Abstract
Transcription of the Neurospora crassa gene con-10 is induced during conidiation and following exposure of vegetative mycelia to light, but light activation is transient due to photoadaptation. We describe mutational analyses of photoadaptation using a N. crassa strain bearing a translational fusion of con-10, including its regulatory region, to a selectable bacterial gene conferring hygromycin resistance (hph). Growth of this strain was sensitive to hygromycin, upon continuous culture in the light. Five mutants were isolated that were resistant to hygromycin when cultured under constant light. Three mutant strains displayed elevated, sustained accumulation of con-10∷hph mRNA during continued light exposure, suggesting that they bear mutations that reduce or eliminate the presumed light-dependent repression mechanism that blocks con-10 transcription upon prolonged illumination. These mutations altered photoadaptation for only a specific group of genes (con-10 and con-6), suggesting that regulation of photoadaptation is relatively gene specific. The mutations increased light-dependent mRNA accumulation for genes al-1, al-2, and al-3, each required for carotenoid biosynthesis, resulting in a threefold increase in carotenoid accumulation following continuous light exposure. Identification of the altered gene or genes in these mutants may reveal novel proteins that participate in light regulation of gene transcription in fungi.
FUNGI use light as an environmental signal in regulating behavior and development, presumably to optimize spore production and dispersal (Corrochano and Galland 2006). The ascomycete Neurospora crassa is a model organism that can be used for studies on the molecular mechanisms of photoreception. Responses to blue light in N. crassa include the following: induction of sporulation and sexual development, induction of synthesis of mycelial carotenoids, and entrainment of the circadian clock, all of which require the products of the wc-1 and wc-2 genes (Linden et al. 1997a; Collett et al. 2002; Lee et al. 2003). WC-1, the product of wc-1, is a zinc-finger protein with a chromophore-binding domain, and it also contains PAS domains used for protein–protein interactions (Ballario et al. 1996; Crosthwaite et al. 1997). The chromophore-binding domain binds the flavin chromophore FAD, allowing WC-1 to act as a photoreceptor (Froehlich et al. 2002; He et al. 2002). WC-2, the product of wc-2, is also a zinc-finger protein; WC-2 interacts with WC-1 (Linden and Macino 1997), to form a white collar complex (WCC). This complex, upon light exposure, binds transiently to the promoters of light-inducible genes, presumably to activate their transcription ( Froehlich et al. 2002; He and Liu 2005; Belden et al. 2007). The structures and functions of the Neurospora WC proteins have been reviewed recently (Liu et al. 2003; Dunlap and Loros 2005). The existence of proteins similar to WC-1 and WC-2 in ascomycetes, basidiomycetes, and zygomycetes, all fungi, has led to the proposal that the WCC arose early in fungal evolution, and was designed to regulate fungal photoresponses by serving both as a photoreceptor and a transcription factor (Purschwitz et al. 2006; Corrochano 2007; Herrera-Estrella and Horwitz 2007).
