ONE of the nicest things anyone ever said about our work was in a (necessarily) anonymous grant review from the early 1990s in which the author commented that our lab had contributed greatly to moving the study of circadian rhythms “out of the era of spoon-bending.” Some years later Bob Metzenberg, who always cherished a well-turned phrase, fessed up to having written this, and it is easy to see his quick wit and word play. I mention it here because it nicely encapsulates the 25 years that I want to cover, a period that extends from the era when belief in intracellular circadian rhythms stretched the credibility of all but devotees to the years when the problem was cracked and rhythms truly entered mainstream science (Science News and Editorial Staffs 1997, 1998). During this time, analysis of rhythms moved from the use of genetics—which opened up the black box and exposed the feedback loops—to molecular biology, where the field is now. Although it is tempting to write about all the vistas that opened up during this time based on work in Neurospora, from clock mechanism to clock output, I have restricted this Perspectives to studies on the circadian mechanism and will leave output to other, highly capable hands (Loros 2008). It is an account of what drew me to rhythms work and to the Neurospora circadian system and of what led our lab to identify the factors and interactions that contributed to the denouement of the question of the molecular bases of circadian rhythms: the assembly, a little over a decade ago, of a complete interconnected regulatory cycle.
EARLY DAYS: THE LURE OF IGNORANCE
I never intended to study rhythms or work on Neurospora. My undergraduate degrees in oceanography and chemistry were aimed at a career in oceanography, but on a whim I also applied to graduate school in biology at Harvard where I ended up. Interactions with J. W. (“Woody”) Hastings led me to bioluminescence in marine organisms, and it was a short step from there to circadian regulation of bioluminescence and to circadian biology. (Why bother to make light during the daytime?) Rhythms struck me as a field in which few were even pursuing the right questions and where an ultimate molecular resolution was nowhere even remotely in sight. This impression was confirmed during a 10-week summer course on rhythms run by Colin Pittendrigh at the Hopkins Marine (dubbed Murine Station, since Pitt then worked on mice) in Pacific Grove, California, in 1977, where nearly an entire generation of rhythms biologists from the United States first met each other. The truly vast biology of the field—from microbial rhythms in bioluminescence to photoperiodism in plants, to activity rhythms in mice, to psychiatric illness in people—was unified by the characteristics of the underlying clock to the extent that one dared to hope that a single mechanism might underlie all of it. Those unifying characteristics, needed to accommodate and explain all this biology, were sufficiently distinct to delineate a field: a circadian rhythm, as the name suggests, has a period of about a day (absent any environmental cues) but can be entrained by environmental cycles to exactly match their periods. Moreover, the period length is close to the same when measured under different ambient temperatures or nutritional conditions (Sweeney 1976). Other biological rhythms—those with extra long or really short period lengths, those whose period changed markedly with temperature, and those measured only under light–dark cycles—were not (and are not) counted as circadian rhythms; this distinction kept research focused on a single mechanism and this focus was crucial to solving the problem.
CIRCADIAN BIOLOGY IN THE PREMOLECULAR ERA
By the late 1970s, genetic approaches to the pursuit of rhythms had virtually ground to a halt, given the near impossibility of pursuing genetic leads at the molecular and biochemical level. An additional and influential factor was the open disbelief in the validity of this approach expressed by leaders in the field of rhythms, whose backgrounds were chiefly in physiology and anatomy. There was still an active community working on microbial circadian clocks at this time, including work on Neurospora, Tetrahymena, Paramecium, Euglena, and Chlamydomonas as well as Gonyaulax (the unicellular eukaryote responsible for “red tides” and “phosphorescent summer seas”), but an undercurrent of opinion was developing (e.g., Menaker et al. 1971) that there would not be a conserved mechanism for rhythms even among higher animals and insects, much less between fungi and people. One reason for this stemmed from experiments done with a favorite neurobiological preparation, the eye of marine mollusks such as Aplysia, where it was known that changes in the pH or ionic composition of the bathing medium would alter by several hours the period length of the circadian rhythm in compound action potentials (e.g., Jacklet 1969). Genetic screens in the early 1970s in both Drosophila (Konopka and Benzer 1971) and Neurospora (Feldman and Waser 1971) had yielded rhythm variants. [The screens were virtually simultaneous, but Konopka and Benzer (1971) published sooner, more prominently, and with greater impact, in part both because flies are animals and because Drosophila was then beginning to be hyped as “the great model system” for behavior and neuro-genetics.] However, the majority in the rhythms community, if they considered these mutants at all, considered it most likely that the chronobiological basis of the rhythm variations would lie in pleiotropic effects on the cellular or intercellular milieu, changes akin to changing the ionic composition of the bathing medium. Also implicit in many discussions, and even in public pronouncements, was the assumption that, although rhythmicity was a cellular property in unicellular organisms, it was more likely to be a property of networks of neurons in animals.
Meanwhile, physiological approaches using metabolic inhibitors, an approach pioneered by J. W. (“Woody”) Hastings and colleagues (Hastings 1960), sought to define the aspects of cellular metabolism required for rhythmicity as well as to pioneer the approach of following the regulatory trail of circadian outputs back to the mechanism. Harvard in the mid-1970s was a great place and time to learn biochemistry and I did so, determining the structure of Gonyaulax luciferin (Dunlap and Hastings 1981a; Dunlap et al. 1981), purifying the luciferase, and determining that it was regulated through daily synthesis and destruction, the first clock-regulated enzyme whose mode of regulation was determined (Dunlap and Hastings 1981b). Meanwhile, although use of inhibitors yielded many blind alleys (Robertson 1975), there emerged one thread of truth in many organisms: protein synthesis on 80s ribosomes at specific times of day was necessary for every eukaryotic clock examined. Phase shifts identified a “sensitive time of day,” generally in the late night to early morning (Jacklet 1977; Dunlap et al. 1980; Dunlap and Feldman 1988), and chronic inhibition stopped the clock at dawn (e.g., Khalsa and Block 1992). Moving beyond this to the proteins involved, however, would obviously be tough, and it required no brilliance to see that genetics was needed to identify the pertinent elements.
Although rhythms (and even some rhythm genetics) were known in Chlamydomonas (e.g., Bruce 1972) and in Paramecium (e.g., Barnett 1966), the two premier organisms with clocks and well-developed genetic systems were flies and Neurospora. And even in 1979, when I finished at Harvard, it was clear to me at least that the answer being universally sought would need to be phrased in the language of genetics and biochemistry. By then, Neurospora could be transformed (Case et al. 1979) but Drosophila could not; biochemical genetics was invented in Neurospora, and biochemistry in flies was a challenge, so the choice of system when I began a postdoc seemed clear. I was aware of the breakthroughs in and using recombinant DNA technologies (libraries!, cloning!, sequencing!, chromosome walks!) that had been developed in part by friends on the third and fourth floors of the Harvard BioLabs, so it seemed natural to develop a postdoctoral fellowship, eventually funded by the Damon Runyon Foundation, around the cloning of clock genes. There were a few labs working on clocks in Neurospora—those of Malcolm Sargent, Stuart Brody, and Jerry Feldman—and, having settled on a genetic and molecular approach, for a postdoc I picked Feldman's, the one in which the clock mutants had been isolated. I set off for Santa Cruz.
NOT A LONG PERIOD, BUT A SLOW PERIOD
By the late 1970s, a handful of rhythm mutants had been identified in Neurospora (Feldman 1982), including alleles of a single gene, frequency (frq), whose spectrum (long, short, and arrhythmic) looked a lot like those mapping to the period locus in Drosophila. I aimed to clone this gene, but unfortunately the postdoctoral years were a dismal period scientifically. Neither molecular tools nor expertise for Neurospora molecular biology were available in the small lab where I toiled, nor were there advanced methods in general. I was, however, unofficially adopted by Harry Noller's lab next door where Joann Kop taught me basic molecular biology; I also met an intellectual peer who turned into a long-term colleague, Jennifer Loros, with whom I have shared many successful projects in both fungal genetics and human heredity ever since. Nonetheless, with no articles from my postdoc (the dozen from graduate school notwithstanding), by today's standards my career would have been over had it not been for unflagging support from Woody Hastings and from within the Neurospora community (especially Rowland Davis and Barry Bowman) and from a single department, biochemistry at Dartmouth, that posted an ad looking for people doing “something new and different.” That was I; rhythms were anything but a hot topic then. Dartmouth's was the only interview and subsequent job offer that I received in 2 years of looking, but you need only one.
