Abstract
The yeast U4/U6⋅U5 pre-mRNA splicing small nuclear ribonucleoprotein (snRNP) is a 25S small nuclear ribonucleoprotein particle similar in size, composition, and morphology to its counterpart in human cells. The yeast U4/U6⋅U5 snRNP complex has been purified to near homogeneity by affinity chromatography and preparative glycerol gradient sedimentation. We show that there are at least 24 proteins stably associated with this particle and performed mass spectrometry microsequencing to determine their identities. In addition to the seven canonical core Sm proteins, there are a set of U6 snRNP specific Sm proteins, eight previously described U4/U6⋅U5 snRNP proteins, and four novel proteins. Two of the novel proteins have likely RNA binding properties, one has been implicated in the cell cycle, and one has no identifiable sequence homologues or functional motifs. The purification of the low abundance U4/U6⋅U5 snRNP from yeast and the powerful sequencing methodologies using small amounts of protein make possible the rapid identification of novel and previously unidentified components of large, low-abundance macromolecular machines from any genetically manipulable organism.
Keywords: pre-mRNA splicing, mass spectrometry
Eukaryotic pre-mRNA must undergo extensive processing in the nucleus before it is converted into the mature mRNA species competent for translation into protein. One important processing step is the excision of intervening sequences, termed introns, which are present within the coding sequences. Removal of introns is effected by the pre-mRNA splicing machinery, which is composed of five phylogenetically conserved small nuclear RNAs (snRNAs) assembled into small ribonucleoprotein particles (snRNPs) and several non-snRNP associated proteins (1). Once assembled onto pre-mRNA, the massive RNP termed the spliceosome removes the intronic RNA sequences by two ordered transesterification reactions ligating the two exons together, forming the mature message. The biochemistry of the mammalian spliceosomal snRNPs has been extensively studied, and many of the protein components have been purified and their genes cloned (2–4). The budding yeast Saccharomyces cerevisiae has been very useful in the study of pre-mRNA splicing, as it is amenable to genetic manipulations. The low abundance of pre-mRNA splicing snRNPs, however, has made their isolation technically challenging. Several yeast proteins involved in splicing have been identified by conditional growth mutations that produce a pre-mRNA splicing defect (5–7) from synthetic lethal analysis (8, 9), identification of second-site suppressors (10–13), two-hybrid analysis (14), and, most recently, from the direct biochemical purification of the yeast U1 snRNP (15, 16).
In yeast, there are an estimated 100–200 copies of each spliceosomal snRNA (17) as compared with 100,000–1,000,000 snRNA copies in mammalian cells (18). This discrepancy in abundance has made the purification of yeast snRNPs much more difficult, especially in the case of the yeast U4/U6⋅U5 tri-snRNP, which exists in equilibrium with the individual U4/U6 and U5 snRNPs and is therefore less abundant than the respective individual snRNPs (19). The U4/U6⋅U5 tri-snRNP undergoes dynamic rearrangements that assemble the particle from individual U4, U5, and U6 snRNPs. U4 and U6 snRNPs associate into a U4/U6 snRNP particle by extensive base pairing mediated by the putative RNA annealing factor Prp24 (21). The U4/U6 snRNP then associates with the U5 snRNP to form the U4/U6⋅U5 snRNP, which sediments at 25S in glycerol gradients (20). The U4/U6⋅U5 snRNP joins the splicing reaction after the formation of the pre-spliceosome, which is composed of the pre-mRNA and the U1 and U2 snRNPs. U6 then replaces U1 at the 5′ splice site in an ATP-dependent, Prp28-mediated exchange (22). The U4 snRNP then is released from the spliceosome, likely in a Brr2 RNA helicase-mediated reaction (23), and the U2, U5, and U6 snRNPs remain associated with the spliceosome. The U5 snRNP is responsible for the alignment of the two exons to be spliced together (24). The U6 and U2 snRNAs undergo dynamic rearrangements to assemble into the presumed catalytic core of the splicing reaction (1). After splicing occurs and the spliceosome dissociates, the U4/U6⋅U5 snRNP recycles, and the process is repeated with a new pre-mRNA. The identity of all of the proteins bound to these rapidly changing particles and which ones may leave and join remain to be determined.
The U1, U2, U4, and U5 snRNPs each contain the canonical Sm proteins B (and B′ in mammals), D1, D2, D3, E, F, and G (25). In yeast, it has not been previously demonstrated that the SmB protein associates with these snRNPs, and, although it was not isolated with purified U1 snRNP, U1 snRNA can be immunoprecipitated by using epitope-tagged SmB (16). U6 snRNA is associated with the Sm-like proteins termed Lsm2-Lsm8 (ref. 26; J. Beggs personal communication) as well as the Prp24 protein (27). Although many U4/U6⋅U5 snRNP proteins have been identified (10, 23, 28–36), the methods used in their discovery have not ruled out the possibility that other proteins may be associated with these snRNPs.