Organisms that sense a wide range of light intensities require mechanisms of range adjustment that allow them to adapt to the changing light intensities in their environment (Galland 1991). Light activation of transcription of some genes of N. crassa is transient, transcription of these responding genes ceases after the illumination time has been extended, and further incubation in the dark is required before they are again transcribed in response to light (Lauter and Yanofsky 1993; Arpaia et al. 1999; Schwerdtfeger and Linden 2001, 2003). This behavioral feature, named “transcriptional adaptation to light” or “photoadaptation,” requires the product of the gene vivid, a small photoreceptor with a flavin-binding domain similar to that found in WC-1 (Heintzen et al. 2001; Schwerdtfeger and Linden 2001, 2003; Shrode et al. 2001; Zoltowski et al. 2007). Photoadaptation might allow a quick transcriptional response after environmental changes in light intensities, like those observed at dawn or sunset in nature, or to allow the precise orientation of reproductive or photosynthetic organs toward a source of bright light in a shaded environment. The response to changes in light intensities, and the absence of a response in an environment with constant light may have an evolutionary advantage for organisms using light as a source of energy or as a source of environmental information. Photoadaptation has been shown to be modified by mutations or inhibitors of protein kinase C (Arpaia et al. 1999; Franchi et al. 2005). In addition, photoadaptation correlates with the phosphorylation status of the photoreceptor WC-1 (He and Liu 2005). Thus, light-dependent activation of gene transcription is observed in mycelia that have been exposed to light for 5 min, but the transient light-dependent phosphorylation of WC-1 is only observed following 10–30 min of light exposure (He and Liu 2005). Since phosphorylation of the WCC reduces its capacity to bind to light-inducible promoters, it has been proposed that light-dependent phosphorylation may play a critical role in photoadaptation (He and Liu 2005). The lack of transcriptional response during adaptation to light can be reversed by increasing the light intensity (Arpaia et al. 1999; Schwerdtfeger and Linden 2001, 2003), suggesting that adaptation to light is probably a consequence of photoreceptor desensitization.
Light regulation of transcription of the N. crassa gene con-10 has been investigated in detail. The gene con-10 is highly expressed during development of conidia, the asexual spores produced by N. crassa during vegetative reproduction (Roberts et al. 1988; Sachs and Yanofsky 1991; Springer and Yanofsky 1992; Springer et al. 1992). Messenger RNA from con-10 accumulates in vegetative mycelia upon light exposure (Lauter and Russo 1991; Lauter and Yanofsky 1993; Corrochano et al. 1995), and con-10's promoter has been shown to contain several developmental, light, and clock-controlled DNA elements (Corrochano et al. 1995; Lee and Ebbole 1998a). The activation of con-10 transcription by light is transient; maximum con-10 mRNA accumulation is observed after 0.5–1 hr of light exposure, whereas con-10 mRNA accumulation is reduced upon longer light exposures (Lauter and Yanofsky 1993). Thus, mycelia exposed to light for ≥4 hr do not have detectable levels of con-10 mRNA, a clear indication of photoadaptation (Lauter and Yanofsky 1993). The function of CON-10 is unknown, and a strain with a deletion in con-10 doesn't show any clear phenotype (Springer et al. 1992). However, con-10 homologs have been described in other fungi and in Escherichia coli (Corrochano et al. 1995; Lee and Ebbole 1998b), suggesting that this protein should have a relevant role in the biology of Neurospora and other microbes.
Photoadaptation has been investigated in the zygomycete fungus Phycomyces blakesleeanus. The transient photoactivation of gene transcription observed for the gene encoding the heat-shock protein HSP100 in P. blakesleeanus is not the result of photoreceptor desensitization, as shown by the absence of light-dependent mRNA accumulation after changes in light intensities. Rather, photoadaptation for the HSP100 gene in P. blakesleeanus seems to be a consequence of a reduction in the level of mRNA for the photoreceptor MADA, a WC-1 homolog, after long light exposures (Rodríguez-Romero and Corrochano 2006). Photoadaptation has also been described for some plant genes (Mochizuki et al. 2004). Thus transient light-dependent activation of gene transcription appears to be a general feature of light perception in many organisms.
Identification of the proteins involved in photoadaptation will aid in our understanding of how light serves as an environmental signal for fungal development and behavior. Isolation of mutants altered in photoadaptation should also help in identifying the proteins involved in regulation of fungal photoreception. In addition, studies with light adaptation mutants may aid in determining the mechanism(s) responsible for transient light activation of gene transcription.
In this report we describe a strategy for the isolation of N. crassa mutants altered in their adaptation of gene transcription to light. By determining its mechanism of regulation we hope to learn more about CON-10 and its functions. We used a strain carrying a translational fusion of the regulatory sequence from the gene con-10 to the hygromycin resistance gene hph, and selected for mutants resistance to hygromycin when cultured with this drug under constant light. These new mutants show altered adaptation to light for only a group of light regulated genes, suggesting the existence of several gene-specific mechanisms for adaptation to light in N. crassa. The phenotypic characterization of these new mutants suggests that they have lost the function of a mechanism that blocks specific gene transcription upon prolonged illumination.
