Abstract
Background
In vertebrates, the primordium of the brain is subdivided by the expression of Otx genes (forebrain/anterior midbrain), Hox genes (posterior hindbrain), and the genes Pax2, Pax5 and Pax8 (intervening region). The latter includes the midbrain/hindbrain boundary (MHB), which acts as a key organizer during brain patterning. Recent studies in Drosophila revealed that orthologous sets of genes are expressed in a similar tripartite pattern in the late embryonic brain, which suggested correspondence between the Drosophila deutocerebral/tritocerebral boundary region and the vertebrate MHB. To gain more insight into the evolution of brain regions, and particularly the MHB, I examined the expression of a comprehensive array of MHB-specific gene orthologs in the procephalic neuroectoderm and in individually identified neuroblasts during early embryonic stages 8–11, at which the segmental organization of the brain is most clearly displayed.
Results and conclusion
I show that the early embryonic brain exhibits an anterior Otx/otd domain and a posterior Hox1/lab domain, but that Pax2/5/8 orthologs are not expressed in the neuroectoderm and neuroblasts of the intervening territory. Furthermore, the expression domains of Otx/otd and Gbx/unpg exhibit a small common interface within the anterior deutocerebrum. In contrast to vertebrates, Fgf8-related genes are not expressed posterior to the otd/unpg interface. However, at the otd/unpg interface the early expression of other MHB-specific genes (including btd, wg, en), and of dorsoventral patterning genes, closely resembles the situation at the vertebrate MHB. Altogether, these results suggest the existence of an ancestral territory within the primordium of the deutocerebrum and adjacent protocerebrum, which might be the evolutionary equivalent of the region of the vertebrate MHB. However, lack of expression of Pax2/5/8 and Fgf8-related genes, and significant differences in the expression onset of other key regulators at the otd/unpg interface, imply that genetic interactions crucial for the vertebrate organizer activity are absent in the early embryonic brain of Drosophila.
Background
In vertebrates, the primordium of the brain is subdivided along the anteroposterior (AP) axis into three basic regions, reflected by the restricted expression of a highly conserved set of developmental genes before these brain regions become morphologically distinct. Otx genes are expressed in the anterior region, which comprises the forebrain and anterior midbrain, Hox genes in the posterior region comprising the hindbrain, and the genes Pax2,Pax5 and Pax8 in the intervening region. The intervening region includes the territory of the midbrain/hindbrain boundary (MHB), which encompasses the posterior part of the midbrain and rhombomere 1 of the hindbrain. The position of the MHB is controlled by the interface between the expression domains of Otx2 and Gbx2. The MHB exerts organizer properties that play an essential role in patterning the midbrain and hindbrain [1,2]. These organizer activities are mediated by fibroblast growth factor 8 (Fgf8) and Wnt1 proteins, which are secreted from the MHB neuroectoderm. The MHB (or isthmic) organizer arises in consecutive developmental steps that are mirrored by the ordered temporal sequence of MHB-specific gene expression. Its development initiates with the formation of an Otx2/Gbx2 interface, where in a second step btd/Sp-related 1 (Bts1), Pax2, Fgf8 and Wnt1 become expressed. In a third step, in addition to the already activated genes, their downstream targets are upregulated, among which are Pax5, Pax8, En1 and En2, whose pathways are mutually dependent with respect to maintaining the boundary [1-5].
The brains of deuterostomes (for example, tunicates and vertebrates) and protostomes (for example, arthropods and annelids) both seem to contain a rostral domain specified by the Otx/otd family, and a caudal domain specified by genes of the Hox family (for example, reviewed by [6-10]). Expression of Pax2/5/8 in the intervening neck region between the Otx and Hox1 domains has been observed in vertebrates and in the closely related ascidian tunicates, suggesting that this tripartite ground pattern of the brain is conserved during evolution within the chordate lineage [11]. Moreover, in the ascidian Ciona, the expression and activation of other crucial MHB determinants in the neck region, such as of Fgf8/17/18 and Engrailed (En) orthologs, are reminiscent of those in vertebrates [12,13], suggesting that the conserved pattern of their expression also pre-dates the splitting of the vertebrates from the chordate lineage. However, since the expression of Pax2/5/8 and En is absent in the intervening neck of appendicularian tunicates [14], and the neck region of another invertebrate chordate, amphioxus, lacks expression of Pax2/5/8, En and Wnt [15-17], this has raised doubt about the existence of a MHB territory in invertebrate chordates (irrespective of whether it includes organizer properties or not) [18,19]. On the other hand, in the late embryonic brain of Drosophila, a tripartite pattern of Otx, Pax2/5/8, and Hox1 expression has been reported, with Pax2/5/8 expression located at the interface between the domains of Otx/otd and Gbx/unpg, and coinciding with the neuromeric border between deutocerebrum and tritocerebrum. These findings have led to the hypothesis that the Drosophila deutocerebral/tritocerebral boundary region and the vertebrate MHB are corresponding structures, and that a basic tripartite regionalization of the brain was existent already in the common ancestor of the bilaterians [20,21].
To broaden the perspective on the evolution of brain regions, and in particular the MHB, I have undertaken a comprehensive analysis of orthologous factors of vertebrate MHB-specific regulatory genes in the Drosophila early embryonic brain. Since the specification of the MHB is one of the earliest decisions in the developing vertebrate brain, taking place before and during the formation of neuroblasts, I focussed on the early period of embryonic brain development. I describe the expression of MHB-specific marker genes at a resolution of identified neuroblasts (NBs) and in relation to the segmental architecture of the brain at stages when it is most clearly displayed. Based on the expression of orthodenticle (otd (oc, Flybase)) and labial (lab), the early brain principally exhibits a tripartite pattern with an anterior otd domain, a posterior Hox (that is, lab) domain, and a territory intervening between both domains. However, the Pax2/5/8 orthologs, D-pax2 (sv, Flybase) and pox neuro, are not expressed in the neuroectoderm and brain NBs of the intervening territory. Moreover, I identified a small interface between the complementary procephalic domains of otd and unplugged (unpg) that is located within the anterior deutocerebrum, corresponding to the anterior border of the intervening zone. The expression of these and further MHB-specific genes (such as the Wnt1 ortholog wingless, the En1,2 ortholog engrailed, and the zebrafish Bts1 ortholog buttonhead), and of dorsoventral (DV) patterning genes (the Msx ortholog muscle specific homeobox (msh (Dr, FlyBase)) and the Nkx2 ortholog ventral nervous system defective) in relation to the otd/unpg interface suggests that the neuroectoderm around this interface may represent an ancestral territory, evolutionarily equivalent to the neuroectodermal region at the MHB in vertebrates. However, in this part of the early embryonic brain, the expression of other MHB-specific markers (the Fgf8-related genes, branchless, pyramus, and thisbe) exhibits profound differences compared to the embryonic MHB domain in vertebrates. This suggests that, for the initial period of neurogenesis, the expression and regulatory interactions of genes, and the accompanying functional properties of the neuroectodermal territory around this interface, have changed during evolution.
Results
In vertebrates, the specification of the MHB is one of the earliest steps in brain development, taking place before and during the formation of NBs [4]. Therefore, in this comparative study in Drosophila I largely focussed on the early developmental period until embryonic stage 11, throughout which the pattern of NBs in the brain and ventral nerve cord (VNC) is fully established [22,23]. Furthermore, stage 11 represents the phylotypic stage of development [24] at which the segmental organization of the brain is most clearly displayed [25], and to which the expression patterns of MHB-specific gene orthologs can most accurately be related.
Expression of Otd and Labial regionalizes the anlagen of the embryonic brain and demarcates an intervening zone
In vertebrates and the closely related invertebrate chordates (that is, urochordates or tunicates), the neuroectoderm exhibits a fundamental tripartite organization already at early developmental stages: Otx is expressed in an anterior domain, Hox1 in a posterior domain, and Pax2/5/8 in an intervening territory [18,19]. In the late embryonic brain of Drosophila, orthologous sets of genes have been shown to be expressed in a tripartite pattern as well [20]. To see if a comparable pattern of gene expression exists in the early anlagen of the Drosophila brain (procephalic neuroectoderm (pNE) and NBs), I investigated the expression of the orthologous genes orthodenticle (otd) and labial (lab). The domain of Otd expression covers the anterior part of the antennal and most of the ocular pNE, and is found in most NBs of the protocerebrum (PC) and some anterior NBs of the deutocerebrum (DC; see also [26]). The posterior border of Otd expression is positioned within the anterior DC (Figure 1a, b). The domain of Lab expression covers the pNE of the intercalary segment and all NBs of the tritocerebrum (TC) as well as two NBs of the DC (Dv2,4; see also [26]). The anterior limit of the Lab domain is tightly linked to the segmental border between TC and DC (Figure 1b).