We have purified the yeast U4/U6⋅U5 snRNP by affinity chromatography of a doubly epitope-tagged SmD3 protein (37), a component of the spliceosomal snRNPs. By using two affinity chromatography steps followed by preparative glycerol gradient sedimentation, the triple snRNP was purified to near homogeneity, and the polypeptides were isolated and identified by mass spectrometry. We show that there are at least 24 proteins stably associated with the U4/U6⋅U5 snRNP, 4 of them novel. This is similar to the ≈20–25 proteins isolated in purified human U4/U6⋅U5 snRNP (38). Previous studies have shown that the ultrastructure of the yeast U4/U6⋅U5 snRNP was very similar to that of the human U4/U6⋅U5 snRNP (20), indicating that a similar protein complement is likely. Tandem mass spectrometry is proving to be a powerful and concise means of identifying the components of low abundance macromolecular complexes from organisms whose genomes have been sequenced.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Molecular Biology.
Yeast strain JWY2445 (37) MATa his1 leu2–1 trp1-Δ101 ura3–52 smd3Δ1∷LEU2 + YCp50-SMD3 was kindly provided by John Woolford (Carnegie Mellon University). Plasmid pSmD3–28 contains an octahistidine epitope followed by two tandem repeats of the polyoma Py epitope (39) at the extreme C terminus of the SmD3 polypeptide introduced by a two step PCR cloning procedure. Plasmid pSmD3–28 was introduced by lithium acetate transformation (41) of strain JWY2445, and transformants were selected by using synthetic yeast medium plates lacking tryptophan. Plasmid shuffle to remove the wild-type YCp50-SmD3 plasmid was performed on synthetic yeast medium lacking tryptophan and containing 5-fluoroorotic acid to create strain YSS201. Bacterial manipulations were according to standard procedures (42). All enzymes were purchased from New England Biolabs or prepared in this laboratory, chemicals were purchased from Sigma, and media ingredients were from Difco.
Yeast Growth and Extract Preparation.
Strain YSS201 was grown in a 200-liter fermentor in yeast extract/peptone/dextrose medium at 30°C to OD at ≈3.0 and was harvested in a continuous flow centrifuge. The cell paste was suspended 1:1 (wt/vol) in buffer A (10 mM Hepes, pH 7.9/10 mM KCl) and was extruded into liquid nitrogen. Extracts were prepared by liquid nitrogen homogenization in a Waring blender essentially as described (43) and were dialyzed into the buffer used to perform the chromatography.
Affinity Chromatography and Glycerol Gradient Sedimentation.
Antibody affinity matrix was prepared by using supernatant from hybridoma cell line AK6967 grown in serum free medium II (GIBCO/BRL). Cell supernatant was incubated for 2 hr with Protein-G Sepharose (Amersham Pharmacia) and was crosslinked with dimethylpimelimidate as described by the manufacturer. Antibody was conjugated to the matrix at a concentration of 1 mg/ml bed volume of resin. Extract dialyzed against buffer D50 or D250 [buffer D (20 mM Hepes, pH 7.9/8% glycerol/10 mM β-mercaptoethanol/0.5 mM phenylmethylsulfonylfluoride/1 μg/ml leupeptin/1 μg/ml pepstatin) containing 50 or 250 mM KCl] was passed over the affinity resin at a flow rate of 5 ml/hr, was washed with 15 column volumes of buffer D50 or D250, and was eluted with 5 column volumes of buffer D50 or D250 containing 100 μg/ml HPLC-purified peptide EYMPME (synthesized at the Caltech Polymer Synthesis Facility, Pasadena, CA) to compete for antibody binding. Ni–nitrilotriacetic acid (NTA) (Qiagen) chromatography was performed on the antibody affinity chromatography eluate. After loading, the column was washed with 15 column volumes of buffer D containing 15 mM imidazole. Elution of snRNPs from the Ni-NTA matrix was effected by incubation with buffer D containing 100 mM imidazole. Linear glycerol gradients (10–30% gradients) were prepared at various KCl concentrations in 20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 0.01% Nonidet P-40, 10 mM β-mercaptoethanol, 1 μg/ml leupeptin, and 1 μg/ml pepstatin and were sedimented at 29,000 rpm for 24 hr at 4°C in a Beckman SW41 rotor. RNA and protein were isolated as described (20) and were stored at −80°C.
RNA Analysis, Protein Analysis, and Mass Spectrometry.
RNA was analyzed on 5% polyacrylamide (19:1), 8.3 M urea, and 1× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) gels and was stained with silver (44). RNA markers were purchased from Ambion. Proteins were separated on 12% SDS/PAGE gels prepared as described (45). Protein markers were purchased from NOVEX (San Diego). Proteins were stained with Coomassie blue R-250 (GIBCO/BRL) and were excised from the SDS/PAGE gels. Protease digestion and sample preparation for nano-electrospray MS/MS mass spectrometry were performed as described (46, 47).
RESULTS
Purification of Total Yeast Spliceosomal snRNPs.
By placing an epitope tag at the SmD3 C terminus that consists of an octahistidine sequence followed by two repeats of an epitope from a polyoma viral protein (39), we were able to purify all of the spliceosomal snRNPs in two steps. First, whole cell extract was passed over an immunoaffinity column to enrich for the epitope tag on the SmD3 protein. Elution was achieved by the incubation of the resin with a molar excess of the epitope in peptide form. A second affinity purification was performed to remove contaminating proteins that copurified on the antibody matrix: The antibody column eluate was passed over a 0.3–0.5 ml Ni-NTA column, was washed extensively, and was eluted by addition of 100 mM imidazole.