Much of the problem with dissecting any problem is learning to think about it in the right way, and the first months in my own lab were spent doing just that—reading and thinking. Then, during a long weekend of 14-hr days, it all poured out in the text for two full grant proposals that described the areas in which I would work for the next decade and beyond. This was a time when the cell cycle was being worked out, and the smart money had it that the circadian cycle would be something like the cell cycle—i.e., a series of events occurring one after the other to create a cycle. In this model, where G1/S cyclins lead to S phase and S phase cyclins, which lead to M phase and M phase cyclins, which lead back to Start, each cyclin defines a phase in the cell cycle and it is impossible to stop at a “phaseless” point. However, this was just what Art Winfree had shown for the circadian oscillator; using light pulses of just the right strength and duration given at just the right time, he could send the circadian cycle to a place corresponding to “no phase” [equivalent to “all phases” (Winfree 1967, 1971; Winfree and Twaddle 1981)]. This suggested that thinking too much along the lines of cell cycles would be a distraction, and this was true to a point: circadian cycles had their own logic. An organizing concept for me was that the clock puzzle was really three different problems in cellular metabolism (e.g., Eskin 1979), each of which could be approached separately:
How do you build a clock—that is, what are the gears and cogs, how do they mesh, what regulates them, and how do they regulate one another so the collective output is a molecular/biochemical cycle with all the circadian characteristics? Underlying our expectations was theory from a number of sources (e.g., Friesen and Block 1984; Brenner et al. 1990), which suggested that a negative feedback loop could yield an oscillation.
Entrainment: how do abrupt and transient changes in the environment, chiefly ambient light or temperature, reset the phase of the clock and thereby bring internal time into line with external time?
Output: how is an intracellular molecular cycle used to regulate the behavior of the cell?
Coming from a microbial background, I was convinced that in all organisms all this happened within single cells, a generalization that came only more recently to be widely adopted by the entire circadian community (e.g., Michel et al. 1993). To get at these molecules, one needed genetics to get at the core mechanism, and one needed molecular biology for output, while knowledge about the mechanism would naturally lead to insights about environmental inputs to the system. My goal then was to explain in genetic and molecular terms how the circadian principles of a compensated, sustained, molecular oscillation of circadian period length, resettable by light and temperature cues, could be assembled in a single cell and used to direct diverse outputs.
IDENTIFICATION OF CLOCK COMPONENTS
In the early 1980s, the plate was clear for studies of mechanism, that is, for studies of how the oscillator at the core of the clock worked. The problem at hand in my own lab was to do what I had been unable to do as a postdoc—clone frq. However, no genetic selections were possible so there was no way beyond brute-force screens to characterize transformants, and physical maps of the genome lay more than a decade in the future. I spent 18 months crossing strains bearing translocations with different breakpoints to generate a minichromosome bearing frq that could be separated by orthogonal field electrophoresis and used to make a library sufficiently enriched in frq that I could identify it by transformation and, indeed, brute-force screening; but the strains proved recalcitrant. We tried a few other too-clever things but settled in the end, with the help of postdoc Rob McClung (who has since gone on to do great things in Arabidopsis clocks, e.g., McClung 2001) and a tech (Barb Fox) on a laborious chromosome walk. Excellent fine-structure genetics from Jennifer Loros's graduate work (Loros et al. 1986) had shown frq9 to be a recessive, arrhythmic, and presumptive null allele of frq and had placed frq on the right arm of chromosome VII, ∼2 cM from oli and 2.5 cM from for. Therefore, we started a walk at the selectable marker oli—perforce a bidirectional walk, since there was no way to orient proximal vs. distal until the target at the other end of the walk was found. This took us to five different chromosomes before we learned to check each step by Southerns; finally, in 1986 we confirmed by transformation/rescue that we had for, thereby proving that we had walked (>200 kb) across frq. Having asserted that rescue by transformation would work for a clock gene as easily as it had for a nutritional gene, we began to screen candidate cosmids by transformation into frq9. Cosmids from one region rescued the circadian rhythm in conidial spore formation (“banding”) on race tubes, thus confirming the location of frq.
Sometime after we started this in 1984, the Drosophila clock gene, period (per), was cloned by the Hall/Rosbash and Young groups who were able to take advantage of the excellent cytogenetics of Drosophila to pinpoint the correct chromosomal region. None of these three researchers were “circadian rhythms biologists,” a by-product in part of the dismissal of genetic approaches within the field. If I felt “scooped” at all by the cloning of per, I do not recall it; for me, it was more than compensated by the arrival of Jeff Hall, who became a mentor and sounding board, into the field. The collective publications on per in Proceedings of the National Academy of Sciences (Bargiello and Young 1984) and in Cell (Reddy et al. 1984) contained less information than we had when we cloned frq 2 years later. Whereas in 1984 it was sufficient just to have cloned an interesting gene, between 1984 and 1986 the bar was rapidly rising: by the time we had frq, we needed transcript and sequence information for a major publication. We dug in and groped toward the rising bar for publication, manually sequencing nearly 9 kb (no mean feat in those days) and mapping transcripts arising from the frq genomic region. This was sufficient to carry it to the level of the vanity journal to which we aspired, Nature (McClung et al. 1989).
OUTPUT
Meanwhile my second basic grant from 1984 was written about circadian output to take advantage of a new technique—subtractive hybridization—based on Bill Timberlake's earlier work from the mid-1970s on identification of sporulation genes in Aspergillus (Zimmermann et al. 1980). Since a wealth of earlier circadian physiological research using inhibitors (cited above) had clearly shown that there had to be protein synthesis for the clock to run—and not just continual synthesis but synthesis specifically in the early morning around subjective dawn—it was a simple step to assert with some conviction that there had to be rhythmic clock-pertinent transcription for the clock to run and that identification of time-of-day specific transcripts could identify the transcripts important for the clock to run. Additionally, it was easy to assert that some output would be derived from daily control of transcription by the clock. We would simply subtract evening cDNAs from morning cDNAs to identify the clock-essential transcripts. It seemed a classic win–win situation, and the National Science Foundation panel agreed. Jennifer Loros then carried out the first systematic screen for circadian-regulated genes and identified clock-controlled gene-1 (ccg-1) and ccg-2. The systematic identification of ccg's, and the realization that clock-controlled transcription likely provided a major conduit for time information from the oscillator to cellular metabolism, afforded a nice way to begin to understand circadian output in all circadian systems and propelled this work into Science (Loros et al. 1989). Analysis of output provided a focus for Loros's subsequent independent work and drew Deborah Bell-Pedersen into the rhythms trade. Studies of this type using ever-advancing technologies will continue in every circadian system into the foreseeable future.
A MECHANISM FOR THE CIRCADIAN CLOCK
The question of the molecular mechanism of the clock dominated the field in the 1970s and 1980s and continued to do so through the 1990s (and some would argue still does). As noted above, the genetics of frq and Drosophila per were similar and consistent with their being core clock components: recessive putative null alleles were arrhythmic, and there were semidominant alleles causing both long- and short-period lengths.