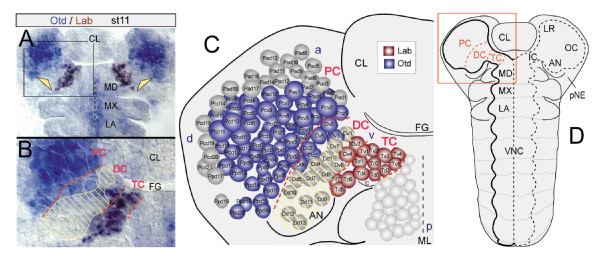
Figure 1.
The brain anlagen in Drosophila is tripartite. (a) Flat preparation of the head of an Otd/Labial antibody double stained embryo at stage 11. (b) Close-up of the region framed in (a) at the level of brain NBs. (c) Schematic summary of the expression of both genes in identified brain NBs in the left hemisphere at late stage 11, corresponding to the boxed area in (d); red dashed lines indicate neuromeric boundaries. An intervening zone can be identified between the domains of Otd and Lab expression, in which both genes are expressed neither in the peripheral ectoderm (yellow arrowheads in (a)) nor in the deriving NBs (hatched area in (b,c)). Note that in the deutocerebrum (DC) some anterior NBs (Dv5,6, Dd2,3,6) express Otd, and some posterior NBs (Dv2,4) express Labial (NB nomenclature according to [23]). (d) Segmental topography of the Drosophila embryo at the phylotypic stage of development (stage 11); flat preparation (anterior to the top), the head capsule has been opened dorsally. The pregnathal (labral (LR), ocular (OC), antennal (AN), intercalary (IC)) and gnathal head segments are indicated on the right side. On the left side, the primordium of the CNS is outlined (PC, protocerebrum; DC, deutocerebrum; TC, tritocerebrum; MD, mandibular, MX, maxillary, and LA, labial neuromere, respectively). Abbreviations: a, d, p, v, anterior, dorsal, posterior, ventral; AN, antennal appendage; CL, clypeolabrum; FG, foregut; ML, midline; VNC, ventral nerve cord.
Thus, the early embryonic Drosophila brain discloses an anterior Otd domain, a posterior Hox domain (that is, of Lab expression), and an 'intervening zone' (IZ) encompassing a fraction of deutocerebral NBs in which neither gene is expressed (Figure 1c).
Expression of Pax2/5/8 orthologous genes is missing in the pNE and NBs of the intervening zone
In the early embryonic brain of vertebrates, expression of genes of the Pax2/5/8 subfamily is indicative for the region of the presumptive mid-hindbrain domain, which is positioned in the intervening region between the Otx/otd and Hoxb1/lab domains [4]. Therefore, I investigated the expression of two orthologous genes in Drosophila, D-pax2 and the closely related pox-neuro (poxn) [27-29]. Expression of D-pax2 initiates at stages 10/11 in a few cells in the truncal peripheral nervous system (most likely including the sensory organ progenitors (SOPs) of the internal and external sensory organs), and in SOPs of the antennal (Dd9,11,12), labral, and ventral hypopharyngeal-/latero-hypopharyngeal (Dv1,3) sensory organs (Figure 2a,b) [23] (see also [30,31]). At later embryonic stages the D-pax2-positive progeny of the two SOPs of the hypopharyngeal-/latero-hypopharyngeal organ are positioned lateroventrally to the foregut and, hence, are clearly separated from the brain. Both SOPs lay ventrally adjacent to the IZ of brain NBs (Figure 2b,c). In the brain, however, I have not found D-pax2 before stage 14/15 (in cells of the DC and PC) (Figure 2c), when it is likewise metamerically expressed in the ventral nerve cord [31]. These D-pax2-expressing cells in the brain and ventral nerve cord originate from pNE and NB(s), which do not express D-pax2. I therefore asked if, instead of D-pax2, the second Pax2/5/8 ortholog, poxn, is expressed in NBs of the IZ. In the trunk at stage 11, Poxn is segmentally expressed first in SOPs of the external sensory organs and slightly later in NB 2–4 (Figure 2d; see also [28]). Likewise, in the head ectoderm, Poxn is first expressed in a few cells of the developing labral and antennal appendages (which presumably contribute to the respective sensory organs) at about the same time it is found in SOPs of the truncal segments (Figure 2d,e). However, in the brain, Poxn expression was first found later (by stage 12/4) in a single cell of the DC (Figure 2f), and by stage 13 in 1–2 protocerebral cells that coexpress En and descend from NBs of the en head spot (hs; Figure 2g). The number of Poxn-positive cells in the DC and PC increases during embryogenesis, but importantly, all these cells develop from Poxn-negative pNE and NBs (Figure 2h).
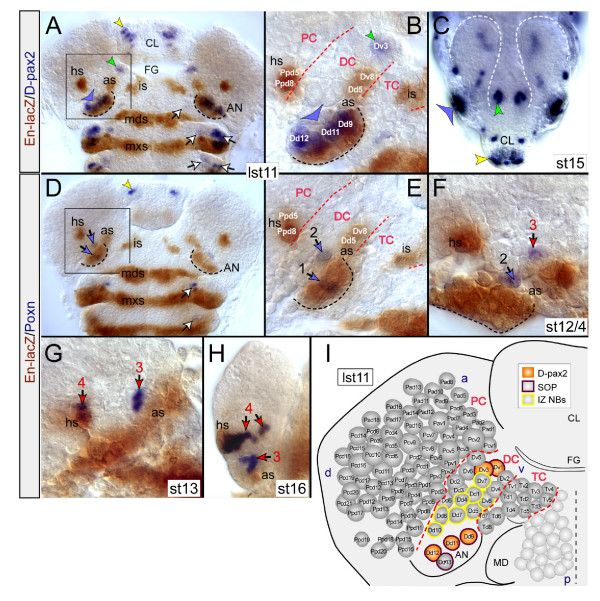
Figure 2.
Expression of Poxn and D-pax2 is lacking in NBs of the IZ. (a,b) D-pax2/En-lacZ double stainings; flat preparations of late stage 11 (lst11) embryos. (b) Magnification of boxed area in (a) at the level of NBs. En-lacZ-positive NBs deriving from the antennal en stripe (as; Dv8, Dd5) and en head spot (hs; Ppd5,8) are indicated. D-pax2 is detected in SOPs of the dorsal organ (blue arrowhead), the hypopharyngeal/latero-hypopharyngeal organ (green arrowhead), and of the labral sensory organ (yellow arrowhead in (a)) [23]. Note that SOPs at positions corresponding to those of the dorsal organ within the developing antennal appendage (AN; outlined by black dashed line) are also found in the appendage of the mandibular and maxillary segment (white arrows in (a)). (c) D-pax2 stained whole mount embryo at stage 15 (st15) with a focus on the brain (outlined by the white dashed lines). D-pax2 is found in the respective sensory organs. (d-h) Poxn/En-lacZ double stained embryos. (d) Late stage 11 (lst11). Few Poxn-positive cells contribute to the labral (yellow arrowhead) and antennal (blue arrows) sensory organs. Poxn sensory founder cells arise at corresponding positions in the antennal, mandibular, maxillary, and labial segments (white arrows), immediately anterior to the respective en stripes. Note that in the CNS, Poxn expression is not yet initiated. (e) Close-up of boxed area in (d). Poxn expression initiates in peripheral ectodermal cells (group 1, 2) positioned in the antennal appendage, which are likely to contribute to the antennal dorsal organ. (f) By stage 12/4, Poxn expression comes up in cells of the DC (group 3) immediately anterior to the antennal en stripe (as). (g) By stage 13, first en-coexpressing cells (group 4), descending from the two head spot (hs) NBs, initiate Poxn expression. Note that the head spot NBs are Poxn-negative. (h) Stage 16. The number of Poxn-positive cells in the deuto- and protocerebral cell cluster is increased. (i) Summary of D-pax2 expression at late stage 11 in SOPs (orange) of the hypopharyngeal organ (Dv1,3) and the dorsal organ (Dd9,11,12). Other abbreviations are as in Figure 1.
Taken together, Poxn and D-pax2 are not expressed in brain NBs but in certain progeny cells at later embryonic stages. Despite this lack of early Poxn and D-pax2 in NBs of the IZ, I suggest the brain anlagen to be tripartite, in the sense of consisting of three spatially distinct regions: an anterior Otd domain, a posterior Lab domain and an IZ, where, at the level of the pNE and brain NBs, neither gene is expressed. In this regard it is worth noting that the D-pax2 expressing SOPs of the hypopharyngeal organ (Dv1,3) are localized immediately ventral, and the D-pax2 expressing SOPs of the dorsal organ (Dd9,11,12,13) immediately dorsal to the NBs of the IZ (Figure 2a,b,i). However, these SOPs do not contribute to the brain. Considering these findings, I propose the IZ to encompass about eight NBs of the DC (Dd1,4,5,7,8,10, Dv7,8; Figure 2i).