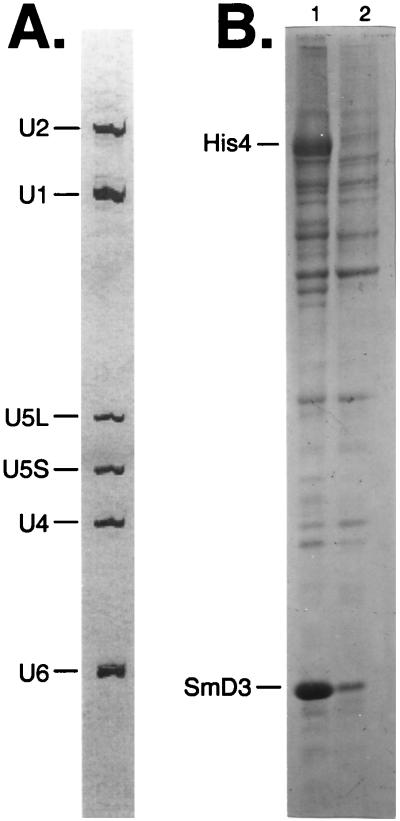
Single affinity chromatography by using either the antibody resin or the Ni-NTA resin did not yield a snRNP fraction of sufficient purity. Using Ni-NTA chromatography alone resulted in the retention of <5% of the snRNPs. Inclusion of two separate blocks of eight consecutive histidine residues was not sufficient to compete for the large number of polyhistidine containing proteins present in yeast (data not shown). In the first lane of Fig. 1B, the bulk proteins eluted from the antibody affinity column are shown. The major band at ≈84 kDa is the His4 protein as determined by mass spectrometry. This protein does not contain sequences related to the antibody epitope but copurifies nonetheless. In Fig. 1B, lane 2, the bulk proteins from a subsequent Ni-NTA chromatography step are shown. The His4 band as well as other likely contaminating minor bands are no longer present; however, many of the proteins remain.
Figure 1.
Total RNA and protein eluted from affinity chromatography purification. (A) Total RNA from the Ni-NTA elution step. Ten micrograms of total snRNPs from the Ni-NTA eluate were phenol-extracted, and RNA was precipitated with ethanol. Resuspended RNA was run on a 5% polyacrylamide gel and was stained with silver. Positions of U1, U2, U4, U5L, U5S, and U6 snRNAs are shown. (B) Total protein from each step of the affinity purification. The organic phase of the phenol extractions from 10 μg of protein from the α-Py (lane 1) and Ni-NTA (lane 2) chromatography steps was precipitated with acetone, and recovered proteins were run on a 12% SDS/PAGE gel. Positions of the epitope-tagged SmD3 protein and the contaminant His4 are indicated.
The RNA species eluting from the Ni-NTA column are shown in Fig. 1A. Note the absence of contaminating tRNA and rRNA in this fraction. The identity of the snRNAs was confirmed by primer extension of the bulk RNA with primers corresponding to individual RNAs yielding products of a defined length (data not shown). For reasons that are not well understood, ≈50–60% of the U1 and U2 snRNPs are present in the antibody column flow through whereas the U4/U6 and U5 snRNPs appear to be quantitatively bound in this step (data not shown). It is possible that either the epitopes on the SmD3 protein are not as readily accessible in these snRNPs, or perhaps the snRNAs in the flow-through are not fully assembled snRNPs.
Purification of the U4/U6⋅U5 snRNP.
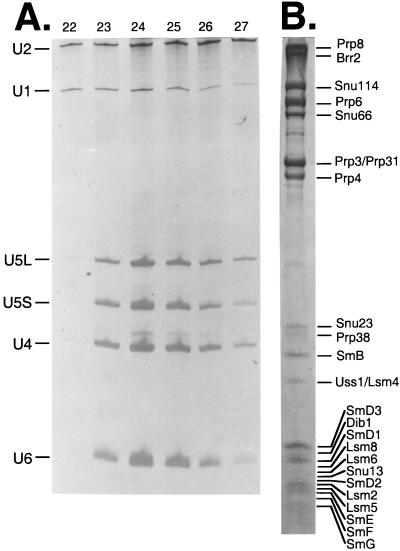
The low abundance of the yeast U4/U6⋅U5 snRNP necessitated the pooling of five purifications to obtain enough material to perform mass spectrometric sequencing of the proteins. A total of 750 ml of yeast whole cell extract was passed over five 1-ml antibody affinity columns (150 ml of splicing extract each) and was eluted with buffer D50 containing 100 μg/ml competitor peptide. The eluates then were passed over five 0.5-ml Ni-NTA columns and were eluted with buffer D50 containing 100 mM imidazole. A total of 350 μg of total spliceosomal snRNPs was layered onto 16 10–30% glycerol gradients. Fractions 22–25 (measured from the top) always contained the U4/U6⋅U5 snRNP with minor amounts of U1 and U2 snRNP (Fig. 2A). Densitometric analysis of serial dilutions of these fractions showed that the U4/U6⋅U5 snRNAs were present in >100-fold molar excess over the U1 or U2 snRNPs.
Figure 2.