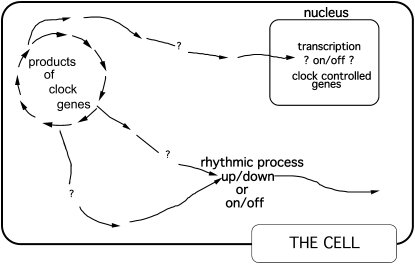
A question that loomed large at the time was whether expression of clock components cycled, as this would clearly implicate the clock genes as encoding elements of mechanism rather than elements that simply enabled a clock. Although we were hard at work on this, molecular confirmation that frq expression was rhythmic was delayed for several years due to a technical problem. Rhythm biochemistry in Neurospora was always done in liquid cultures having little added glucose (Nakashima 1981; Loros et al. 1989), ostensibly to slow growth and development, leaving only rhythm-pertinent metabolism. Indeed, the clock runs well under these conditions, but frq is expressed only at extremely low levels, levels virtually below detection. Although Jennifer had produced convincing data showing rhythmic frq expression by the time of a meeting that Bambos Kyriacou ran in Leicester, United Kingdom, in early 1992, it was not until Ben Aronson, perhaps inadvertently, tried high glucose for culture and analysis of frq that it became possible to routinely follow frq mRNA levels over time (Aronson et al. 1994a). However, efforts by Kathy Siwicki in Jeff Hall's Drosophila lab led the way in the identification of cycling clock components. Rhythm biochemistry in the fly was given a giant leg up by the fact that the cells in the fly eyes have clocks, so the heads (which could be isolated as a quasi-pure fraction by freezing and shaking) represented an enormous enrichment for clock molecules, even though the clock molecules in the eyes had no role in maintaining the free-running behavioral rhythms. In 1988, using antipeptide antisera for immunocytochemistry, Kathy provided the first evidence for rhythmic expression of a putative clock molecule (Drosophila PER; Siwicki et al. 1988), and Paul Hardin in the Rosbash lab provided the logical and essential extension of this 2 years later with the demonstration that per mRNA was rhythmically expressed in the head with a period length appropriate to the per allele (Hardin et al. 1990). The facile (and ultimately correct) interpretation of the rhythmic expression of PER and (later) frq was that they controlled their own expression at the core of the clock, although the possibility remained that PER or FRQ fed back in some different and perhaps remote way, for example, in regulating a behavioral or physiological output (rather than being a core element) that would feed back to affect the pace of the oscillator or to affect input. In this scenario, a null mutant could be arrhythmic at the overt, whole-organism level (no output), and alleles defective in feedback to the core clock would result in altered period lengths. [Years later, prd-4 provided exactly this precedent: a mutant that changes period length by affecting input but that is not a part of the clock (Pregueiro et al. 2006).] Also contributing to the uncertainty in interpretation was the observation that simple expression of per from a (presumably constitutive) heat-shock promoter was sufficient to rescue arrhythmicity in per-null flies (Ewer et al. 1988): maybe the transcriptional rhythms were not really necessary after all. Figure 1 provides a “clock model” from this era. One could put the products of clock genes “in the loop,” but it was not clear where or how.
Figure 1.—
An early 1990s view of the cellular clock. “The application of genetics in the context of clocks has largely been targeted to filling in the arrows within the feedback cycle itself; the output arrows, even within the cell, have been approached only via molecular genetics and biochemistry. However, both for questions concerning how the clock works and how time information is transduced within the cell, there is the hope that the problem will not need to be independently solved for each different clock system.” (Figure and quotation are from Dunlap 1993, p. 688.)
PROOF OF NEGATIVE FEEDBACK IN THE SERVICE OF THE CLOCK
The logical way out of this interpretational conundrum was to begin to manipulate the pertinent molecules (e.g., FRQ or PER) to demonstrate that they, in fact and directly, regulated themselves (or the genes that encoded them) and that both the period length and the phase of the molecular rhythm in expression, not just the behavioral rhythm, could be controlled by timed and regulated expression of the putative component. To execute an experiment like this at the time was a tall order and had not been done in any circadian system. The experiment required turning up frq expression at a known time and seeing how this affected phase or driving expression at a high level before releasing it and following the subsequent phase of the clock. Because the experiment was based on the knowledge that there was rhythmic expression of the clock molecules, and assumed that overexpression at some times would have an effect and at other clock phases would not have one, a key aspect was knowing the clock phase at which the increased level of FRQ appeared. For this reason, both rhythms theory and common sense made it a logical necessity that the means used to drive expression of the clock component (the inducer) could not itself affect the clock; otherwise, the results would be logically uninterpretable since you could not know the real clock phase of the treatment, or if the inducer, the clock molecule, or both synergistically were having the effect. This precluded the use of available heat-shock or light-regulated promoters, since both temperature and light were known to rapidly shift all clocks, and no nutritionally regulated promoters had yet been tamed for heterologous expression in Neurospora. To find one for taming, we cast about and settled on the quinic acid-2 promoter in a gene cluster that had been intensively studied by Norman Giles and colleagues (Giles et al. 1985); we showed that a fragment of the promoter could be used to drive frq expression up to a 100-fold by addition of the gratuitous inducer quinic acid, a compound that we showed had no effect on the clock. We knew by early 1992 that constant frq expression was not sufficient to rescue overt rhythmicity (and reported so at the above-mentioned meeting in Leicester, UK). But rhythmicity per se was only a part of the question; the other part was phase.
In the field of rhythms, phase refers to the time in the cycle when something happens. Two rhythms can have identical periods but be different if they are “out of sync,” i.e., if they have different phases. We were developing theory and tests to show that frq encoded a component rather than an enabler of the clock and that daily expression of frq was a part of the clock. In this concrete model, the phase of the cycle at which frq expression rises and falls has real biological meaning. Chronic overexpression of frq could stop the rhythm and release from overexpression restored the rhythm, but if frq encoded a component, then the phase that the clock assumed upon release had to be dictated by when frq levels fell. We took extra time to look carefully at frq rhythmicity and phase following release from inducer, as well as to develop methods for gene replacements (Aronson et al. 1994c) so as to engineer a definitive null mutant of frq (Aronson et al. 1994b) as a recipient for transformations. Armed with these new tools, Ben Aronson carried out the experiment(s) that demonstrated negative feedback of proteins (FRQ) on the genes that encode them (frq) in the service of a circadian oscillator. We showed that (1) overexpression of frq from a heterologous promoter reduced expression of frq from its native promoter in both a frq+ and a frq null background (i.e., neither overt rhythmicity nor output was needed to mediate feedback); (2) continued overexpression resulted in arrhythmicity (i.e., rhythmic frq expression, not simply expression, was needed for the clock to run); and (3) the phase of the rhythm was determined by the time at which the cell was released from overexpression: autoregulation of frq expression controlled rhythmicity as well as both period and phase. We wrote, “The amount of frq transcript in the cell is regulated by the clock, frq mRNA encodes FRQ, and point mutations in FRQ set the period of the clock; frq must therefore determine the timing of its own expression through regulation of transcript synthesis or turnover. The characteristic loss of stable rhythmicity in frq loss-of-function mutations shows that this gene is critical for circadian rhythmicity, and the observation of high amounts of transcript in frq9 [a frameshifted null mutant] suggests autoregulation by negative feedback” (Aronson et al. 1994a, p. 1581). Rhythms in per expression were already known, and we inferred that the other untested aspect of this story in Drosophila would also be would true, namely that “These transcript and protein cycles provide a ready explanation for the universal effectiveness of transcriptional and translational inhibitors in clock resetting. This universality, in turn, suggests that the clock-gene transcript and protein cycling seen and implied here [in Neurospora] may be a universal feature of circadian oscillators”—as indeed they seem to be (Aronson et al. 1994a, p. 1583). Figure 2 is taken from Aronson et al. (1994a).
Figure 2.—
“A schematic diagram of a cell expressing a circadian rhythm. The frq gene encodes the FRQ protein that has multiple roles, of which the best established is to regulate, probably through intermediates, the amount of frq mRNA as a part of the negative feedback loop that constitutes the circadian oscillator… . clock-controlled genes (ccgs) … CCRE, the circadian clock responsive element or “clock box” is the DNA element that mediates the action of clock-controlled transcription factors …” FRQ is now in the core loop but the loop retains notions of elements acting in sequential steps. (Figure and quotation are from Aronson et al. 1994a, p. 1582.)