Common interface of otd and unpg domains corresponds to the anterior border of the intervening zone
Otx2 and Gbx2 are expressed in the region of the presumptive vertebrate midbrain/hindbrain domain. Their mutual repressive interaction results in a clear interface between the posterior Gbx2 and the adjacent anterior Otx2 domain, which has an important role in positioning the isthmic organizer [4]. In the late embryonic Drosophila brain (stage 14/15), the expression domains of the Otx2 ortholog, otd, and the Gbx2 ortholog, unplugged (unpg) have been reported to form a common interface at the segmental boundary between TC and DC [20]. Since in vertebrates both genes are the first factors expressed in the presumptive midbrain/hindbrain domain (from gastrulation onwards), I was interested to see if such an otd/unpg interface is established in the early anlagen of the embryonic brain, at the level of NBs. By stage 11, unpg-lacZ (as revealed in the enhancer trap line 1912) is expressed in the antennal and adjacent ocular pNE, as well as in most NBs in the DC and adjacent PC, as previously shown [26]. Consequently, unpg-lacZ and Otd are not complementarily expressed, as they exhibit an overlap within the posterior PC and anterior DC. This prompted me to investigate if the lacZ pattern reliably represents the expression of the unpg gene. In situ hybridizations showed that, in the pNE, the pattern of unpg transcripts does not fully match the pattern of unpg-lacZ (data not shown). Detectable levels of unpg mRNA do not become visible before stage 11 and then only in the part of the antennal ectoderm (and deutocerebral NBs) where unpg-lacZ is expressed strongest (Figure 3c). Accordingly, I found unpg mRNA only in the DC, and in a significantly smaller subset of NBs (Dd1, Dv7), immediately anterior to those deriving from the en antennal stripe (Figure 3d). Interestingly, Dd1 and Dv7 exactly abut the posterior limit of the domain of Otd expressing NBs (Figure 3a–c).
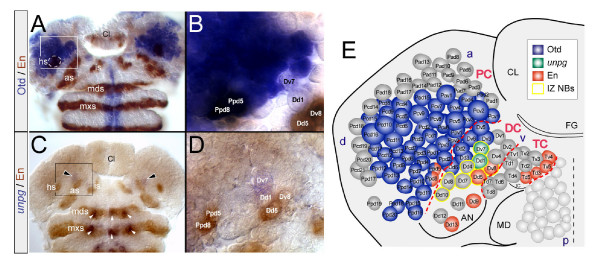
Figure 3.
Otd and unpg form a common interface within the IZ. (a,b) Otd/En-lacZ double labelling. (c,d) unpg mRNA/En-lacZ double labelling in the peripheral head ectoderm (a,c) and deriving brain NBs at stage 11 (b,d). (b,d) Close-ups of regions framed in (a,c), respectively. (b) Note that Dd1 and Dv7 do not express Otd. (d) In the brain, unpg mRNA is detected in only two deutocerebral NBs (Dd1, Dv7) immediately anterior to the antennal en stripe (as) and bordering the Otd domain (b). (e) Summary of the expression of Otd, unpg, and En according to the colour code. Note the complementary expression of Otd and unpg, and that Dd1, Dv7 are part of the NBs of the IZ (IZ NBs). Red dashed lines indicate segmental borders. Other abbreviations are as in Figure 1.
Taken together, procephalic Otd and unpg (mRNA) are complementarily expressed, exhibiting a small common interface at the anterior border of the IZ, which is positioned within the anterior half of the deutocerebral anlagen. These data suggest that the IZ and the anterior adjacent pNE, which are separated by the otd/unpg interface, represent an ancestral ectodermal territory, evolutionarily equivalent to the early embryonic vertebrate midbrain/hindbrain domain (including the Otx2/Gbx2 border).
Expression of other vertebrate MHB-specific orthologs in the region around the Drosophila otd/unpg interface
Early in embryonic development, the region of the vertebrate MHB is characterised by the expression of several other genes, among which are En and Bts1, as well as the secreted factors Wnt1 and Fgf8. These factors have been shown to be involved in the patterning and differentiation of the evolving structures of the midbrain and anterior hindbrain [1,3,4,32]. I was interested to explore how far the expression of orthologous genes is conserved in the pNE and NBs around the otd/unpg interface in Drosophila.
In vertebrates, Fgf8 is expressed in a narrow domain immediately posterior to the border between the developing mid- and hindbrain [33]. Three Fgf8-related genes have been described in Drosophila: branchless (bnl) [34], and the more closely related pyramus (pyr) and thisbe (ths) [35]. Using in situ hybridizations I found that, during embryogenesis, the pattern of bnl [34], pyr and ths transcripts [35] is the same as described previously. By stage 11, bnl transcripts were found at dorsal-most sites of the ocular pNE and in a few corresponding protocerebral NBs (Figure 4a). In the examined period until stage 11, detectable levels of bnl mRNA were not observed in the remainder of the brain anlagen. pyr transcripts are prominently expressed during stages 10/11 in one protocerebral NB, but are not found in the region of the otd/unpg interface (Figure 4b). ths mRNA is found in some protocerebral NBs during stages 8/9, but becomes largely downregulated by stage 10, when it is still found in low concentrations at the level of the en head spot. By stage 11, ths mRNA is restricted to a single anterior protocerebral NB (Figure 4c,d), at a position comparable to that of the pyr expressing NB (Figure 4b). Importantly, all procephalic ths expression domains lie anterior to the otd/unpg interface and partly within the Otd domain, which is dissimilar to the situation in vertebrates. At later embryonic stages, none of the Fgf8-related genes exhibit detectable levels of transcripts in the vicinity of the otd/unpg interface (data not shown). Hence, the expression of all three Fgf8-related genes in Drosophila is in striking contrast to the expression of Fgf8 at the embryonic vertebrate Otx2/Gbx2 border. In vertebrates, Wnt1 is expressed in a narrow domain just anterior to the Otx2/Gbx2 border and overlaps with expression of Otx2 [1,32]. Comparably, Drosophila Wingless (Wg) is coexpressed with otd in a protocerebral domain anterior to the otd/unpg interface; additionally, Wg is found in a deutocerebral domain located immediately posterior to the otd/unpg interface (Figure 4e,f,i) [26]. En is found in two protocerebral NBs (Ppd5,8) deriving from the en head spot immediately anterior to the otd/unpg border, and in two deutocerebral NBs (Dd5, Dv8) deriving from the antennal en stripe in the posterior vicinity of the otd/unpg border (Figure 4i,m) [25]. This is similar to the expression of En1,2, the domains of which in vertebrates span large parts of the neuroectoderm adjacent to the Otx2/Gbx2 border. In the zebrafish, Bts1, a member of the Sp gene family, is one of the earliest genes expressed at the presumptive midbrain/hindbrain domain [3]. In Drosophila, I detected transcripts of the closely related cephalic gap gene buttonhead (btd) in the same pattern as described previously [36,37]. By stage 11, btd mRNA was found in the dorsal antennal pNE and in one to two corresponding deutocerebral NBs located at the same AP level as the otd/unpg interface (Figure 4g,h,i). This again corresponds to the situation at the vertebrate MHB.
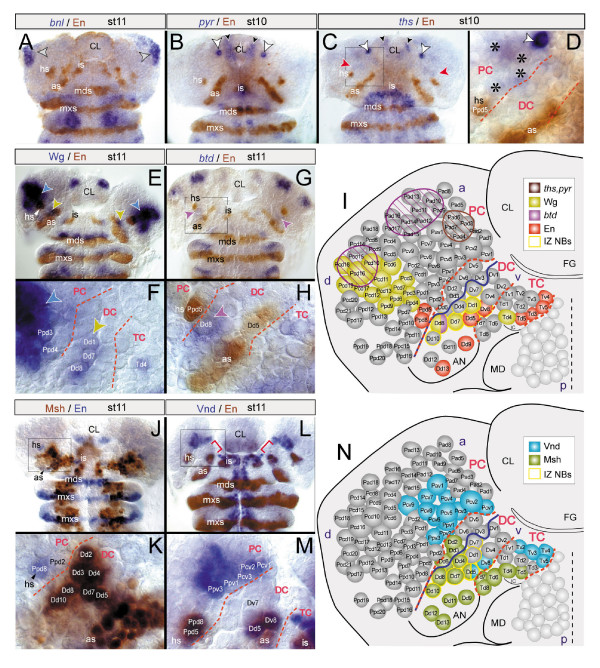
Figure 4.
