). A similar spatial nuclear organization has also been suggested as a global regulator of developmental gene expression in higher eukaryotes (de Laat and Grosveld, 2003). More recent studies have demonstrated that there are indeed physical interactions between distant regulatory regions with the BX-C (Cleard et al., 2006), although the details remain to be fully elucidated. This model would explain the necessity of a functional PTE in the BX-C, as the disruption of this element would prevent the formation of the chromatin loop structures essential for promoter-enhancer communication, leaving the Abd-B target promoter inactive.
Implications for regulatory specificity in gene complexes
Promoter-tethering elements represent a precise mechanism for regulating specific enhancer-promoter interactions in gene complexes. Other mechanisms of cis-regulation may be less flexible. Recent studies have identified enhancers in the Drosophila genome capable of interacting only with distinct sub-sets of promoters. Some enhancers will only interact with DPE-containing promoters, whereas others only interact with TATA-containing promoters (Butler and Kadonaga, 2001). At the BX-C this may not be a feasible method for regulating enhancer-promoter interactions as the addition of a TATA box to one promoter, for example, may result in recruitment of all the enhancers in the complex. However, the PTE may function similarly to promoter competition in some respects, as the specific IAB5-Abd-B interaction it mediates appears to predominantly prevent the enhancer from activating other promoters.
The existence of a tethering element capable of specifically recruiting the distal T1 enhancer to the Scr gene promoter at the antennapedia Hox gene complex in Drosophila (Calhoun et al., 2002) suggests that promoter-tethering elements may represent a common mechanism for regulating enhancer-promoter interactions at complex loci. The ability of the IAB5 enhancer to activate Abd-B across insulator DNAs provides an intriguing model for tethering activities at other gene complexes. An example is the well characterized insulator at the mouse H19 imprinting control region which separates 5′ enhancers from the H19 promoter (Bell and Felsenfeld, 2000; Drewell et al., 2002b; Hark et al., 2000; Szabo et al., 2000). It is possible that a tethering element is required to selectively recruit these 5′ enhancers to the target promoter. The identification of an enhancer-containing global control region at the mouse Hoxd complex (Spitz et al., 2003) raises the possibility that promoter tethering over long distances may also be required at mammalian Hox genes.
Acknowledgments
We thank Sumio Ohtsuki for unpublished DNA constructs, Cory Olsen and Viet Ngo for technical assistance and Tom Kidd for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health to R.A.D. (HD54977).
References
- Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev Biol. 2006;293:294–304. doi: 10.1016/j.ydbio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Busturia A, Bienz M. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Sharma S, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–4286. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Lin Q, Zhou J. Multiple Promoter Targeting Sequences exist in Abdominal-B to regulate long-range gene activation. Dev Biol. 2005;286:629. doi: 10.1016/j.ydbio.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM The FlyBase Consortium. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Drewell RA, Brenton JD, Ainscough JF, Barton SC, Hilton KJ, Arney KL, Dandolo L, Surani MA. Deletion of a silencer element disrupts H19 imprinting independently of a DNA methylation epigenetic switch. Development. 2000;127:3419–3428. doi: 10.1242/dev.127.16.3419. [DOI] [PubMed] [Google Scholar]
- Drewell RA, Arney KL, Arima T, Barton SC, Brenton JD, Surani MA. Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development. 2002b;129:1205–1213. doi: 10.1242/dev.129.5.1205. [DOI] [PubMed] [Google Scholar]
- Drewell RA, Bae E, Burr J, Lewis EB. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002a;99:16853–16858. doi: 10.1073/pnas.222671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hogga I, Mihaly J, Barges S, Karch F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell. 2001;8:1145–1151. doi: 10.1016/s1097-2765(01)00377-x. [DOI] [PubMed] [Google Scholar]
- Karch F, Weiffenbach B, Peifer M, Bender W, Duncan I, Celniker S, Crosby M, Lewis EB. The abdominal region of the bithorax complex. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Martin CH, Mayeda CA, Davis CA, Ericsson CL, Knafels JD, Mathog DR, Celniker SE, Lewis EB, Palazzolo MJ. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development. 1991;111:437–449. doi: 10.1242/dev.111.2.437. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Peterson AJ, Burr J, Simon JA, O’Connor MB. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev Biol. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- Sipos L, Gyurkovics H. Long-distance interactions between enhancers and promoters. The case of the Abd-B domain of the Drosophila bithorax complex. FEBS J. 2005;272:3253–3259. doi: 10.1111/j.1742-4658.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- Sipos L, Mihaly J, Karch F, Schedl P, Gausz J, Gyurkovics H. Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics. 1998;149:1031–1050. doi: 10.1093/genetics/149.2.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Tang SHE, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development. 1999;126:3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]