HOW LIGHT RESETS THE CLOCK
As noted above, we had assumed that solid identification of a clock component would lead the way to an understanding of input and the means of entrainment; and, with the proof that FRQ fulfilled the role of a clock component, understanding light resetting seemed within reach. The whole concept of circadian light resetting and entrainment was inextricably wrapped up with the phase response curve (PRC), which maps the response of the clock to light perceived at different times of day. If you imagine a circadian system in a light–dark cycle, a part of the cycle will naturally correspond to the daytime (the day phase) and a part to nighttime. Even if the lights never come back on, the clock will continue to cycle in constant darkness, and under these conditions, the part of the cycle that used to happen in the day phase is now referred to as subjective day and the night phase as subjective night. The PRC is the graphical depiction of the truism that, for a photic stimulus to entrain a clock (that is, for an internal clock to be brought into phase with an external light–dark cycle), light perceived late at night has to advance the clock to early subjective day, and light perceived early in the night must delay the clock to the previous day, while having little or no effect when seen in the subjective day. [A testament to the fascination that this concept held in the field is that, although the light PRC must be in this form for entrainment to occur, decades of work from many labs went into showing that this was true for >50 different species (Johnson 1990).] Originally performed by Hastings and Sweeney in the 1950s and later used extensively by Pittendrigh and colleagues (e.g., Pittendrigh 1967, 1993), the light PRC played an absolutely central role in the development of the complex multi-oscillator models for the circadian system that dominated the field in the premolecular era [including those of Winfree (1967, 1971) cited above] and in development of the modeling formalisms through which circadian clocks are still described (e.g., Johnson et al. 2004). Because overt circadian output could be influenced by many factors in addition to the circadian clock itself, an axiom of the rhythms field was that the PRC was the only true measure of the progress of the central circadian oscillator through time; simple overt rhythmicity was never deemed sufficient. And because it was the only accepted mirror of this Holy Grail, the means by which light acted on the clock appeared shrouded by unassailable complexity and dissecting it a problem of mythic proportions. Also contributing to the aura of complexity was pharmacological evidence from animals that delays were mechanistically different from advances (Ralph and Menaker 1985).
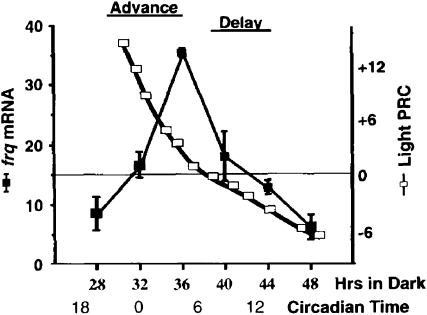
In hindsight, however, it is hard to see why this was perceived as so difficult since in its resolution the answer was so simple. A part of the barrier to apprehending the simplicity lay in the fact that the very idea of intracellular transcription/translation feedback loops as underlying all organismal eukaryotic clocks was still very novel (and not universally accepted). Instead, resetting mechanisms were envisioned as complicated affairs in which a photoreceptor in one tissue would trigger a signal that would transit to another tissue wherein magical things would transpire. Also, the cycle in frq mRNA peaked in the morning, whereas per (and later, tim), thought to encode the components in the Drosophila clock analogous to frq, peaked at night. If the oscillators worked in a similar manner, how could these be 180° out of phase? The first hint that frq expression was light responsive came from some fishing experiments by Jennifer Loros in the days before Ben Aronson discovered how to nutritionally boost frq expression; using large quantities of tissue and very (very) hot probes, Jennifer thought that she saw light-induced frq expression. This emboldened Sue Crosthwaite, a year or so later when we could see frq mRNA more easily, to do a time course of frq expression following light exposure; she found a lovely >10-fold induction within just 5 min after lights on. In the context of all the anticipated complexity surrounding the nature of the resetting mechanism, our initial observation that frq was rapidly and strongly light induced did not immediately suggest a global mechanism for resetting. Instead, we focused on proving that the threshold and fluence response for frq induction corresponded to that for clock resetting and that the clock effect was really mediated by transcriptional induction of frq and not some other ancillary effect such as message stabilization. However, while messing around with Sue's data, in particular plotting the daily cycle of frq transcript levels with the PRC on the same time axis (as in Figure 3), I was led to one of the few actual eureka moments that I can recall: the realization that all of light resetting, both advances and delays, in large part could be explained simply by the fact that there was a daily cycle in frq transcript level and that light induced an abrupt upswing of frq expression. If light induced frq mRNA when it was on its way to the peak (late night to morning), it would bring the RNA level to peak faster, resulting in an advance; if frq mRNA was declining, light would increase the levels, countering the decline, and cause a delay. This unexpectedly straightforward explanation implied that the inherent molecular mechanisms behind advances and delays were not distinct, as some had supposed, but were exactly the same. Light induced frq, and it was the dynamics of the clock that made the effects of light different at different times. In Crosthwaite et al. (1995, p. 1009), we wrote, “Together these data can explain … how a simple unidirectional physical or molecular signal and response (the light induction of frq) can be turned into a bidirectional clock response (clock phase-specific advances and delays).”
Figure 3.—
“The response of frq to light predicts the shape of the Neurospora PRC. The endogenous molecular cycling in the level of expression of the frq locus (thin line, relative level of frq mRNA plotted as mean ± SEM, n = 3) is shown in time correlated with the response of the clock to light (thick line, the Neurospora PRC … In broad outline, the PRC shows two regions, advances in the late [subjective] night through early morning and delays in the late [subjective] day through early evening. The response of the frq locus to light suggests an explanation for the general shape and timing of the PRC.” (Figure and legend are from Crosthwaite et al. 1995, p. 1006.)
The impact of the answer went beyond Neurospora because the real differences among organisms allowed distinct predictions. For example, remembering that per cycled out of phase to frq, we predicted that “light might be expected to entrain the Drosophila oscillator by acting rapidly to depress the level or activity of per or PER” (Crosthwaite et al. 1995, p. 1010). In this way, a mechanism similar to Neurospora's or its converse (light-mediated decay of the clock component) could explain resetting in all organisms; and later work in Drosophila (e.g., Hunter-Ensor et al. 1996) and in mammals (e.g., Shigeyoshi et al. 1997) has shown this to be the universal mechanism for light resetting. “We set out to determine the initial clock-specific event in light-induced resetting of the clock, and determined the light induction of frq to be a likely candidate. The data have in addition provided a substantial and specific confirmation of the major molecular predictions of [Pittendrigh's] model for entrainment, which predicted entrainment via [rapid] light-induced changes in a state variable of the oscillator. Since circadian physiological data, in a wide variety of organisms, has been consistent with the behavioral predictions of this model, it seems likely that most circadian clocks will be reset in a manner similar to that reported here for frq and Neurospora through rapid light-induced changes in an oscillator state variable” (Crosthwaite et al. 1995, p. 1010). The cover of the journal in which Sue's work appeared showed a photo of the Neurospora banding rhythm phase shifted by light and was captioned “How Light Resets the Clock.”
WHERE THE HELL'S THE PROTEIN?
The single, obvious aspect of the story that was missing in Sue's otherwise revealing analysis of resetting was knowledge of FRQ protein levels. It was known that cycloheximide blocked light resetting (Johnson and Nakashima 1990), and we had used inhibitors to show that translation of the induced mRNAs was important; but our inability to follow the protein was becoming an embarrassment. As with the frq message, the problem was subtle and the resolution surprising. As a graduate student, Norm Garceau had chosen the problem of making antisera to FRQ. Because we could not get soluble expression of full-length constructs, Norm raised a series of antisera against the N-terminal portion of FRQ, but we just could not see the protein in cell extracts. After 3 years, he retooled to get full-length expression in baculovirus, and antisera to this FRQ worked well. Surprisingly, however, the Westerns were rather busy and revealed not only changes in the amount, but also in the phosphorylation status of FRQ, and when Norm used λ-phosphatase to resolve the phosphorylations, we were surprised to see two FRQ bands, a long and a short form (Garceau et al. 1997). He and postdoc Yi Liu went on to show that these were the result of temperature modulation of both the choice of starting ATG and the overall amount of FRQ, eliciting a series of studies as to how and why this happens. Temperature-modulated FRQ expression provides a mechanism for temperature resetting of the clock (Liu et al. 1998) and means by which the effective range for rhythmicity could be extended (Liu et al. 1997), the period length fine tuned, and the daily waveform sculpted in a changing environment (Diernfellner et al. 2007). In hindsight, we saw that Norm had been making his extracts from cultures grown at a temperature where relatively little of the long form of FRQ containing his N-terminal antigen was made, which is why he could not see it. In any case, with the ability to see FRQ, we could begin to come to closure on mechanism.