Expression of other vertebrate MHB-specific genes in the early Drosophila brain. Head flat preparations. (a-d) mRNA expression of FGF8-related genes branchless (bnl; (a); stage 11), pyramus (pyr; (b); stage 10), or thisbe (ths; (c), stage 10) in relation to Engrailed (En)-lacZ. (a) In the pNE, bnl transcripts are confined to most dorsal sites of the ocular pNE (arrowheads). (b) pyr expression is detected in the labral ectoderm (black arrowheads) and in one protocerebral NB in each hemibrain (white arrowheads). (c,d) Expression of ths, similar to pyr, is found in the labral ectoderm (black arrowheads) and in one protocerebral NB (white arrowheads). ths is faintly detected in a further pNE domain (red arrowheads), slightly anterior to the en head spot (hs) and the Ppd5 (in (d)). (d) Higher magnification of framed area in (c). Protocerebral NBs (asterisks) emanating from that area express ths very faintly. (e,g,h) Expression of Wingless protein (Wg) (e) or buttonhead mRNA (btd) (g,h) in combination with En-lacZ, or (f) only Wg. (e,f) Wg is expressed in the ocular (blue arrowheads) and deutocerebral pNE (brown arrowheads) and (f) in the corresponding protocerebral and deutocerebral NBs (Dd1,7,8), respectively. (g,h) btd is weakly expressed in a deutocerebral pNE domain ((g), purple arrowheads) and in the deutocerebral Dd8 (h) of the IZ. (i) Scheme summarizing the expression of Wg, btd, ths,pyr, and En in brain NBs at stage 11. Red dashed lines indicate neuromeric boundaries, the blue line the posterior border of the otd domain, which corresponds (at least partially) to the otd/unpg interface. Coloured hatched areas indicate neuroectodermal regions in which yet unidentified NBs express btd (purple), or ths or pyr (brown). (j-n) Expression of Muscle segment homeobox (Msh-lacZ) (j,k) or Ventral nervous system defective (Vnd) (l,m) and En (Inv in (j,k); En-lacZ in (l,m)). (j,k) The anteriomost limit of Msh expression abuts the anterior border of the deutocerebrum (DC; red line), which roughly coincides with the anterior border of the IZ. (k) Msh is expressed in dorsal NBs of the IZ (Dd4,5,7,8,10).(l,m) Vnd expression is specifically lacking in the ventral pNE (indicated by red brackets in (l)) and in all ventral NBs of the deutocerebrum (m), including Dv7 (of the IZ) at the otd/unpg interface (blue line in (n)). (n) Scheme summarizing the expression of Msh and Vnd in brain NBs at stage 11.
Taken together, btd, en, and wg are expressed in the immediate vicinity of the otd/unpg interface, corresponding to the expression of orthologous genes at the vertebrate Otx2/Gbx2 border. However, the Fgf8-related genes pyr and bnl are not expressed in the area of the otd/unpg interface, and ths is activated only transiently at low levels and at an improper position in relation to the otd/unpg interface and other MHB-specific marker genes, indicating crucial differences to the situation in the vertebrate embryo.
Discontinuous expression of DV patterning genes msh and vnd at the otd/unpg interface
In the vertebrate neural tube, the order of expression of DV patterning genes of the Nkx and Msx gene families along the DV axis is analogous to that of the orthologs ventral nervous system defective (vnd) and muscle specific homeobox in the Drosophila neuroectoderm: Nkx/vnd are expressed in ventral regions, and Msx/msh in dorsal regions [38,39]. Along the AP axis, Nkx2.2 and Msx3 (which presumably represents the ancestral Msx/msh gene) have been reported to be discontinuously expressed at the MHB. Nkx2.2 exhibits a gap of expression specifically at the MHB [40]. Moreover, the anteriomost sharp limit of Msx3 abuts exactly the MHB [41,42]. In Drosophila, the AP expression patterns of Vnd and Msh exhibit striking similarities. Until stage 11, I observed a lack of Vnd expression specifically at the AP level of the otd/unpg interface (Figure 4l,m,n). In addition, Msh, which is expressed in the dorsal NBs of the TC and DC [25], exhibits an anterior limit that largely coincides with the AP level of the otd/unpg interface (Figure 4j,k,n). Thus, the discontinuous expression of Vnd and Msh at the otd/unpg interface is similar to Nkx2.2 and Msx3 at the Otx2/Gbx2 border. This lends further support to the proposed ancient evolutionary origin of the pNE anteriorly and posteriorly adjacent to the otd/unpg interface.
Discussion
This comprehensive expression analysis of factors orthologous to key regulatory genes of the embryonic vertebrate MHB was aimed at clarifying whether the early embryonic brain anlagen in Drosophila reveal a tripartite regionalization, contain a conserved Otx/Gbx border and include an ectodermal territory that shares similarities with the anlagen of the vertebrate MHB. I have focussed my study mainly on the early phase of embryonic brain development, because positioning and establishment of the MHB region is a very early decision in vertebrate central nervous system (CNS) development (taking place in the neuroectoderm before and during the formation of NBs). In addition, in Drosophila, the segmental organization of the brain is most clearly displayed in this phase and the examination can be done at the highest resolution, at the level of individually identifiable NBs.
Is a tripartite regionalization of the anterior CNS, based on Otx, Pax2/5/8, and Hox1 orthologous domains, conserved in bilaterians?
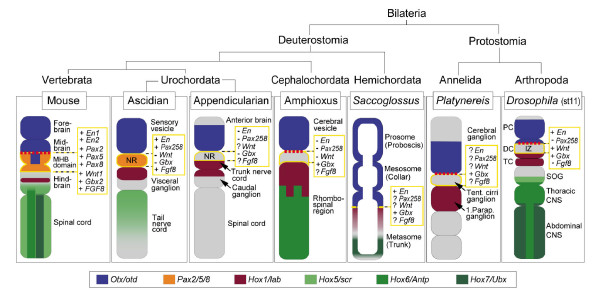
Deuterostomes comprise the hemichordate/echinoderm clade and the chordates, which include vertebrates and the closely related invertebrates, amphioxus (cephalochordates) and tunicates (urochordates) (Figure 5). On the basis of the expression of highly conserved regulatory genes, the anterior part of the early vertebrate CNS is considered to exhibit a fundamentally tripartite regionalization along the AP axis: Otx is expressed in the forebrain/midbrain, Hox genes in the hindbrain/spinal cord, and Pax2/5/8 genes in the intervening domain that constitutes the MHB region [1] (Figure 5). This tripartite organization of gene expression is also found in ascidian tunicates, that is, in Halocynthia and Ciona. Tunicates, together with amphioxus, represent the closest relatives of vertebrates [43]. Expression of a Pax2/5/8 ortholog has been reported for the ascidian neck region between the Otx and Hox1 domains [11,44] (Figure 5), leading to the speculation that the neck region is orthologous to the vertebrate MHB [11,19]. This is supported by the findings that Pax2/5/8 is also expressed in the neck region of Ciona savingyi [45], and Pax2/5/8, Fgf8/17/18, and En in Ciona intestinalis [13]. However, accumulating data show that the expression of MHB-specific markers is already diverse among the tunicates. In contrast to the ascidians, for example, the expression of Pax2/5/8 in the appendicularian tunicates (that is, in Oikopleura) is lacking in the neck [14]. Furthermore, whereas in C. savignyi En is found in the Pax2/5/8 expressing neck region [45], in C. intestinalis En is expressed in two domains, anteriorly and posteriorly adjacent to the Pax2/5/8 expressing neck [13]. Wnt orthologous genes seem to be generally lost in the ascidian genome [46,47] and, therefore, are not expressed in the neck region. Since it has been difficult to deduce the ancestral pattern, the existence of a MHB homologous region in tunicates is controversial [14,48]. In the intervening region of the protochordate amphioxus, expression of Pax2/5/8 as well as of Wnt1 and En is lacking, similar to the situation in the appendicularian tunicate Oikopleura [15-17] (Figure 5). However, a tripartite organization seems basically conserved, considering the existence of an anterior Otx domain, a posterior Hox1 domain, and an intervening region in which neither of these genes is expressed [16].
Figure 5.
Comparison of CNS regions and gene expression domains among chordates, hemichordates, annelids, and arthropods. Focus is on the anteroposterior regionalization of the anterior CNS, including the brain. The schemes reflect the situation in the CNS of the vertebrate mouse (at about embryonic day 10–12.5), the ascidians Ciona and Halocynthia [11, 13, 43, 44, 54], the appendicularian Oikopleura (at the late hatchling stage) [14], the amphioxus Branchiostoma (at the 10–13 somite stages) [15, 16, 45, 76], the hemichordate Saccoglossus (at the one gill slit stage) [48, 49], the polychaete annelid Platynereis (neuroectoderm at the metatrochophora stage) [46, 47], and the arthropod Drosophila (at stage 11, st11; this study). Expression of Otx/otd, Pax2,5,8, Hox1/lab, Hox5/scr, Hox6/Antp, and Hox7/Ubx genes is indicated according to the colour code. The dashed line in red indicates the interface between Otx/otd and Gbx/unpg expression domains. The expression of further genes within the gap (encircled in yellow) between the anterior Otx/otd and posterior Hox1/lab domains is noted: '+' indicates expression of the respective gene; '-' absence of expression; and '?' expression is not yet determined. The phylogenetic tree is based on [76]. For further details, see the text. IZ, intervening zone; MHB domain, midbrain/hindbrain boundary domain; NR, neck region; Parap., Parapodial; Tent., Tentacular.