Glycerol gradient purification of the U4/U6⋅U5 snRNP. (A) RNA extracted from fractions 22–27 from a 10–30% glycerol gradient was run on a 5% polyacrylamide gel and was stained with silver. Positions of the snRNAs are indicated. (B) Proteins associated with the U4/U6⋅U5 snRNP. Proteins from 64 glycerol gradient fractions were precipitated with acetone, were run on a 12% high TEMED SDS/PAGE gel, and were stained with Coomassie blue. Positions of the 24 stably associated U4/U6⋅U5 snRNP proteins are indicated.
Fractions 22–25 of the 16 gradients were phenol extracted, and the organic phase was precipitated with acetone. The precipitated proteins were resuspended in SDS/PAGE gel loading buffer and were separated electrophoretically on a 12% high-TEMED gel (45). In Fig. 2B, the proteins of the yeast U4/U6⋅U5 snRNP are indicated. The 24 proteins identified in the yeast U4/U6⋅U5 snRNP are listed in Table 1. The minor contaminating band migrating at 35-kDa between Prp4 and Snu23 is glyceraldehyde-3-phosphate dehydrogenase isoforms 1, 2, and 3, the bulk of which occurs at the top of the gradient (data not shown). This protein represents 20% of the total soluble protein in yeast (48) and also was found to be a contaminant in the U1 snRNP purification protocol (15). Two possible explanations for the copurification of this enzyme with the U1 and U4/U6⋅U5 snRNPs are (i) nonspecific interaction with the carbohydrate based matrices involved in the affinity purification steps and/or (ii) nonspecific interaction with the many phosphate moieties in the U4, U5, and U6 snRNAs. The other minor contaminating bands were not analyzed by mass spectrometry, but, by size, some can be correlated to the known U1 snRNP proteins, which are likely to be present as shown by RNA analysis of the fractions used (Fig. 2A).
Table 1.
Mass spectrometric identification of U4/U6⋅U5 snRNP proteins
| Protein name | Predicted molecular mass | SGD ORF name | No. of peptides sequenced | Putative homology/functions |
|---|---|---|---|---|
| Prp8 | 280 kDa | YHR165C | 8 | Unknown |
| Brr2 | 246 kDa | YER172C | 25 | RNA helicase |
| Snu114 | 114 kDa | YKL173W | 19 | GTPase |
| Prp6 | 104 kDa | YBR055C | 18 | Zinc finger/TPR |
| Snu66 | 66 kDa | YOR308C | 13 | Unknown |
| Prp31 | 56 kDa | YGR091W | 8 | Spliceosome assembly |
| Prp3 | 56 kDa | YDR473C | 11 | Spliceosome assembly |
| Prp4 | 52 kDa | YPR178W | 8 | WD repeats |
| Prp38 | 28 kDa | YGR075C | 4 | snRNP stability |
| Snu23 | 23 kDa | YDL098C | 10 | Zinc finger |
| SmB | 22 kDa | YER029C | 4 | Sm motifs |
| Uss1/Lsm4 | 21 kDa | YER112W | 4 | Sm motifs |
| Dib1 | 17 kDa | YPR082C | 6 | Cell cycle |
| SmD1 | 16 kDa | YGR074W | 3 | Sm motifs |
| Lsm8 | 15 kDa | YJR022W | 1 | Sm motifs |
| SmD3 | 15 kDa | YLR147C | 4 | Sm motifs |
| Lsm6 | 14 kDa | YDR378C | 1 | Sm motifs |
| Snu13 | 13 kDa | YEL026W | 4 | Nhp2 homology |
| SmD2 | 12 kDa | YLR275W | 6 | Sm motifs |
| Lsm2 | 11 kDa | YBL026W | 3 | Sm motifs |
| Lsm5 | 10.4 kDa | YER146W | 2 | Sm motifs |
| SmE | 10.4 kDa | YOR159C | 2 | Sm motifs |
| SmF | 9.6 kDa | YPR182W | 2 | Sm motifs |
| SmG | 8.5 kDa | YFL017W-A | 2 | Sm motifs |
SGD, Saccharomyces Genome Database (Stanford, CA); TPR, tetratrico-peptide repeat.
Mass Spectrometry Identification of Purified U4/U6⋅U5 snRNP Proteins.
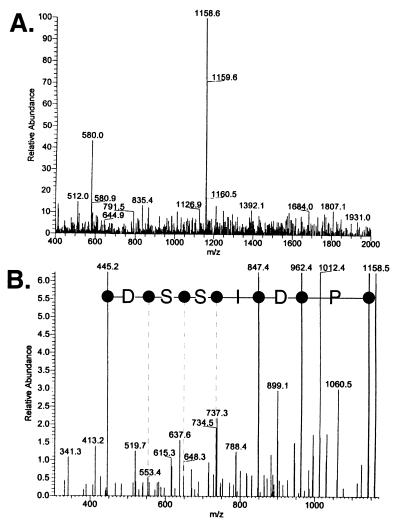
We analyzed all of the proteins extracted from the SDS/PAGE gels slices by nano-electrospray tandem mass spectrometry. After in-gel digestion with trypsin, the peptide fragments were extracted from the gel matrix and were separated by HPLC. The individual peptides were identified by mass and were isolated and fragmented. An example is shown in Fig. 3. The 1,158-dalton tryptic peptide from Brr2 was isolated (Fig. 3A) and fragmented by high energy collision with argon gas. The resulting fragments were analyzed in a second mass spectrometer (Fig. 3B) and the B and Y′′ ions corresponding to the fragmented masses from the N and C termini, respectively, were determined. The protein sequence PDISSD was determined by inverting the sequence shown in Fig. 3B because these ions correspond to single amino acid truncations from the C terminus. The peptide sequence tags derived from all of the analyzed peptides were compared against a theoretical trypsin digestion of all of the proteins in the owl database (Rockefeller Univ., New York). For each of the U4/U6⋅U5 snRNP proteins, a single yeast ORF could be identified. This procedure was repeated for all of the U4/U6⋅U5 snRNP proteins.