THE CIRCADIAN LOOP AS A CYCLE OF FRQ EXPRESSION, ACTION, AND TURNOVER
At this point, the circadian clock mechanism was still generally imagined as a multi-step process (Figure 1) where FRQ (or in Drosophila PER, now with TIM) would act on other components, which would act on still other components, which, after additional steps, would eventually impact frq (or per/tim) expression. To begin to describe the kinetics of these steps, we (chiefly postdoc Martha Merrow) used Norm's new antisera in a reconstruction experiment to separate the circadian feedback loop into two parts: a repressive phase where frq expression was turned off by expression of frq and FRQ from a heterologous promoter and an activating phase where this repression was lifted.
The time required for each phase could be actively modeled in vivo by using the inducible qa-2 promoter to drive a frq+ transgene in strains bearing the frameshifted null mutant frq9. Lacking negative feedback from FRQ, frq9 mRNA is normally expressed at a high level (Aronson et al. 1994a), but induction of FRQ in such a cell would shut down frq9 mRNA expression; the time required for this shutdown would provide a limit for how long the normal repressive phase should take in the cycle. Subsequently, when the inducer was removed, we could ask how long it took for the heterologous FRQ to decay away and for expression of frq9 mRNA to return to its normal high levels; this would provide a limit for how long the activating phase would normally take.
In a model for the circadian cycle analogous to the cell cycle in which many different subsequent steps and proteins were involved, the steps required for turning frq expression on and off should have occupied only a part of the day, in the same way that the events involved in activation and inactivation of any one cyclin involve only a part of the cell cycle. However, much to our surprise, we saw that repression took ∼3–6 hr and decay of FRQ another ∼14–18 hr, a long period during which de novo FRQ synthesis has stopped. In other words, instead of the expected necessity to invoke a series of events to make up the long ∼22-hr time constant of the clock, it appeared instead that the entire cycle might be described by the simple self-regulated expression and subsequent turnover of FRQ, with the long circadian time constant largely attributable just to the factors controlling inactivation/turnover of FRQ. We surmised that “this circadian oscillator can be largely described by the events and molecules surrounding activation and repression of frq alone” (Merrow et al. 1997, p. 3882). Even in hindsight, this seems a pretty good guess, and later work (Liu et al. 2000; Ruoff et al. 2005) clearly showed that changes in FRQ phosphorylation impacted its turnover and dictated circadian period length. Merrow et al. (1997), with the separate but nearly coincident demonstration of a daily cycle of synthesis, processive phosphorylation, and ultimate precipitous turnover of FRQ (Garceau et al. 1997; Liu et al. 1997), provided a plausible molecular basis for the circadian-cycle-as-an-FRQ-cycle idea, which has subsequently been shown to be true. In Neurospora, as in other eukaryotic circadian systems, phosphorylation-mediated turnover of the negative elements of the clock loop are principal factors in determining the length of the circadian day. More generally, the kinetics of the circadian cycle in Neurospora, in Drosophila, and in mammals is described principally as a cycle in the expression of clock components, their action in turning off their own expression and their subsequent turnover in a single feedback loop. Although in the ensuing decade we have learned a great deal more about additional interlocked loops that affect this core, it remains that such a core loop is the only aspect of the clock that appears essential for expression of all overt true circadian rhythms in eukaryotes.
MOVING FROM MODELS TO MECHANISMS
At this point in time, between 1996 and 1997, it was largely agreed within the rhythms community that the core of the circadian oscillator in the systems where it was at all understood (flies and fungi) was a transcription–translation-based feedback loop. Ongoing Drosophila genetics in Mike Young's lab had provided an additional essential player in the fly clock, timeless (Sehgal et al. 1994), whose molecular rhythms in expression were interdependent with those of per. Thus, data showed rhythms in transcript levels from all the genes encoding essential clock proteins (PER, FRQ, and TIM), and there was compelling evidence that this daily rhythmic transcription both was needed and set the phase of the clock. The model accounted well for the early physiological data describing the period and phase-altering effects of inhibitors of translation (e.g., Jacklet 1977; Dunlap et al. 1980; Dunlap and Feldman 1988) and provided explanations for how light resetting (Crosthwaite et al. 1995) and later temperature resetting (Liu et al. 1998) worked. However, and despite all the apparent progress and developing consensus, it was still impossible to associate any biochemical activity with any known clock molecule. Several missteps very early in the game had briefly led to claims that “The period clock locus of D. melanogaster codes for a proteoglycan” (the title of Reddy et al. 1986) or that it affected cell–cell coupling (Bargiello et al. 1987), but both claims sunk under more careful analysis; the series of errors as well as triumphs from this era have been candidly (Hall 1995) and exhaustively (Hall 2003) reviewed elsewhere. The truth was that the primary sequences of FRQ or PER or TIM did not really suggest anything about what they might do, and there was only a very (in hindsight, pathetically) weak bit of sequence similarity between FRQ and PER in regions of each having repeats of (Thr/Ser)Gly (McClung et al. 1989). And yet, the commonality of cycling clock transcripts was consistent with assumptions about common mechanisms, and it was expected that at some point identifiably similar molecules with identifiably similar functions would emerge.
The unexpected collapse of models that invoked sequential action of proteins in many steps focused our interest on what was actually driving frq expression in the cell. It now seemed as if this activator might be the only remaining component in the clock loop. Identifying it might be sufficient to close the loop. And it might turn up the first example of a clock protein with an identifiable biochemical function, a function that would nail down the core mechanisms as well as provide a candidate for phylogenetic conservation.
A BIOCHEMICAL ACTIVITY FOR A CLOCK MOLECULE: CLOSING THE CIRCADIAN FEEDBACK LOOP
The data from Martha's enlightening qa-2 expression and reconstruction experiments (Merrow et al. 1997) implied that FRQ acted upon a single element or complex to regulate its own expression, but the identity of this positive element in Neurospora, as in all other circadian systems, was completely unknown. We came upon it in an unexpected manner in a collaborative project with Jennifer Loros through simply following up the light-resetting work.
Neurospora is normally yellow/orange in color due to the presence of carotenoids whose production in vegetative tissue is induced by light; mutational analyses many decades earlier had identified genes required for this light induction, white collar-1 (wc-1) and wc-2 (Perkins et al. 1962). [They were so named because of their appearance on slants. Normally, mycelia of Neurospora will cover an agar slant, eventually giving rise to aerial hyphae that differentiate to yield asexual spores (conidia), so a side view of the top of a slant on a lab bench will reveal a ring of agar covered by yellow mycelia topped by yellow spores. Carotenoid production in spores is constitutive, but in mycelia it is induced by light. As a result, in agar slants of blind mutants viewed from the side, white mycelia will form a ring (a collar) around the top of a slant beneath the yellow spores; hence the name white collar-1 and white collar-2 for the two genes whose mutation yielded blind mutants.] Since the wc mutants were blind, it was natural for us to ask whether these wc genes were required for the light induction of frq, and to no one's surprise, they were. However, we were surprised to see that even in the dark, the levels of frq mRNA in both wc-1 and wc-2 mutant strains were quite low. Moreover, the strains bearing these mutations were actually overtly arrhythmic (Crosthwaite et al. 1997). This had been observed before but shrugged off on the assumption that, because the mutant strains were known to be blind, the clocks in these cells were probably not inherently arrhythmic but simply asynchronous and incapable of being synchronized by the light that they could not see. The absence of light induction of frq was consistent with this, but the low level of frq mRNA in both mutants suggested another possibility: that WC-1 and WC-2 actually had an additional and qualitatively distinct function in that they were required in the dark to activate frq expression. In other words, these mutants would define the missing link in the circadian feedback loop. The controls to test the arrhythmicity-via-asynchronicity assumption were simple: we could use a temperature pulse, which works independently of photoreceptors, to reset the clock and initiate rhythmicity. The temperature treatments failed to elicit rhythms, and molecular analyses confirmed our hunch in that the wc mutants lacked both overt and molecular rhythms in frq or FRQ expression: wc-1 and wc-2 were essential clock genes. [The article, even in hindsight, makes interesting reading to me as much because of what is correct or correctly predicted as because of what is not. The alleles of wc-1 and wc-2 that we used had been described as nulls, but subsequent molecular and physiological analyses showed that they were in fact only extreme hypomorphs (Collett et al. 2002; Lee et al. 2003); the resulting residual activity played out in some ways that required some explanatory gymnastics in the prose, but in the end this did not interfere with reaching the right answer. With the ready availability of engineered whole-gene deletions now in Neurospora (Colot et al. 2006), this need never again be a problem.] Taken altogether, the evidence was consistent with WC-1 and WC-2 encoding the molecular activator of frq, the first positive element in any circadian feedback loop. But what did they do?