The situation in amphioxus and Oikopleura is comparable to the findings made in Drosophila in this study (Figure 5). The early embryonic brain can be subdivided into an anterior domain of Otx/otd expression, encompassing most of the PC and an adjacent part of the DC, and a posterior domain of Hox1/lab expression, encompassing the TC. Both domains are separated by an IZ, covering part of the deutocerebral pNE and eight deutocerebral NBs, in which neither gene is expressed. The two Pax2 genes (D-pax2 and poxn) are not expressed in the early embryonic IZ, which is a significant difference to the situation in vertebrates and ascidian tunicates (Halocynthia and Ciona). However, both Pax2 genes are expressed during later embryonic brain development [20], when they are also expressed in segmentally repetitive neuronal clusters in the VNC. Importantly, at that time, AP patterning (that is, segmentation) and early neurogenesis (that is, formation/specification of NBs) are already completed. Thus, the later phase of expression of both genes is presumably involved in specification/differentiation of neural progeny cells, but is not compatible with an early function in patterning or specification of the pNE or NBs at the IZ. Accordingly, no obvious brain phenotype has been observed in poxn or D-pax2 mutants [20]. On the other hand, early expression of poxn and D-pax2 is found in progenitors of the peripheral nervous system, some of which are placed in immediate vicinity of NBs of the IZ, suggesting an early function of both genes in the development of head sensory structures. This is in accordance with findings made in the trunk, where poxn and D-pax2 are first expressed in the precursors of the developing peripheral nervous system [28,31] (Figure 2a,d), and later on in the ventral nerve cord.
Similarly, in the neuroectoderm of another protostomia, the polychaete annelid Platynereis dumerilii, an anterior Otx domain [49] seems to be spatially separated from a posterior Hox1 domain (Figure 5) [50]; although expression of a Pax2/5/8 ortholog has been reported in the trunk nerve cord [51], it has not yet been described for the brain. Hemichordates, distant deuterostomes, which do not have an internalized CNS but a body-encircling basiepithelial nerve net, reveal an anterior Otx and a posterior Hox1 expression domain, comparable to the situation in chordates, Platynereis, and Drosophila (Figure 5). A Poxn ortholog has been identified in the hemichordate Saccoglossus kowalevskii, but it is distinct from the Pax2/5/8 group of genes; expression of Pax2/5/8 orthologs has not yet been characterized [52]. Lastly, the MHB-specific marker En is expressed in the intervening region between the Otx and Hox1 domains [53].
Taken together, the data suggest that a tripartite ground plan characterizing the development of the chordate (and perhaps polycheate and hemichordate) brain is basically also present in the insect brain, which is in agreement with Hirth et al. [20]. However, in the early embryonic Drosophila brain expression of Pax2/5/8 orthologs is absent in the IZ. Nevertheless, the presence of a brain region that expresses neither otd nor labial indicates that a domain regionally homologous to the vertebrate MHB domain may also exist in Drosophila (see also the following discussion). Thus, it is tempting to speculate that a tripartite ground pattern (which lacks early Pax2/5/8 expression) was acquired already in the bilaterian ancestor.
Is the Otx/Gbx border an ancestral condition in bilaterians?
In vertebrates,Otx2 and Gbx2 are among the earliest genes expressed in the nervous system [33,54]. The establishment of a border between the complementary neuroectodermal domains of Otx2 (anterior) and Gbx2 (posterior) is the initial step in MHB development. The border, which forms due to mutual repression of these genes, is crucial for the proper positioning of the MHB [1,2]. In the early amphioxus embryo, Gbx and Otx domains abut the cerebral vesicle and hindbrain; therefore, this border is likely to be homologous to the vertebrate MHB (although it seems unlikely that this boundary has organizer properties) [48]. Yet there are no Gbx genes in urochordates, suggesting that they have been lost secondarily in this lineage [14]. In hemichordates by contrast, the domains of Otx and Gbx overlap considerably, indicating that these genes do not antagonize each other [53]. This is reminiscent of the initial phase of Otx and Gbx expression in amphioxus, when the domains of both genes slightly overlap, although they sharply abut later on [48].
Similar to the situation in vertebrates and amphioxus, in Drosophila a common border can be recognized between the procephalic expression domains of otd and unpg (the Gbx2 ortholog). However, expression of unpg initiates significantly later (by stage 11) than that of otd (by stage 6), different to the situation in vertebrates (Additional file 1). As shown at the level of NBs, the common border between both expression domains is positioned in the anterior DC. Thus, the otd/unpg interface is located more anterior and does not coincide with the boundary between the TC and DC as supposed previously [20]. Nevertheless, in the late embryonic brain, there is evidence that otd and unpg negatively regulate each other, similar to the genetic interaction of their vertebrate orthologs [20]. In this regard it is important to note that in the anterior neuroectoderm of the polychaete Platynereis, a boundary between the anterior domain of Otx expression and a posterior domain of Gbx expression has also been reported recently [49], supporting its ancient origin.
In summary, the border between the Otx/otd and Gbx/unpg domains found in chordates, polychaetes, and Drosophila (but not in hemichordates) suggests a homologous use of both genes in AP patterning. The Otx2/Gbx2 border in vertebrates determines the position of the MHB. Considering that the otd/unpg border can be identified in the early embryonic brain, and that the genetic interactions of otd and unpg appear to be conserved [20], it seems likely that at least the machinery for positioning the MHB might already be existent in Drosophila. Therefore, the border between the Otx/otd and Gbx/unpg domains may represent an ancestral condition in bilaterians.
Does the pNE surrounding the otd/unpg interface share homology with the vertebrate MHB organizer?
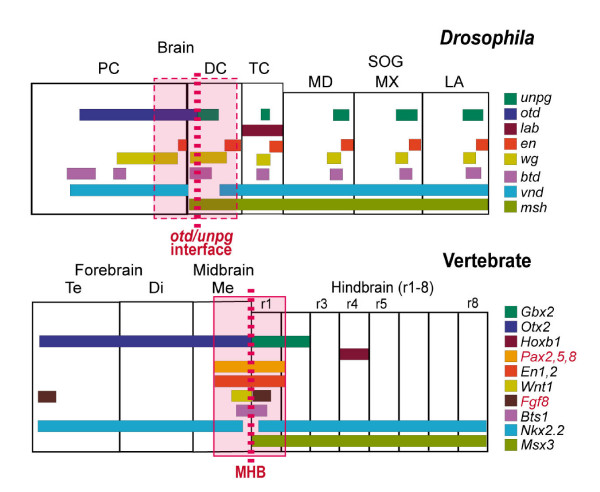
After the Otx2/Gbx2 border in vertebrates has been formed at the prospective MHB, several other key genetic factors, such as Pax2, En, Wnt1, and Fgf8 are expressed in an ordered spatial and temporal mode [1,2,4,5]. A comparison of the spatial and temporal expression of those gene orthologs at the vertebrate MHB and the Drosophila otd/unpg interface is given in Figure 6 and Additional file 1, respectively. As discussed above, the formation of the otd/unpg interface within the IZ already indicates that the corresponding neuroectodermal domain in Drosophila might share basic similarities with the vertebrate MHB. Moreover, comparable to the situation in vertebrates, en is expressed at the otd/unpg border, in a deutocerebral and in a protocerebral domain. However, in contrast to vertebrates [55,56], early expression of en cannot be activated by Pax2 genes in Drosophila since the latter are not expressed in the early brain. Nevertheless, from embryonic stage 13 onwards, some of the protocerebral en cells coexpress poxn, suggesting possible genetic interactions at those later stages (Figure 2g,h); interestingly, coexpression of both genes is not observed elsewhere in the CNS. In Ciona, two En domains have been recognized to be positioned similarly to those in Drosophila, in the immediate vicinity of the neck region [13]. Since neither of the En domains coexpresses Pax2/5/8, both in Drosophila and Ciona, Pax2/5/8 is unlikely to activate En expression.
Figure 6.
Comparison of brain organization and expression of key developmental genes involved in CNS regionalization and MHB formation in vertebrates and Drosophila. Data for Drosophila reflect the situation at stage 11, data for vertebrates reflect the situation in mouse at about embryonic day 10 (E10; see [1]; Nkx2.2, [40]; Msx3, [41]; the data for Bts1 are from a comparable developmental stage in zebrafish [3]). The boxed area coloured in light red indicates the neuroectodermal region around the MHB in vertebrates, according to the expression domains of Pax2,5 and 8 and En1 and 2. Although early Pax2/5/8 gene expression is lacking in Drosophila, the corresponding box may indicate a similar domain around the otd/unpg border. Identical colours indicate orthologous genes. Vertebrate gene names in red indicate those genes that are not or differently expressed at the Drosophila otd/unpg interface. For details, see the text. PC, DC, TC; proto-, deuto-, tritocerebrum; MD, MX, LA, mandibular, maxillary, labial neuromere, respectively; SOG, subesophageal ganglion. Di, Me, Te, Dien-, Mesen-, Telencephalon, respectively; r1–8, rhombomeres 1–8.