Figure 3.
Mass spectrometry microsequencing spectrum from a Brr2 peptide. (A) Representative spectrum from an ≈1,158-Da tryptic peptide derived from Brr2. (B) MS/MS mass spectrometry analysis of fragmented 1,158-Da peptide. Masses calculated for the resulting ions were correlated to the entire nonredundant protein database. The labeled mass peaks for the Y′′ ions represent the protein sequence PDISSD from the Brr2 protein corresponding to amino acids 958–963.
All Canonical Sm Core Proteins of Spliceosomal snRNPs Are Present in the Yeast U4/U6⋅U5 snRNP.
Many of the lower molecular weight proteins in the U4/U6⋅U5 snRNP (Fig. 2B) correspond to the Sm core proteins present in spliceosomal snRNPs. The D1, D2, D3, E, F, and G polypeptides are all present in the U4/U6⋅U5 snRNP, as was shown to be the case in the U1 snRNP (15). Note that the SmD3 protein is shifted to a higher relative molecular weight because of the presence of the affinity tag. The SmB protein, which was not stoichiometrically represented in the purified U1 snRNP (16), is present in the U4/U6⋅U5 snRNP. It appears by Coomassie blue staining to be present in twice the amount of closely migrating proteins, as SmB should be stably associated with both the U4 and U5 snRNPs.
A Set of U6-Specific Sm Proteins Are Present in the Yeast U4/U6⋅U5 snRNP.
U6 snRNA does not contain a canonical Sm binding site for the assembly of the Sm core onto the RNA (49). It does, however, have a set of proteins similar to the canonical Sm proteins (J. Beggs personal communication). The Lsm4/Uss1 polypeptide has been shown to associate with the U6 snRNP (26) and is present in the isolated U4/U6⋅U5 snRNP. The mass of the protein in the band indicates that it is likely to be present in one copy per U4/U6⋅U5 snRNP. Lsm2, Lsm5, Lsm6, and Lsm8 were not visible by Coomassie staining of the gel but were fortuitously identified as proteins comigrating with the canonical Sm protein bands. Lsm3 and Lsm7 were not identified in this analysis because we did not analyze the regions of the gel in which they migrated. Indeed, tagged Lsm3 or Lsm7 extracts co-immunoprecipitate U4, U5 and U6 snRNAs, indicating their presence in the U4/U6⋅U5 snRNP (J. Beggs, personal communication).
Proteins Previously Shown to Associate with U4/U6⋅U5 snRNP.
The Prp8, Brr2, and Snu114 proteins are known to associate with the U5 as well as the U4/U6⋅U5 snRNP. Prp8, which is calculated to have a molecular mass of 280 kDa migrates at ≈260 kDa, Brr2 (calculated molecular mass = 246 kDa) migrates aberrantly at ≈205 kDa, and Snu114 (calculated molecular mass = 114 kDa) migrates normally. Five additional proteins have been shown to be associated with the U4/U6⋅U5 snRNP. The Prp6, Prp31, Prp3, Prp4, and Prp38 proteins migrate close to their predicted molecular mass (104, 56, 56, 52, and 28 kDa, respectively; refs. 28–30, 35, and 36). The ability of this technique to identify Prp31 and Prp3 in a single band demonstrates that, if other proteins were comigrating, they likely would have been identified.
Novel Proteins Associated with the U4/U6⋅U5 snRNP.
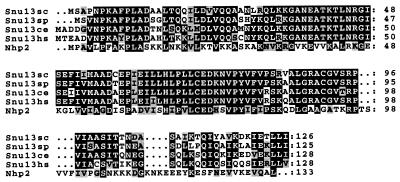
Four novel proteins were isolated in the gradient purified U4/U6⋅U5 snRNP. In keeping with the nomenclature for other isolated snRNP proteins, the novel U4/U6⋅U5 snRNP proteins have been designated Snu (SNUrp associated) proteins with the corresponding molecular weight. Snu66 is a novel 66-kDa protein that migrates aberrantly at ≈80 kDa. There are sequence homologues in humans and C. elegans with homology constrained to the N and C termini. There are, however, no identifiable functional sequence motifs. Snu23 is a 23-kDa protein that migrates aberrantly at 30 kDa. Snu23 contains a putative zinc finger motif, and there exist in the database weakly homologous proteins from human and fly containing regions of homology restricted to the zinc finger motif, but nothing that clearly resembles a Snu23 homologue. Dib1 is a 17 kDa protein that previously has been shown from budding yeast and fission yeast (50) to be a likely cell cycle component (see below). Its presence in stoichiometric amounts, along with the fact that it copurifies through two chromatography steps and sediments at 25S in a glycerol gradient, is good evidence that it is a U4/U6⋅U5 snRNP component. The fourth novel protein associated with the U4/U6⋅U5 snRNP is a 13-kDa protein termed Snu13. There are homologues of this protein present in several organisms (Fig. 4), all with a high degree of homology. Snu13 is homologous to the Nhp2 protein, a basic protein with homology to ribosomal proteins (51), indicating that Snu13 may have RNA binding capacity. Indeed, Nhp2 is a component of the H BOX/ACA motif small nucleolar RNPs, suggesting that Snu13 and Nhp2 may have a similar function in their respective RNPs.