Fortuitously, while Sue's work was being completed, Giuseppe Macino and colleagues had cloned wc-1 (Ballario et al. 1996) and wc-2 (Linden and Macino 1997) and had shown them to act together as a “white collar complex.” These protein–protein interactions were mediated in each protein by PAS domains, a motif originally, if unwittingly, discovered in PER (Huang et al. 1993). Moreover, WC-1/WC-2 bound to the DNA of pertinent promoters (Carattoli et al. 1994) and had sequences consistent with their functions as card-carrying dyed-in-the-wool GATA-family DNA-binding transcription factors, complete with zinc-finger DNA-binding domains, nuclear localization signals, and Gln-rich or acidic activation domains (reviewed in Linden et al. 1999).
So here, then, was the missing link that tied up many a crucial loose end. In a circadian feedback loop believed to be based on daily transcription and translation, two novel clock proteins were identified as transcription factors: a biochemical function for a clock molecule wholly consistent with the anticipated molecular basis of the clock. As transcription factors, the means by which they could provide the anticipated activating function in the loop by driving frq expression was obvious, so now the predicted activating and repressing components of the loop were known. The identification of PAS domains in the WC proteins provided the long-anticipated example of a motif that was phylogenetically conserved among diverse clock proteins and led us to predict that “the association between clock molecules and PAS domains may extend well beyond the fungi and invertebrates” (Crosthwaite et al. 1997, p. 768), as indeed it does: PAS domains are found in the activators of every circadian feedback loop known in the fungi and animals—from Neurospora to Drosophila to mice, fish, and humans. In each of these systems, two proteins interact via PAS domains to make a heterodimeric complex (such as WC-1/WC-2) that drives rhythmic expression of the negative element(s) of the feedback loop: WC-1/WC-2 in Neurospora drives the negative element frq; CYC/CLK in Drosophila drives per and tim; BMAL1/CLOCK and BMAL1/NPAS2 in mammals drive the analogous negative elements. WC-1/WC-2 was the first of these to be described.
Another aspect of the sequence conservation among PAS domains also struck us: among the many proteins having PAS domains were five photoreceptors, and in fact so many have since been identified that they now describe a specific subclass of PAS domains, the LOV domain (for light/oxygen/voltage sensing; Huala et al. 1997). wc-1 and wc-2, of course, had been identified first as essential for photoreception, but now we had associated them with a novel and exclusively dark function: maintenance of a sustained circadian oscillation. Observing more broadly, there was a well-appreciated and universal association of circadian rhythmicity with the capacity for photoreception—in animals often in the same tissues and now in Neurospora in the same proteins. For a photoreceptor to transduce a gradual lights-on dawn signal into a pulse rather than a ramp or step function, there has to be a light-triggered deactivation of the photoreceptor molecule—a negative feedback loop. Given this, along with the molecular conservation of PAS domains in clock molecules and in photoreceptors, the possibility occurred to us that, perhaps, clock feedback loops first arose from modification of negative feedback loops associated with photoperception. As we wrote, “the widespread occurrence of PAS domains in light- and clock-associated proteins, combined with the predicted ancient origins of circadian rhythmicity, suggest that circadian clocks may have arisen from the cellular processes associated with the perception of the daily light-dark cycle and the transduction of this information in the cell to regulate metabolism in response to light” (Crosthwaite et al. 1997, p. 768). The prediction that PAS domains might recur in clock proteins was borne out later when PAS domains turned up in the first mammalian clock protein, CLOCK (King et al. 1997), a similarity Michael Rosbash described as “a molecular link” between the mammalian clock and those of lower organisms (Barinaga 1997). Extended sequence similarity between WC-1 and BMAL1 (Lee et al. 2000) is consistent with this common origin for rhythm generators, as is the striking similarity in the overall design of the transcription–translation feedback loop. It was and remains a nice idea for the origin of circadian clocks. The beauty of such theories about how processes evolve is that they are as impossible to prove as they are to disprove.
THE END OF THE BEGINNING
When WC-1 and WC-2 surfaced in 1997 as clock proteins and activators in the feedback loop, they were the only game in town. Within 15 months, however, the clock field more resembled a convention. As noted above, mammalian CLOCK appeared soon after, all by itself, but its PAS domains along with the precedents of WC-1/WC-2 and other nonrhythm-associated PAS heterodimers suggested that it likely had a partner. In studies appearing a bit over a year later (but initiated years before), classic unbiased forward genetic screens for rhythm mutants in Drosophila identified a CLOCK homolog (Clk; Allada et al. 1998) as well as its partner in the heterodimeric complex, CYCLE (Rutila et al. 1998). In a manner completely analogous to the wc-1 and wc-2 mutants, loss of either cycle or Clk abrogated behavioral rhythms as well as flattened transcription of per and tim, thus satisfyingly confirming through phenotype-driven genetics the central importance of these PAS-PAS heterodimers to overt behavioral circadian rhythms. Molecular biological analyses of the interactions and partners continued apace: low-stringency annealing using probes derived from CLOCK (Darlington et al. 1998) as well as screening of EST databases (Bae et al. 1998) co-identified Drosophila Clk, and the clone was used to show in vitro that the corresponding protein acted to induce transcription of per and tim, the genes encoding their own inhibitors. The partner for CLOCK in mammals was later identified through molecular biology, both via a yeast two-hybrid screen of hamster hypothalamic cDNAs and when an open-ended screen to catalog interactions among bHLH-PAS proteins turned up a strong interaction between MOP3 (a.k.a. BMAL1) and CLOCK (Gekakis et al. 1998; Hogenesch et al. 1998). All of this appeared between mid-1997 and late 1998.
During June of 1998, coincident in the journal with several of these articles, a Perspectives about these various accomplishments prepared by me appeared under the title “An End in the Beginning” (Dunlap 1998). I used this title both because it brought to mind the idea of a cycle and because of where I thought the data and insights had placed the rhythms field. Physiological studies, beginning in the 1940s (and before) and reaching a crescendo in the 1970s, had defined and refined the definition of circadian rhythmicity until it was sufficiently explicit to allow informative genetic screens. This era yielded the restricted vocabulary (circadian vs. diurnal, free-running, phase response curve, entrainment, limit cycle, etc.) with which I have had to burden the reader, but that allowed the phenomenon to be precisely discussed and mutants to be characterized. This era also revealed the importance of critical daily episodes of clock protein translation. Reports of mutant screens in Drosophila and Neurospora in the 1970s, followed by cloning of the pertinent genes in the 1980s, were all directed toward developing a picture of how a clock could be assembled as much as toward identifying individual components. By mid-1997, in Neurospora, with the identification of both repressing and activating factors in the negative feedback loop, knowledge of their biochemical activities and proof that these cycling activities did not just reflect the clock but were the clock, this picture was largely in place, and by mid-1998 the picture (Figure 4) had been fleshed out in other model systems, flies, and mice.
Figure 4.—
Circadian cycles as negative feedback loops. Transcription factors are heterodimers: two proteins interacting with each other via mutual PAS domains. These heterodimers drive expression of proteins that act as negative elements, turning down the activity of their activators. E-box is the cis-acting element through which trans-acting heterodimers act; P signifies phosphorylation. Several elements/aspects were educated guesses. (Figure is from Dunlap 1998.)
It was easy to assert (Dunlap 1998) that there would be a great may additional things to discover as indeed there have been; the number of “circadian” articles continues to grow exponentially. But qualitatively the nature of the questions has changed. Once the pattern was elucidated on the basis of work from Neurospora and Drosophila, additional components could be much more easily identified through molecular biological rather than genetic routes. Indeed, although dozens of clock components and ancillary factors (kinases, transcriptional cofactors, etc.) have been reported in fungi, flies, and mammalian systems since 1998, nearly all have arisen first or simultaneously from biochemical or molecular biological studies rather than from genetic screens, although reverse genetic analysis of putative components remains the gold standard for discerning significance. The year 1998 is a good point to break as I am not sure that I have sufficient perspective on the past decade yet to do it justice.