As shown above, the expression data for other conserved key factors, such as wg,btd, and the DV patterning genes msh and vnd, suggest that the pNE surrounding the otd/unpg interface may represent an ancestral territory that shares molecular similarities with the region around the vertebrate MHB. Important differences are that, in Drosophila, expression of Fgf8-related genes, bnl and pyr, and early expression of Pax2/5/8 is not found at the otd/unpg interface. In agreement with that, no obvious brain phenotype has been observed in bnl mutant embryos [20]. Only ths expression is detected, but it is very faint, transient, and anterior to the otd/unpg interface. Since ths seems to be colocalized with wg expression in this region, it is unlikely to serve the same function as Fgf8 at the vertebrate MHB. These differences in the expression of Fgf8-like genes in Drosophila are most significant, as Fgf8 is the key player at the vertebrate MHB, eliciting the expression of other organizer genes and exerting most of the organizer functions [2].
In addition, the temporal gene expression profile of this brain territory and the vertebrate MHB domain reveals distinctions (Additional file 1); for example, unpg is expressed significantly later than otd. Altogether, the spatial and temporal dissimilarities indicate that during the early period of neurogenesis, basic regulatory interactions among these genes, crucial to exert organizer properties, do not exist or are modified. In the light of these data, it is therefore very unlikely that the pNE surrounding the otd/unpg interface represents a functional homolog of the MHB organizer. This supports the assumption (see work on Amphioxus [48]) that during evolution the Otx/Gbx border was established before it became equipped with organizer function. Since there is also no convincing evidence for an MHB organizer in any tunicate, it presumably first evolved in the vertebrate lineage.
Equivalents of 'hindbrain' and 'spinal cord' in Drosophila?
In the vertebrate neural tube, the border between hindbrain and spinal cord is supposed to be approximately indicated by the anterior limit of Hoxb5 expression [57-59]. This molecular regionalization may be conserved among chordates, since in the tunicates Ciona and Oikopleura the restricted expression of a Hox5 ortholog is also suggested to coincide with the anterior border of the spinal chord [14,19,60,61] (Figure 5). However, recent data in vertebrates [59,62-64] show that the Hoxb5 domain, although initially confined to the spinal cord, extends rostrally into the developing posterior hindbrain. Unlike Hoxb5, the rostral borders of Hoxb6 and Hoxb7 domains finally come to lie close to the transition between hindbrain and spinal cord [65], and might, therefore, be more suitable indicators to distinguish between both CNS domains. In Drosophila, the anterior border of the Scr/Hoxb5 domain maps within the maxillary neuromere, those of Antp/Hoxb6 and Ubx/Hoxb7 domains within the labial and third thoracic neuromere, respectively (Additional file 2). Assuming that, in analogy to vertebrates [66], the posterior border of otd expression (within the anterior DC) sets the anterior limit, the hindbrain equivalent would comprise four neuromeres, when considering the anterior border of Scr expression as its posterior limit, or, what is more likely, up to eight neuromeres, when considering the anterior Ubx expression border as the posterior limit. Interestingly, a comparable number of seven to eight rhombomeres comprises the vertebrate hindbrain [67]. In this perspective, the CNS region posterior to the anterior limit of the Scr or Ubx domain would be equivalent to the vertebrate spinal cord.
The vertebrate hindbrain is clearly subdivided into segmental units [67], as opposed to the mid- and forebrain in which the underlying metamerism is unclear [40,68,69]. Accordingly, the segmented part of the anterior CNS is separated from the less overt segmented part by the Otx2/Gbx2 border. The situation in Drosophila exhibits similarities. Whereas the segmental characteristics of DC, TC and ventral nerve cord are obvious, they are cryptic in the PC [26,25]. Considering the position of the otd/unpg border within the anterior DC, this border, as in vertebrates, separates the segmented part of the CNS (including the posterior DC, the TC, the gnathal, thoracic and abdominal CNS) from an anterior part (the PC) in which the metameric identity is less obvious.
Conclusion
Molecular characterization of the neuroectoderm and of individually identified neural stem cells in the early Drosophila embryo, indicate the existence of a non-segmental Otx/Gbx orthologous interface located within the anterior DC. Furthermore, my data support the idea that the area surrounding this interface (encompassing the anterior DC/posterior PC) may represent an ancestral territory that shares molecular similarities with the region around the vertebrate MHB. Otherwise, lack of expression of Pax2/5/8 and Fgf8-related genes, and significant differences in the expression onset of other key regulators at the otd/unpg interface, imply that genetic interactions crucial to exert vertebrate organizer activity do not exist or are modified in the early embryonic brain of Drosophila.
Materials and methods
Drosophila strains
The following fly strains were used: Oregon R (wild type), unplugged-lacZ [22], engrailed-lacZ [70].
Staging and mounting of embryos
Staging of the embryos was done according to [71]; additionally, the trunk NB pattern [22] was used as a further reference for staging. Flat preparations of the head ectoderm of stained embryos and mounting were done as described previously [23].
Antibodies and immunohistochemistry
Embryos were dechorionated, fixed and immunostained according to previously published protocols [72]. The following primary antibodies were used: mouse-anti-Antennapedia (1:20; DSHB, Iowa City, IA, USA), rabbit-anti-Atonal (1:5000; A Jarman, Edinburgh, UK), anti-DIG-AP (1:1,000; Roche, Mannheim, Germany), mouse-anti-Engrailed/Invected (1:4; DSHB), mouse-anti-β-galactosidase (1:500; Promega, Madison, WI, USA), rabbit-anti-β-galactosidase (1:2,500; Cappel, Costa Mesa, CA, USA), rat-anti-Labial (F Hirth, London, UK), rabbit-anti-Muscle specific homeobox (1:500; MP Scott, Palo Alto, CA, USA), rabbit-anti-Pax2 (1:50; M Noll, Zürich, Switzerland), mouse-anti-Pox-neuro (1:100; C Dambly-Chaudiere, Montpellier, France), rabbit-anti-Sex comb reduced (1:1,000, T Kaufman, Bloomington, IN, USA), mouse-anti-Ultrabithorax (1:20; DSHB), rabbit-anti-Ventral nervous system defective (1:500; CQ Doe, Eugene, OR, USA), mouse-anti-Wingless (1:10; DSHB). The secondary antibodies (all from Dianova, Hamburg, Germany) were either biotinylated (goat anti-mouse, goat anti-rabbit) or alkaline phosphatase-conjugated (goat anti-mouse, goat anti-rabbit, goat anti-rat) and diluted 1:500.
Whole mount in situ hybridization
Dioxigenin (DIG)-labelled buttonhead RNA probe was synthesized using HindIII linearised pBKS-btd [73] as a template with T7 polymerase. Other DIG-labelled RNA probes were synthesized using oligonucleotide primers amplified by PCR on wild-type genomic DNA: branchless (1,209 bp; forward primer CAGAACTACAACACTTACTCCTCC, reverse primer CTCGTAGCTCGCATCTTCTAGG); pyramus (2,020 bp, forward primer GGCAATCAGAACTTTAGTAGCG, reverse primer CAGACCACCATCGTTATGATTC); thisbe (2,284 bp, forward primer GCCCAATGTCAGCCACATCGG, reverse primer GTCGAGGTGGGCAGGAACC); unplugged (908 bp, forward primer GTGTCTGCTCGGGAACA-GAAACG, reverse primer GTCCATCTCGCCGTTGTAGTTCC). In all cases, the resulting PCR fragment was used as a template. All DIG-labelled RNA probes were prepared using a DIG-RNA-labelling mix (Roche) according to the manufacturer's protocol. The hybridization on embryos was performed as described previously [74,75].
Documentation
Embryos were viewed under a Zeiss Axioplan equipped with Nomarski optics using 40×, 63× and 100× oil immersion objectives. Pictures were digitized with a CCD camera (Contron progress 3012) and different focal planes were combined using Adobe Photoshop 7.0. Semi-schematic presentations are based on camera lucida drawings.
Competing interests
The author(s) declare that they have no competing interests.
Supplementary Material
Expression onset of MHB-specific key developmental genes in three different vertebrate species compared to orthologous factors around the otd/unpg interface in Drosophila. In vertebrates, the temporal order of genetic interactions among MHB-specific genes is closely reflected by the temporal order of the onset of their expression. Otx2 and Gbx2 are the first genes expressed (from gastrulation onwards), succeeded by the expression of further genes in the following order: Bts1 [3], Pax2, En1/2, Wnt1 and FGF8 [4]. In Drosophila, comparable to vertebrates, otd is the first gene expressed at the presumptive region of the otd/unpg interface, already before gastrulation (stage 6). In contrast, unpg is expressed significantly later than otd, not before stage 11, when it is also found in the TC and in a segmental pattern in the ventral nerve cord. Similar to otd, the head gap gene btd is expressed before gastrulation; in the pNE and NBs at the later otd/unpg interface it is not detected before stages 7/8. Furthermore, wg and en are expressed in the region at about stage 8, thus later than otd and btd, but before unpg. This indicates that head segments can be distinguished already before unpg expression initiates. In contrast to vertebrates, D-pax2 and poxn are expressed significantly later than en, but neither in the pNE nor in brain NBs (indicated by the stippled line for poxn). Moreover, expression of the Fgf8-related genes bnl and pyr is lacking around the otd/unpg interface. Transient expression of ths (indicated by stippled line), is unlikely to reflect MHB-specific expression of Fgf8. The temporal onset of gene expression is in part reminiscent of the situation in vertebrates (for example, of otd,btd,en,wg), but otherwise discloses significant differences (for example, unpg,D-pax2,poxn,Fgf8-related genes), implying that the chronological order of possible genetic interactions at the otd/unpg interface seems to be different from those at the vertebrate MHB. Data for mouse, zebrafish and chick are according to Figure 1 in [4] and references therein. Mouse: HDF, head fold stage; s, somite stage. Zebrafish: tb, tail bud period; s, segmentation period; h, hatching period. Chick: HH, stages after Hamburger and Hamilton. n.e., not expressed in the investigated period until stage 13; n.d., not determined.