Figure 4.
Snu13 is highly conserved from yeast to man. Sequences with high degrees of identity to budding yeast Snu13(sc) were found from Schizosaccharomyces pombe (Snu13sp), C. elegans (Snu13ce), and human (Snu13hs). Regions of homology also were found with budding yeast Nhp2. Sequences were aligned with the pileup program (GCG) and were aligned in boxshade (K. Hofman and M. Baron, Chemin des Boveresses, Lausanne, Switzerland). Identical amino acids are shaded black, with similar amino acids shaded gray.
DISCUSSION
Purification of the Yeast U4/U6⋅U5 snRNP.
We have purified the yeast U4/U6⋅U5 snRNP to near homogeneity and have identified 24 proteins that copurify through two affinity chromatography steps and cosediment with the U4, U5, and U6 snRNAs in glycerol gradients. Isolation of U4/U6⋅U5 snRNP at 50 mM KCl or 250 mM KCl produced essentially identical results (data not shown) with only minor variations in the abundance of some proteins. The identification by mass spectrometry of the known proteins Prp8, Brr2, Snu114, Prp6, Prp3, Prp31, Prp4, Prp38, and the core Sm proteins indicates that this purified 25S complex is indeed the yeast U4/U6⋅U5 snRNP. At least some of the faintly staining protein bands present in these fractions are likely to be U1 and U2 snRNP proteins (Fig. 2A). Because of the absence of any mass spectrometric data indicating the proteins identified in this study are known U1 or U2 snRNP proteins, it is likely that the novel proteins we have identified are indeed associated with the U4/U6⋅U5 snRNP.
Purified human U4/U6⋅U5 snRNP also contains ≈20–25 proteins. The human Prp8 homologue (52), the Brr2 homologue (32), the Snu114 homologue (33), the Prp3 and Prp4 homologues (4, 53), and the Sm core proteins (54) all have been shown to be associated with the human U4/U6⋅U5 snRNP. It will be very interesting to see whether the remaining uncharacterized human U4/U6⋅U5 snRNP proteins correspond to any of the new proteins identified in this study.
U6 Specific Sm Proteins.
The Lsm proteins are Sm-like proteins associated with U6 snRNA. Although we have not identified a complete set of Lsm2-Lsm8, the two that were not identified by mass spectrometry have been shown by others to immunoprecipitate U4/U6⋅U5 snRNP (refs. 26 and 49; J. Beggs personal communication). Further studies should show whether these U6 Sm proteins are as stably bound to U6 snRNA as the canonical Sm proteins are to their snRNAs, as well as the sequence to which the U6 Sm core binds.
Four Novel Proteins Are Associated with the Yeast U4/U6⋅U5 snRNP.
We have identified four new proteins in the purified U4/U6⋅U5 snRNP. The inessential Snu66 does not have any known homologues or any sequence motifs that would allow us to speculate as to what its function may be. There are, however, two similarly sized, uncharacterized proteins in the isolated human U4/U6⋅U5 snRNP to which it may correspond (38). As a completely unknown protein, further biochemical and genetic studies will help to gain insight into its function.
The essential Snu23 protein has a putative zinc finger domain, a common nucleic acid binding motif. Although this does not help to elucidate its function in the context of pre-mRNA splicing, it demonstrates that it likely interacts with either a snRNA or the pre-mRNA.
Dib1 is a 17-kDa yeast protein previously shown to be essential in budding yeast (50). An incomplete human homologue exists in the database, but its exact size remains unknown. Because there exist two potential human U4/U6⋅U5 snRNP proteins of similar size that are as yet uncharacterized, it is possible that the human homologue is also associated with the U4/U6⋅U5 snRNP. The isolation of Dib1 in the yeast U4/U6⋅U5 snRNP poses far more questions that it answers. In fission yeast, its homologue dim1 was isolated as a second-site mutation that reduced the restrictive temperature of a cdc2 mutation. This implicated the fission yeast gene product in the cell cycle, and, indeed, the depletion of this protein leads to lethal arrest in G2. Many connections have been found in fission yeast between pre-mRNA processing and the cell cycle (55, 56). For instance, the fission yeast pre-mRNA processing prp8 gene (coding for a putative RNA helicase) is allelic with the cell cycle cdc28 gene (57). Overexpression of a portion of the fission yeast prp4 gene leads to a cell cycle defect (58). In budding yeast, the PRP20 gene first was characterized as a temperature-sensitive mutant with a defect in pre-mRNA splicing (59). Further analysis showed that this mutation has pleiotropic effects ranging from improper mRNA 3′ end formation and mRNA transport defects to mitotic and chromosome condensation defects in fission yeast and mammalian homologues. It functions biochemically in nuclear trafficking as the Ran guanosine release factor (60). Alleles of PRP8 and PRP22 in budding yeast also have been shown to have cell cycle phenotypes (61, 62) but are clearly involved in pre-mRNA splicing as well. It is difficult to separate these effects to determine which are primary and which are secondary. The demonstration that Dib1 is an integral U4/U6⋅U5 snRNP component is good evidence that, in this case, its primary function may be in pre-mRNA splicing, with the cell cycle phenotypes arising from secondary effects. A likely hypothesis is that some genes coding for cell cycle components contain introns; the inability to splice these pre-mRNAs during the critical juncture in their expression in the cell cycle would make the production of its gene product impossible, thereby arresting the cells at that point in the cell cycle. Further biochemical and genetic analysis will need to be done to demonstrate what the role of Dib1 is in pre-mRNA splicing.