MATERIALS AND METHODS
Strains and culture conditions:
Strain CH10 contains the con-10 regulatory region followed by a translational fusion of part of the con-10 ORF to the hygromycin phosphotransferase gene (hph), integrated following the his-3 locus (Madi et al. 1994). The fusion contained the con-10-associated sequence from nucleotides 1139–1935 (Roberts et al. 1988), corresponding to base pair −517 (relative to the con-10 transcription start site) to codon 40 of the con-10 ORF. The con-10 upstream region contains the sequences responsible for light-dependent gene transcription (Corrochano et al. 1995). The segment of the con-10 coding region was fused in frame to the second codon of hph, with the remainder of the hph coding region remaining intact. The transcription termination region for trpC of Aspergillus nidulans was placed after the hph ORF to ensure mRNA termination (Madi et al. 1994). To facilitate integration, the plasmid containing the con-10∷hph fusion also included a 5′ truncated copy of the his-3 gene that allowed reconstitution of an intact copy of the his-3 gene by homologous integration of the circular plasmid at his-3. Integration resulted in duplicated his-3 sequences flanking plasmid sequences and the con-10∷hph fusion (Madi et al. 1994). Strain CHZ10 was obtained by cotransformation of strain CH10 with plasmids pDE1559 and p3SR2. Plasmid pDE1559 contains con-10 regulatory DNA from position −1559 (relative to the transcription start site) to codon 40 of the con-10 ORF fused to the eighth codon of the E. coli lacZ gene; the trpC transcription termination region from A. nidulans was placed after the lacZ ORF to ensure correct transcription termination (Corrochano et al. 1995). Plasmid p3SR2 contains the A. nidulans acetamidase gene, amdS, and allows the selection of transformants by growth on acetamidase as a nitrogen source (Yamashiro et al. 1992). The presence of the con-10∷lacZ fusion in the CHZ10 strain was detected by the blue color of CHZ10 colonies plated at high density after growth in the dark on sorbose agar supplemented with X-gal (5-Bromo-4-chloro-3-indoyl-β-d-galactopyranoside). N. crassa colonies grown at high density express the con-10∷lacZ fusion, presumably due to starvation and other environmental factors that regulate con-10 expression (Lee and Ebbole 1998b). DNA hybridization analyses demonstrated that several copies of the con-10∷lacZ fusion had been integrated in the genome of CHZ10, presumably at ectopic positions since we couldn't select for integration after his-3 (not shown). We used the standard N. crassa wild-type strain 74-OR23-1VA (FGSC 2489 mat A) and a vivid mutant strain (FGSC 7852 vvdaub mat A) from the Fungal Genetics Stock Center. All strains were maintained by growth in Vogel's minimal media with 1.5% sucrose as a carbon source. Sorbose agar was used to promote colonial growth when required. Strain manipulation and growth media preparation followed standard procedures and protocols (Davis 2000). See also, the Neurospora protocol guide (http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm).
Mutagenesis and selection of hygromycin-resistant mutants exposed to continuous light:
Cells of CHZ10 do not grow on minimal agar supplemented with 2 mg/ml hygromycin in the presence or absence of light. Mutants altered in their photoadaptation should grow in illuminated plates containing hygromycin if their con-10∷hph fusion is capable of being continuously expressed. To perform this mutagenesis/selection procedure, freshly harvested conidia from strain CHZ10 (1–5 × 106) were exposed to UV light to reduce survival to 60–70%. Mutagenized conidia (about 106 per plate) were mixed with 15 ml of sorbose agar containing 2 mg/ml hygromycin and poured into a standard petri dish. Once the agar solidified, an additional 15 ml of the same hygromycin agar medium mixture were added and the plates incubated overnight at 34° in the dark to prevent photoreactivation. The two layers of agar ensured that germinated conidia were not exposed to air, an environmental signal that promotes conidiation and would result in constitutive transcription of con-10 (Springer 1993). The plates with the mutagenized conidia were then incubated under continuous light at room temperature (22°–29°). Continuous white light was provided by a set of white fluorescent bulbs (10 W/m2 blue light). Colonies (∼5 of 106 conidia per plate) that reached the surface of the top agar after 7 days of growth, were picked for purification and characterization.