The years 1997–1998 marked “the end of the beginning” phase of rhythms research, the era when the field succeeded in figuring out the big picture, the basic molecular logic underlying the oscillator and the aspect(s) of metabolism associated with it. I was blessed to fall upon a terrific and molecularly untouched problem, backstopped by decades of biology, at a time when it was possible to develop the right tools to answer the right questions. It was also a gift to work with some remarkable students and postdocs who were equally attracted to the problem and saw, in the same way that I did, the virtues of this marvelous system. Many of them have since gone on to run labs of their own in Neurospora. It was an exciting period, this time when the problem of how clocks work was cracked, and work on Neurospora played an undeniably prominent role in this story.
Acknowledgments
I thank Jennifer Loros and Jeff Hall for their careful reading the draft in the expectation that they will forgive me for not including all their suggestions. Thanks go to Jim Crow and Bill Dove for useful feedback as well as for exceptionally patient encouragement while this piece evolved. I remain grateful to the students and postdocs, named and unnamed, upon whose insights and hard work this story is built, and I look forward to seeing their perspectives on these and later events, all in the fullness of time. Our work has been supported by grants from the National Science Foundation and the National Institutes of Health, (GM34985, GM68087, and GM83336).
References
- Allada, R., N. E. White, W. V. So, J. C. Hall and M. Rosbash, 1998. A mutant Drosophila homolog of mammalian CLOCK disrupts circadian rhythms and transcription of period and timeless. Cell 93: 805–814. [DOI] [PubMed] [Google Scholar]
- Aronson, B., K. Johnson, J. J. Loros and J. C. Dunlap, 1994. a Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science 263: 1578–1584. [DOI] [PubMed] [Google Scholar]
- Aronson, B. D., K. A. Johnson and J. C. Dunlap, 1994. b The circadian clock locus frequency: a single ORF defines period length and temperature compensation. Proc. Natl. Acad. Sci. USA 91: 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson, B. D., K. M. Lindgren, J. C. Dunlap and J. J. Loros, 1994. c An efficient method of gene disruption in Neurospora crassa and with potential for other filamentous fungi. Mol. Gen. Genet. 242: 490–494. [DOI] [PubMed] [Google Scholar]
- Bae, K., C. Lee, D. Sidote, K.-Y. Chuang and I. Edery, 1998. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 18: 6142–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo et al., 1996. White collar-1, a central regulator of blue-light responses in Neurospora crassa, is a zinc-finger protein. EMBO J. 15: 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Bargiello, T. A., and M. W. Young, 1984. Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. USA 81: 2142–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiello, T. A., L. Saez, M. K. Baylies, G. Gasic, M. W. Young et al., 1987. The Drosophila clock gene per affects intercellular junctional communication. Nature 328: 686–691. [DOI] [PubMed] [Google Scholar]
- Barinaga, M., 1997. New clues found to circadian clocks—including mammals'. Science 276: 1030–1031. [DOI] [PubMed] [Google Scholar]
- Barnett, A., 1966. A circadian rhythm of mating type reversals in Paramecium multimicronucleatum, syngen 2, and its genetic control. J. Cell. Physiol. 67: 239–270. [DOI] [PubMed] [Google Scholar]
- Brenner, S., W. Dove, I. Herskovitz and R. Thomas, 1990. Genes and development: molecular and logical themes. Genetics 126: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, V. G., 1972. Mutants of the biological clock in Chlamydomonas reinhardtii. Genetics 70: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli, A., C. Cogoni, G. Morelli and G. Macino, 1994. Molecular characterization of upstream regulatory sequences controlling the photoinduced expression of the al-3 gene of Neurospora crassa. Mol. Microbiol. 13: 787–795. [DOI] [PubMed] [Google Scholar]
- Case, M. E., M. Schweizer, S. R. Kushner and N. H. Giles, 1979. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. USA 76: 5259–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett, M. A., N. Garceau, J. C. Dunlap and J. J. Loros, 2002. Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics 160: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H., G. Park, J. Jones, G. Turner, K. Borkovich et al., 2006. High throughput knockout of transcription factors in Neurospora reveals diverse phenotypes. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite, S. C., J. J. Loros and J. C. Dunlap, 1995. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81: 1003–1012. [DOI] [PubMed] [Google Scholar]
- Crosthwaite, S. C., J. C. Dunlap and J. J. Loros, 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276: 763–769. [DOI] [PubMed] [Google Scholar]
- Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Stankis, N. Gekakis et al., 1998. Closing the circadian loop: CLOCK induced transcription of its own inhibitors, per and tim. Science 280: 1599–1603. [DOI] [PubMed] [Google Scholar]
- Diernfellner, A., H. V. Colot, O. Dintsis, J. J. Loros, J. C. Dunlap et al., 2007. Long and short isoforms of Neurospora clock protein FRQ support temperature compensated circadian rhythms. FEBS Lett. 581: 5759–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., 1993. Genetic analysis of circadian clocks. Annu. Rev. Physiol. 55: 683–728. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., 1998. An end in the beginning. Science 280: 1548–1549. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., and J. F. Feldman, 1988. On the role of protein synthesis in the circadian clock of Neurospora crassa. Proc. Natl. Acad. Sci. USA 85: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., and J. W. Hastings, 1981. a Biochemistry of dinoflagellate bioluminescence: purification and characterization of dinoflagellate luciferin from Pyrocystis lunula. Biochemistry 20: 983–989. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., and J. W. Hastings, 1981. b The biological clock in Gonyaulax controls luciferase activity by regulating turnover. J. Biol. Chem. 256: 10509–10518. [PubMed] [Google Scholar]
- Dunlap, J. C., W. R. Taylor and J. W. Hastings, 1980. The effects of protein synthesis inhibitors on the Gonyaulax clock I: phase shifting effects of cycloheximide. J. Comp. Physiol. 138: 1–8. [Google Scholar]
- Dunlap, J. C., J. W. Hastings and O. Shimomura, 1981. Dinoflagellate luciferin is structurally related to chlorophyll. FEBS Lett. 135: 273–276. [Google Scholar]
- Eskin, A., 1979. Identification and physiology of circadian pacemakers. Fed. Proc. 38: 2570–2572. [PubMed] [Google Scholar]
- Ewer, J., M. Rosbash and J. C. Hall, 1988. An inducible promoter fused to the period gene of Drosophila conditionally rescues adult per-mutant arrhythmicity. Nature 333: 82–84. [DOI] [PubMed] [Google Scholar]
- Feldman, J. F., 1982. Genetic approaches to circadian clocks. Annu. Rev. Plant Physiol. 33: 583–608. [Google Scholar]
- Feldman, J. F., and N. Waser, 1971. New mutations affecting circadian rhythmicity in Neurospora, pp. 652–656 in Biochronometry, edited by M. Menaker. National Academy of Sciences, Washington, DC.
- Friesen, O., and G. Block, 1984. What is a biological oscillator? Am. J. Physiol. 246: R847–R851. [DOI] [PubMed] [Google Scholar]
- Garceau, N., Y. Liu, J. J. Loros and J. C. Dunlap, 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476. [DOI] [PubMed] [Google Scholar]
- Gekakis, N., D. Stankis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher et al., 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–1569. [DOI] [PubMed] [Google Scholar]
- Giles, N. H., M. E. Case, J. Baum, R. Geever, L. Huiet et al., 1985. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol. Rev. 49: 338–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J. C., 1995. Tripping along the trail to the molecular mechanisms of biological clocks. Trends Neurosci. 18: 230–240. [DOI] [PubMed] [Google Scholar]
- Hall, J. C., 2003. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv. Genet. 48: 1–280. [DOI] [PubMed] [Google Scholar]
- Hardin, P. E., J. C. Hall and M. Rosbash, 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Hastings, J. W., 1960. Biochemical aspects of rhythms: phase shifting by chemicals. Cold Spring Harbor Symp. Quant. Biol. 25: 131–143. [DOI] [PubMed] [Google Scholar]
- Hogenesch, J. B., Y.-Z. Gu, S. Jain and C. A. Bradfield, 1998. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 95: 5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen et al., 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123. [DOI] [PubMed] [Google Scholar]
- Huang, Z. J., I. Edery and M. Rosbash, 1993. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature 364: 259–262. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor, M., A. Ousley and A. Sehgal, 1996. Regulation of the Drosophila protein TIMELESS suggests a mechanism for resetting the circadian clock by light. Cell 84: 677–685. [DOI] [PubMed] [Google Scholar]
- Jacklet, J. W., 1969. Circadian rhythm of optic nerve impulses recorded in darkness from isolated eye of Aplysia. Science 164: 562–563. [DOI] [PubMed] [Google Scholar]
- Jacklet, J. W., 1977. Neuronal circadian rhythm: phase shifting by a protein synthesis inhibitor. Science 198: 69–71. [DOI] [PubMed] [Google Scholar]
- Johnson, C. H., 1990. An atlas of phase response curves for circadian and circatidal rhythms. Vanderbilt University, Nashville, TN. http://www.cas.vanderbilt.edu/johnsonlab/prcatlas/index.html.