The fourth novel protein identified in this study has been termed Snu13. Although its function has not yet been determined, it is essential, and homologues from fission yeast, worm, and man exist in the database (Fig. 4). Because of the extensive identity of protein sequence, it is likely that these are functional homologues, and, indeed, there exist similarly sized proteins in the human U4/U6⋅U5 snRNP. Because there is homology to the highly basic Nhp2 protein, which itself is a snRNP component that has homology to two ribosomal subunits, it is possible that this is an RNA binding protein. Further studies should shed light on what the function of this protein is and with which proteins or RNAs it interacts.
Proteins Absent from the U4/U6⋅U5 snRNP.
Two proteins previously shown to be associated with the U4/U6⋅U5 snRNP were not identified in this study. Prp18 is a 25-kDa protein known to be associated with the U5 snRNP and incorporated into the yeast U4/U6⋅U5 snRNP (63) and with the human U4/U6⋅U5 snRNP (64). Spp381 is a 34-kDa protein that was identified as a suppressor of prp38–1 and also has been shown to be associated with the U4/U6⋅U5 snRNP. Prp18 antiserum immunoprecipitated U4, U5, and U6 snRNAs at salt concentrations up to 200 mM, and hemagglutinin-tagged Spp381 immunoprecipitated U4, U5, and U6 snRNAs at salt concentrations up to 100 mM. By comparison, the U4/U6 and U4/U6⋅U5 snRNP component Prp4 is stably associated with U4/U6⋅U5 snRNP at salt conditions at which the triple snRNP dissociates and is stably associated with U4/U6 snRNP to even higher salt concentrations (29). Even though Prp18 and Spp381 are not present in preparations of U4/U6⋅U5 snRNP isolated at 50 mM KCl, it is likely that the inability to identify these two proteins is attributable to their relatively weaker interactions with the U4/U6⋅U5 snRNP. It is also possible that there exist other proteins that, because of their behavior in our purification scheme, were not present in our preparation. Further genetic and biochemical experimentation may elucidate other potential U4/U6⋅U5 snRNP proteins, as was shown for the U1 snRNP (16).
In the purification of the U1 snRNP, it was shown that a 42-kDa protein copurified with the free U5 snRNP (15). This protein also was not isolated in our studies, which indicated that either the 42-kDa protein is not stably associated with the U4/U6⋅U5 snRNP or, possibly, that it may leave the U5 snRNP on assembly of the U4/U6⋅U5 snRNP.
The human U4/U6⋅U5 snRNP also was shown to have associated with it a 20-kDa cyclophyllin protein that was shown to have proline isomerase activity (3, 53). Although there exist sequence homologues in the yeast proteome, it was not isolated in our purification. Immunoprecipitation studies using the yeast homologues may prove whether it is indeed U4/U6⋅U5 snRNP-associated.
We have shown that the preparative purification and characterization of snRNPs from yeast present in lower amounts than U1 snRNP is feasible. Selection of the proper protein as an affinity handle and the use of sensitive electrospray tandem mass spectrometry of the isolated polypeptides has proven to be a powerful technique for the characterization of large multicomponent complexes from a variety of organisms. The mass spectrometry data presented here and data presented previously show that there are at least 28 proteins associated with the yeast U4/U6⋅U5 snRNP. Further experimentation on the proteins of the yeast U4/U6⋅U5 snRNP should help to shed light on the mechanism of pre-mRNA splicing, as well as to help identify the interconnection between the cell cycle and mRNA processing.
Acknowledgments
We thank John Woolford, Amy Kistler, and Christine Guthrie for providing strains and hybridomas. We are grateful to Tony Moreno, Mary Young, and Terry Lee at the City of Hope Beckman Research Institute for their excellent mass spectrometry work (supported by National Institutes of Health Grant CA33572) and to Manny Ares, Tracy Johnson, Stephen Rader, and Hong Li for critical reviewing the manuscript. S.W.S. was supported by a postdoctoral fellowship PF4447 provided by the American Cancer Society. This work was supported by National Institutes of Health Grant GM32627 to J.A.