Mapping of a 'hindbrain-like' CNS domain in Drosophila. Expression of (a,b) Scr, (c,d) Antp, or (e,f) Ubx in combination with En-lacZ (En) at late stage 11. (b,d,f) Close-ups of the domains boxed in (a,c,e) at the level of NBs. Scr, Antp, and Ubx are parasegmentally expressed. (a,b) The anterior limit of the Scr expression coincides with the anterior border of the maxillary en stripe (mxs); the Scr domain covers the posterior compartment of the maxillary and the anterior compartment of the labial neuromere (see also [77]). (c,d) The anterior limit of the Antp domain coincides with the anterior border of the labial en stripe (las); the Antp domain covers the posterior compartment of the labial, all thoracic, as well as weakly all abdominal neuromeres (see also [78,79]). (e, f) The anterior limit of the Ubx domain corresponds to the anterior border of the en stripe in the third thoracic neuromere (t3s); Ubx is found in the posterior compartment of the third thoracic neuromere and in the abdominal neuromeres 1–8. (g) Model of the extension of a hindbrain-like CNS domain in Drosophila (corresponding NBs are encircled in grey). The expression domains of En, Otd, Scr, as well as the anterior domains of Antp and Ubx are indicated. Additionally, the SOPs of the dorsal organ and hypopharyngeal/latero-hypopharyngeal organ are indicated. For further details, see the text. T1–T3, first to third thoracic neuromeres; A1, first abdominal neuromere. Other abbreviations are as in the figures.
Acknowledgments
Acknowledgements
This project was carried out in the lab of GM Technau and I am grateful for the fruitful discussions and his general support. I also thank D Volland for excellent technical assistance, GM Technau and A Rogulja-Ortmann for critical reading of the manuscript, and C Dambly-Chaudiere, CQ Doe, F Hirth, A Jarman, T Kaufman, M Noll, MP Scott and E Wimmer for providing reagents. This work was supported by grants from the Deutsche Forschungsgemeinschaft to RU and GM Technau.
References
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Divide et Impera – the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 2004;27:727–734. doi: 10.1016/j.tins.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Wilm TP, Crozatier M, Pfeffer P, Wassef M, Bally-Cuif L. The zebrafish buttonhead-like factor Bts1 is an early regulator of pax2.1 expression during mid-hindbrain development. Development. 2001;128:4021–4034. doi: 10.1242/dev.128.20.4021. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Brand M. The midbrain-hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11:34–42. doi: 10.1016/S0959-4388(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Millet S, Bloch-Gallego E, Alvarado-Mallart RM. Specification of the meso-isthmo-cerebellar region: the Otx2/Gbx2 boundary. Brain Res Brain Res Rev. 2005;49:134–149. doi: 10.1016/j.brainresrev.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Bruce AE, Shankland M. Expression of the head gene Lox22-Otx in the leech Helobdella and the origin of the bilaterian body plan. Dev Biol. 1998;201:101–112. doi: 10.1006/dbio.1998.8968. [DOI] [PubMed] [Google Scholar]
- Arendt D, Nübler-Jung K. Comparison of early nerve cord development in insects and vertebrates. Development. 1999;126:2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- Hirth F, Reichert H. Conserved genetic programs in insect and mammalian brain development. Bioessays. 1999;21:677–684. doi: 10.1002/(SICI)1521-1878(199908)21:8<677::AID-BIES7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Saiga H. Organization of Hox genes in ascidians: present, past, and future. Dev Dyn. 2005;233:382–389. doi: 10.1002/dvdy.20374. [DOI] [PubMed] [Google Scholar]
- Wada H, Saiga H, Satoh N, Holland PW. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100:189–203. doi: 10.1016/S0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Region specific gene expressions in the central nervous system of the ascidian embryo. Mech Dev. 2002;119:S275–277. doi: 10.1016/S0925-4773(03)00128-X. [DOI] [PubMed] [Google Scholar]
- Canestro C, Bassham S, Postlethwait J. Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Kene M, Williams NA, Holland ND. Sequence and embryonic expression of the amphioxus engrailed gene (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development. 1997;124:1723–1732. doi: 10.1242/dev.124.9.1723. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Holland ND, Kalousova A, Paces J, Schubert M, Holland LZ. Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development. 1999;126:1295–1304. doi: 10.1242/dev.126.6.1295. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Holland NN, Schubert M. Developmental expression of AmphiWnt1, an amphioxus gene in the Wnt1/wingless subfamily. Dev Genes Evol. 2000;210:522–524. doi: 10.1007/s004270000089. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Holland ND. Chordate origins of the vertebrate central nervous system. Curr Opin Neurobiol. 1999;9:596–602. doi: 10.1016/S0959-4388(99)00003-3. [DOI] [PubMed] [Google Scholar]
- Wada H, Satoh N. Patterning the protochordate neural tube. Curr Opin Neurobiol. 2001;11:16–21. doi: 10.1016/S0959-4388(00)00168-9. [DOI] [PubMed] [Google Scholar]
- Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H. An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development. 2003;130:2365–2373. doi: 10.1242/dev.00438. [DOI] [PubMed] [Google Scholar]
- Reichert H. A tripartite organization of the urbilaterian brain: developmental genetic evidence from Drosophila. Brain Res Bull. 2005;66:491–494. doi: 10.1016/j.brainresbull.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains, and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Hartenstein V. The terminal regions of the body pattern. In: Bate CM, Martinez-Arias A, editor. The Development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory press; 1993. pp. 687–746. [Google Scholar]
- Urbach R, Technau GM. Segment polarity and D/V patterning gene expression reveals segmental organization of the Drosophila brain. Development. 2003;130:3607–3620. doi: 10.1242/dev.00532. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Bopp D, Jamet E, Baumgartner S, Burri M, Noll M. Isolation of two tissue-specific Drosophila paired box genes, Pox meso and Pox neuro. EMBO J. 1989;8:3447–3457. doi: 10.1002/j.1460-2075.1989.tb08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Jamet E, Burri M, Bopp D, Basler K, Hafen E, Dumont N, Spielmann P, Ghysen A, Noll M. The paired box gene pox neuro: a determinant of poly-innervated sense organs in Drosophila. Cell. 1992;69:159–172. doi: 10.1016/0092-8674(92)90127-X. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437X(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Ghysen A, O'Kane C. Neural enhancer-like elements as specific cell markers in Drosophila. Development. 1989;105:35–52. doi: 10.1242/dev.105.1.35. [DOI] [PubMed] [Google Scholar]
- Czerny T, Bouchard M, Kozmik Z, Busslinger M. The characterization of novel Pax genes of the sea urchin and Drosophila reveal an ancient evolutionary origin of the Pax2/5/8 subfamily. Mech Dev. 1997;67:179–192. doi: 10.1016/S0925-4773(97)00119-6. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain-hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr Opin Cell Biol. 2000;12:736–741. doi: 10.1016/S0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/S0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer EA, Jäckle H, Pfeifle C, Cohen SM. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature. 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Green P, Liaw GJ, Rudolph K, Lengyel J, Hartenstein V. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol. 1997;182:270–283. doi: 10.1006/dbio.1996.8475. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Ohlen TV. Vnd/nkx, ind/gsh, and msh/msx: conserved regulators of dorsoventral neural patterning? Curr Opin Neurobiol. 2000;10:63–71. doi: 10.1016/S0959-4388(99)00049-5. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Dorso-ventral patterning of the brain: A comparative approach. In: Technau GM, editor. Brain Development in Drosophila. Texas: Landes Bioscience; 2007. [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, McKay IJ, Sharpe PT. The murine homeobox gene Msx-3 shows highly restricted expression in the developing neural tube. Mech Dev. 1996;55:201–210. doi: 10.1016/0925-4773(96)00505-9. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X, Xu H, Lufkin T. Msx3: a novel murine homologue of the Drosophila msh homeobox gene restricted to the dorsal embryonic central nervous system. Mech Dev. 1996;58:203–215. doi: 10.1016/S0925-4773(96)00562-X. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Katsuyama Y, Wada S, Saiga H. Homeobox genes exhibit evolutionary conserved regionalization in the central nervous system of an ascidian larva. Zoolog Sci. 1996;13:479–482. doi: 10.2108/zsj.13.479. [DOI] [PubMed] [Google Scholar]
- Jiang D, Smith WC. An ascidian engrailed gene. Dev Genes Evol. 2002;212:399–402. doi: 10.1007/s00427-002-0255-y. [DOI] [PubMed] [Google Scholar]