- Johnson, C. H., and H. Nakashima, 1990. Cycloheximide inhibits light-induced phase shifting of the circadian clock in Neurospora. J. Biol. Rhythms 5: 159–167. [DOI] [PubMed] [Google Scholar]
- Johnson, C. H., J. Elliott, R. Foster, K.-I. Honma and R. Kronauer, 2004. Fundamental properties of circadian rhythms, pp. 67–105 in Chronobiology: Biological Timekeeping, edited by J. C. Dunlap, J. J. Loros and P. Decoursey. Sinauer Associates, Sunderland, MA.
- Khalsa, S. B. S., and G. D. Block, 1992. Stopping the biological clock with inhibitors of protein synthesis. Proc. Natl. Acad. Sci. USA 89: 10862–10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, D., Y. Zhao, A. Sangoram, L. Wilsbacher, M. Tanaka et al., 1997. Positional cloning of the mouse circadian CLOCK gene. Cell 89: 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka, R. J., and S. Benzer, 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., J. J. Loros and J. C. Dunlap, 2000. Interconnected feedback loops in the Neurospora circadian system. Science 289: 107–110. [DOI] [PubMed] [Google Scholar]
- Lee, K., J. C. Dunlap and J. J. Loros, 2003. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., and G. Macino, 1997. White collar-2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 16: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., P. Ballario, G. Arpaia and G. Macino, 1999. Seeing the light: news in Neurospora blue light signal transduction. Adv. Genet. 41: 35–54. [DOI] [PubMed] [Google Scholar]
- Liu, Y., N. Garceau, J. J. Loros and J. C. Dunlap, 1997. Thermally regulated translational control mediates an aspect of temperature compensation in the Neurospora circadian clock. Cell 89: 477–486. [DOI] [PubMed] [Google Scholar]
- Liu, Y., M. Merrow, J. J. Loros and J. C. Dunlap, 1998. How temperature changes reset a circadian oscillator. Science 281: 825–829. [DOI] [PubMed] [Google Scholar]
- Liu, Y., J. Loros and J. C. Dunlap, 2000. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl. Acad. Sci. USA 97: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros, J. J., J. C. Dunlap, A. Mehra, M. Shi, L. F. Larrondo et al., 2008. Circadian output, input and intracellular oscillators — insights into the circadian systems of single cells. Cold Spring Harb. Symp. Quant. Biol. 72: (in press). [DOI] [PMC free article] [PubMed]
- Loros, J. J., A. Richman and J. F. Feldman, 1986. A recessive circadian clock mutant at the frq locus in Neurospora crassa. Genetics 114: 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros, J. J., S. A. Denome and J. C. Dunlap, 1989. Molecular cloning of genes under the control of the circadian clock in Neurospora. Science 243: 385–388. [DOI] [PubMed] [Google Scholar]
- McClung, C. R., 2001. Circadian rhythms in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 139–162. [DOI] [PubMed] [Google Scholar]
- McClung, C. R., B. A. Fox and J. C. Dunlap, 1989. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature 339: 558–562. [DOI] [PubMed] [Google Scholar]
- Menaker, M., D. S. Farner, E. Gwinner, J. W. Hastings, F. B. Salisbury et al., 1971. Preface, pp. v–vi in Biochronometry, edited by M. Menaker. National Academy of Sciences, Washington, DC.
- Merrow, M., N. Garceau and J. C. Dunlap, 1997. Dissection of a circadian oscillation into discrete domains. Proc. Natl. Acad. Sci. USA 94: 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, S., M. Geusz, J. Zaritsky and G. D. Block, 1993. Circadian rhythm in membrane conductance expressed in isolated neurons. Science 259: 239–241. [DOI] [PubMed] [Google Scholar]
- Nakashima, H., 1981. A liquid culture system for the biochemical analysis of the circadian clock of Neurospora. Plant Cell Physiol. 22: 231–238. [Google Scholar]
- Perkins, D. D., M. Glassey and B. A. Bloom, 1962. New data on markers and rearrangements in Neurospora. Can. J. Genet. Cytol. 4: 187–205. [Google Scholar]
- Pittendrigh, C. S., 1967. Circadian rhythms. 1. The driving oscillator and its assay in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 58: 1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C. S., 1993. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55: 17–54. [DOI] [PubMed] [Google Scholar]
- Pregueiro, A., Q. Liu, C. Baker, J. C. Dunlap and J. J. Loros, 2006. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science 313: 644–649. [DOI] [PubMed] [Google Scholar]
- Ralph, M. R., and M. Menaker, 1985. Bicuculline blocks circadian phase delays but not advances. Brain Res. 325: 362–365. [DOI] [PubMed] [Google Scholar]
- Reddy, P., W. A. Zehring, D. A. Wheeler, V. Pirrotta, C. Hadfield et al., 1984. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38: 701–710. [DOI] [PubMed] [Google Scholar]
- Reddy, P., A. C. Jacquier, N. Abovich, G. Petersen and M. Rosbash, 1986. The period clock locus of D. melanogaster codes for a proteoglycan. Cell 46: 53–61. [DOI] [PubMed] [Google Scholar]
- Robertson, M., 1975. Circadian rhythms. Nature 258: 291. [Google Scholar]
- Ruoff, P., J. J. Loros and J. C. Dunlap, 2005. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc. Natl. Acad. Sci. USA 102: 17681–17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila, J. E., V. Suri, M. Le, W. V. So, M. Rosbash et al., 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93: 805–813. [DOI] [PubMed] [Google Scholar]
- Science News and Editorial Staffs, 1997. Breakthrough of the year. The runners-up. Science 278: 2039–2042. [PubMed] [Google Scholar]
- Science News and Editorial Staffs, 1998. Breakthrough of the year. The runners-up. Science 282: 2157–2161. [PubMed] [Google Scholar]
- Sehgal, A., J. Price, B. Man and M. Young, 1994. Loss of circadian behavioral rhythms and per oscillations in the Drosophila mutant timeless. Science 263: 1603–1606. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi, Y., K. Taguchi, S. Yamamoto, S. Takeida, L. Yan et al., 1997. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Siwicki, K. K., C. Eastman, G. Petersen, M. Rosbash and J. C. Hall, 1988. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1: 141–150. [DOI] [PubMed] [Google Scholar]
- Sweeney, B. M., 1976. Circadian rhythms: definition and general characterization, pp. 77–83 in The Molecular Basis of Circadian Rhythms, edited by J. W. Hastings and H.-G. Schweiger. Dahlem Konferenzen, Berlin.
- Winfree, A. T., 1967. Biological rhythms and the behavior of populations of coupled oscillators. J. Theor. Biol 16: 15–42. [DOI] [PubMed] [Google Scholar]
- Winfree, A. T., 1971. Corkscrews and singularities in fruitflies: resetting behaviour of the circadian eclosion rhythm, pp. 81–106 in Biochronometry, edited by M. Menaker. National Academy of Sciences, Washington, DC.
- Winfree, A. T., and G. M. Twaddle, 1981. The Neurospora mycelium as a two-dimensional continuum of coupled circadian clocks, pp. 237–249 in Mathematical Biology, edited by T. A. Burton. Pergammon Press, New York.
- Zimmermann, C. R., W. C. Orr, R. F. Leclerc, E. C. Barnard and W. E. Timberlake, 1980. Molecular cloning and selection of genes regulated in Aspergillus development. Cell 21: 709–715. [DOI] [PubMed] [Google Scholar]