ABBREVIATIONS
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- NTA
nitrilotriacetic acid
References
- 1.Staley J P, Guthrie C. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 2.Laggerbauer B, Achsel T, Luhrmann R. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teigelkamp S, Achsel T, Mundt C, Gothel S F, Cronshagen U, Lane W S, Marahiel M, Luhrmann R. RNA. 1998;4:127–141. [PMC free article] [PubMed] [Google Scholar]
- 4.Lauber J, Plessel G, Prehn S, Will C L, Fabrizio P, Groning K, Lane W S, Luhrmann R. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- 5.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 6.Noble S M, Guthrie C. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maddock J R, Roy J, Woolford J L. Nucleic Acids Res. 1996;24:1037–1044. doi: 10.1093/nar/24.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank D, Patterson B, Guthrie C. Mol Cell Biol. 1992;12:5197– 5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D M, Field D J, Tang S J, Moris A, Bobechko B P, Friesen J D. Mol Cell Biol. 1998;18:2055–2066. doi: 10.1128/mcb.18.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lybarger S, Beickman K, Brown V, Dembla-Rajpal N, Morey K, Seipelt R, Rymond B C. Mol Cell Biol. 1999;19:577–584. doi: 10.1128/mcb.19.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy J, Kim K, Maddock J R, Anthony J G, Woolford J L. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson D J, Beggs J D. Mol Microbiol. 1991;5:805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 13.Chapon C, Legrain P. EMBO J. 1992;11:3279–3288. doi: 10.1002/j.1460-2075.1992.tb05406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FromontRacine M, Rain J C, Legrain P. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 15.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Seraphin B, Rosbash M, Luhrmann R, et al. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 17.Wise J A, Tollervey D, Maloney D, Swerdlow H, Dunn E J, Guthrie C. Cell. 1983;35:743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- 18.Busch H, Reddy R, Rothblum L, Choi Y C. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- 19.Cheng S C, Abelson J. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 20.Fabrizio P, Esser S, Kastner B, Luhrmann R. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 21.Raghunathan P L, Guthrie C. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 22.Staley J P, Guthrie C. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 23.Raghunathan P L, Guthrie C. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 24.Teigelkamp S, Newman A J, Beggs J D. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper M, Johnston L H, Beggs J D. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon K W, Guthrie C. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 28.Anthony J G, Weidenhammer E M, Woolford J L. RNA. 1997;3:1143–1152. [PMC free article] [PubMed] [Google Scholar]
- 29.Banroques J, Abelson J N. Mol Cell Biol. 1989;9:3710–3719. doi: 10.1128/mcb.9.9.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abovich N, Legrain P, Rosbash M. Mol Cell Biol. 1990;10:6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto A L, Steitz J A. Proc Natl Acad Sci USA. 1989;86:8742– 8746. doi: 10.1073/pnas.86.22.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Luhrmann R. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 33.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Luhrmann R. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horowitz D S, Abelson J. Mol Cell Biol. 1993;13:2959–2970. doi: 10.1128/mcb.13.5.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie J, Beickman K, Otte E, Rymond B C. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidenhammer E M, RuizNoriega M, Woolford J L. Mol Cell Biol. 1997;17:3580–3588. doi: 10.1128/mcb.17.7.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy J, Zheng B H, Rymond B C, Woolford J L. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrens S E, Luhrmann R. Genes Dev. 1991;5:1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- 39.Schneider K R, Smith R L, O’Shea E K. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 40.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 41.Geitz R D, Scheistel R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Labroatory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 43.Ansari A, Schwer B. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blum H, Hildburg B, Gross H J. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 45.Lehmeier T, Foulaki K, Luhrmann R. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis M T, Lee T D. J Am Soc Mass Spectrom. 1997;8:1059–1069. [Google Scholar]
- 47.Stahl D C, Swiderek K M, Davis M T, Lee T D. J Am Soc Mass Spectrom. 1996;7:532–540. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 48.Krebs E G. J Biol Chem. 1953;200:471–478. [PubMed] [Google Scholar]
- 49.Seraphin B. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry L D, Gould K L. J Cell Biol. 1997;137:1337–1354. doi: 10.1083/jcb.137.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolodrubetz D, Burgum A. Yeast. 1991;7:79–90. doi: 10.1002/yea.320070202. [DOI] [PubMed] [Google Scholar]
- 52.Anderson G J, Bach M, Luhrmann R, Beggs J D. Nature (London) 1989;342:819–821. doi: 10.1038/342819a0. [DOI] [PubMed] [Google Scholar]
- 53.Horowitz D S, Kobayashi R, Krainer A R. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- 54.Will C L, Behrens S E, Luhrmann R. Mol Biol Rep. 1993;18:121–126. doi: 10.1007/BF00986766. [DOI] [PubMed] [Google Scholar]
- 55.Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. Mol Biol Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Curr Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 57.Imamura O, Saiki K, Tani T, Ohshima Y, Sugawara M, Furuichi Y. Nucleic Acids Res. 1998;26:2063–2068. doi: 10.1093/nar/26.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gross T, Lutzelberger M, Wiegmann H, Klingenhoff A, Shenoy S, Kaufer N F. Nucleic Acids Res. 1997;25:1028–1035. doi: 10.1093/nar/25.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forrester W, Stutz F, Rosbash M, Wickens M. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 60.Bischoff F R, Ponstingl H. Nature (London) 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 61.Hwang L H, Murray A W. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shea J E, Toyn J H, Johnston L H. Nucleic Acids Res. 1994;22:5555–5564. doi: 10.1093/nar/22.25.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horowitz D S, Abelson J. Mol Cell Biol. 1993;13:2959–2970. doi: 10.1128/mcb.13.5.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horowitz D S, Krainer A R. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]