- Hino K, Satou Y, Yagi K, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev Genes Evol. 2003;213:264–272. doi: 10.1007/s00427-003-0318-8. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Ueno N, Gojobori T. A genome-wide survey of the genes for planar polarity signaling or convergent extension-related genes in Ciona intestinalis and phylogenetic comparisons of evolutionary conserved signaling components. Gene. 2003;317:165–185. doi: 10.1016/S0378-1119(03)00700-5. [DOI] [PubMed] [Google Scholar]
- Castro LF, Rasmussen SL, Holland PW, Holland ND, Holland LZ. A Gbx homeobox gene in amphioxus: insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Dev Biol. 2006;295:40–51. doi: 10.1016/j.ydbio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Steinmetz PR, Zelada-Gonzales F, Burgtorf C, Wittbrodt J, Arendt D. Polychaete trunk neuroectoderm converges and extends by mediolateral cell intercalation. Proc Natl Acad Sci USA. 2007;104:2727–2732. doi: 10.1073/pnas.0606589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova M, Bakalenko N, Novikova E, Cook CE, Eliseeva E, Steinmetz PR, Kostyuchenko RP, Dondua A, Arendt D, Akam M, Andreeva T. Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa) Dev Genes Evol. 2007;217:39–54. doi: 10.1007/s00427-006-0119-y. [DOI] [PubMed] [Google Scholar]
- Denes AS, Jékely G, Steinmetz PR, Raible F, Snyman H, Prud'homme B, Ferrier DE, Balavoine G, Arendt D. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/S0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Lewandoski M, Campbell K, Joyner AL, Rubenstein JL, Martinez S, Martin GR. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development. 1997;124:2923–2934. doi: 10.1242/dev.124.15.2923. [DOI] [PubMed] [Google Scholar]
- Song DL, Chalepakis G, Gruss P, Joyner AL. Two Pax-binding sites are required for early embryonic brain expression of an Engrailed-2 transgene. Development. 1996;122:627–635. doi: 10.1242/dev.122.2.627. [DOI] [PubMed] [Google Scholar]
- Lun K, Brand M. A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development. 1998;125:3049–3062. doi: 10.1242/dev.125.16.3049. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R. Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature. 1989;341:405–409. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- Hunt P, Gulisano M, Cook M, Sham MH, Faiella A, Wilkinson D, Boncinelli E, Krumlauf R. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353:861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Gionti M, Ristoratore F, Di Gregorio A, Aniello F, Branno M, Di Lauro R. Cihox5, a new Ciona intestinalis Hox-related gene, is involved in regionalization of the spinal cord. Dev Genes Evol. 1998;207:515–523. doi: 10.1007/s004270050142. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Holland PWH. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrate origins of midbrain development. Development. 2004;131:3285–3294. doi: 10.1242/dev.01201. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- Oosterveen T, Meijlink F, Deschamps J. Expression of retinaldehyde dehydrogenase II and sequential activation of 5' Hoxb genes in the mouse caudal hindbrain. Gene Expr Patterns. 2004;4:243–247. doi: 10.1016/j.modgep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–4719. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- Nolte C, Krumlauf R. Expression of Hox genes in the nervous system of vertebrates. In: Papageorgiou S, editor. HOX gene expression. Texas: Landes Bioscience; 2006. pp. 1–28. [Google Scholar]
- Millet S, Bloch-Gallego E, Simeone A, Alvarado-Mallart RM. The caudal limit of Otx2 gene expression as a marker of the midbrain/hindbrain boundary: a study using in situ hybridisation and chick/quail homotopic grafts. Development. 1996;122:3785–3797. doi: 10.1242/dev.122.12.3785. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2. Berlin, Heidelberg, New York: Springer Verlag; 1997. [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg E, editor. Methods in Cell Biology Drosophila melanogaster: Practical Uses in Cell Biology. Vol. 44. New York: Academic press; 1994. [DOI] [PubMed] [Google Scholar]
- Wimmer EA, Cohen SM, Jäckle H, Desplan C. buttonhead does not contribute to a combinatorial code prosposed for Drosophila head development. Development. 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- Plickert G, Gajewski M, Gehrke G, Gausepohl H, Schlossherr J, Ibrahim H. Automated in situ detection (AISD) of biomolecules. Dev Genes Evol. 1997;207:362–367. doi: 10.1007/s004270050124. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Schubert M, Holland ND, Laudet V, Holland LZ. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev Biol. 2006;296:190–202. doi: 10.1016/j.ydbio.2006.04.457. [DOI] [PubMed] [Google Scholar]
- Rogers BT, Peterson MD, Kaufman TC. Evolution of the insect body plan as revealed by the Sex combs reduced expression pattern. Development. 1997;124:149–157. doi: 10.1242/dev.124.1.149. [DOI] [PubMed] [Google Scholar]
- Wirz J, Fessler LI, Gehring WJ. Localization of the Antennapedia protein in Drosophila embryos and imaginal discs. EMBO J. 1986;5:3327–3334. doi: 10.1002/j.1460-2075.1986.tb04647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142X.2002.02034.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression onset of MHB-specific key developmental genes in three different vertebrate species compared to orthologous factors around the otd/unpg interface in Drosophila. In vertebrates, the temporal order of genetic interactions among MHB-specific genes is closely reflected by the temporal order of the onset of their expression. Otx2 and Gbx2 are the first genes expressed (from gastrulation onwards), succeeded by the expression of further genes in the following order: Bts1 [3], Pax2, En1/2, Wnt1 and FGF8 [4]. In Drosophila, comparable to vertebrates, otd is the first gene expressed at the presumptive region of the otd/unpg interface, already before gastrulation (stage 6). In contrast, unpg is expressed significantly later than otd, not before stage 11, when it is also found in the TC and in a segmental pattern in the ventral nerve cord. Similar to otd, the head gap gene btd is expressed before gastrulation; in the pNE and NBs at the later otd/unpg interface it is not detected before stages 7/8. Furthermore, wg and en are expressed in the region at about stage 8, thus later than otd and btd, but before unpg. This indicates that head segments can be distinguished already before unpg expression initiates. In contrast to vertebrates, D-pax2 and poxn are expressed significantly later than en, but neither in the pNE nor in brain NBs (indicated by the stippled line for poxn). Moreover, expression of the Fgf8-related genes bnl and pyr is lacking around the otd/unpg interface. Transient expression of ths (indicated by stippled line), is unlikely to reflect MHB-specific expression of Fgf8. The temporal onset of gene expression is in part reminiscent of the situation in vertebrates (for example, of otd,btd,en,wg), but otherwise discloses significant differences (for example, unpg,D-pax2,poxn,Fgf8-related genes), implying that the chronological order of possible genetic interactions at the otd/unpg interface seems to be different from those at the vertebrate MHB. Data for mouse, zebrafish and chick are according to Figure 1 in [4] and references therein. Mouse: HDF, head fold stage; s, somite stage. Zebrafish: tb, tail bud period; s, segmentation period; h, hatching period. Chick: HH, stages after Hamburger and Hamilton. n.e., not expressed in the investigated period until stage 13; n.d., not determined.
Mapping of a 'hindbrain-like' CNS domain in Drosophila. Expression of (a,b) Scr, (c,d) Antp, or (e,f) Ubx in combination with En-lacZ (En) at late stage 11. (b,d,f) Close-ups of the domains boxed in (a,c,e) at the level of NBs. Scr, Antp, and Ubx are parasegmentally expressed. (a,b) The anterior limit of the Scr expression coincides with the anterior border of the maxillary en stripe (mxs); the Scr domain covers the posterior compartment of the maxillary and the anterior compartment of the labial neuromere (see also [77]). (c,d) The anterior limit of the Antp domain coincides with the anterior border of the labial en stripe (las); the Antp domain covers the posterior compartment of the labial, all thoracic, as well as weakly all abdominal neuromeres (see also [78,79]). (e, f) The anterior limit of the Ubx domain corresponds to the anterior border of the en stripe in the third thoracic neuromere (t3s); Ubx is found in the posterior compartment of the third thoracic neuromere and in the abdominal neuromeres 1–8. (g) Model of the extension of a hindbrain-like CNS domain in Drosophila (corresponding NBs are encircled in grey). The expression domains of En, Otd, Scr, as well as the anterior domains of Antp and Ubx are indicated. Additionally, the SOPs of the dorsal organ and hypopharyngeal/latero-hypopharyngeal organ are indicated. For further details, see the text. T1–T3, first to third thoracic neuromeres; A1, first abdominal neuromere. Other abbreviations are as in the figures.