Light induction experiments:
Cultures of isolated mutants were grown and mycelia were illuminated for the times indicated to measure regulation of gene expression by light and to detect changes in the phosphorylation status of the WC-1 protein. Cultures were prepared by inoculating ∼106–107 viable conidia into 25 ml of liquid Vogel's minimal medium containing 0.2% Tween 80 as a wetting agent. Liquid medium was used to prevent conidiation and to allow the growth of N. crassa as submerged vegetative hyphae. The plates were incubated in the dark for 24 hr (34°) inside a dark box and were then exposed to white light provided by a set of fluorescent bulbs (10 W/m2 of blue light). Light exposure with different intensities was obtained using an illumination chamber that allowed the simultaneous irradiation of three plates for 30 min in a temperature-controlled room set at 22°. The illumination chamber was a black wooden box with plate holders placed at the bottom. Light from a quartz halogen lamp installed in a slide projector passed through an upper window carrying a filter holder with two heat filters and neutral-density filters, as required to obtain the desired light intensity. After light exposure, mycelia were collected with the help of a tweezer, dried on filter paper, wrapped in aluminum foil, frozen in liquid nitrogen, and stored at −80°, unless otherwise indicated. Control cultures were kept in the dark prior to collection. All the manipulations in the dark were performed under red light. Light intensities were measured with a calibrated photodiode.
RNA isolation and hybridizations:
Neurospora mycelia were disrupted by two 0.5-min pulses in a minibeadbeater (BioSpec, Bartlesville, OK), in an RNA extraction buffer, with 1.5 g of zirconium beads (0.5 mm diameter) in 1.9-ml screw-cap tubes. The samples were cooled on ice for 4 min after the first pulse of the minibeadbeater. The extracts in screw-cap tubes were clarified by centrifugation in a microcentrifuge (13,000 rpm) for 5 min prior to RNA purification. Total RNA from mycelia was obtained using the perfect RNA eukaryotic mini kit (Eppendorf, Westbury, NY). RNA hybridizations were performed following standard procedures (Sambrook and Russell 2001). Ten micrograms of each RNA sample were separated by electrophoresis on an agarose gel (12 g/liter agarose), transferred to a nylon membrane, and probed with DNA segments from genes hph, con-10, con-6, al-2, wc-1, wc-2, and vvd labeled with [32P]dCTP. The DNA probes were prepared by PCR or after restriction-enzyme digestion of plasmid DNAs prepared in our laboratory. For mRNA normalization and loading controls the filters were stripped of radioactivity and reprobed with a segment of the β-tubulin gene (tub-2) labeled with [32P]dCTP. The hybridization signal was quantified using a phosphor imaging plate in a fluorescent image analyzer (FLA-3000, Fujifilm) and the program Image Gauge (Fujifilm) or by scanning the exposed film and quantification of the hybridization signal with the program ImageJ (http://rsb.info.nih.gov/ij/). Each hybridization signal was normalized to the corresponding tub-2 signal to correct for loading errors. Then, the hybridization signal for each filter was normalized to the RNA sample from mycelia exposed for 30 min to light, unless otherwise indicated.
Quantitative PCR:
Quantitative PCR experiments were performed to determine relative mRNA abundance using one-step RT–PCR, using 25 μl 1× power SYBR green PCR master mix (Applied Biosystems, Foster City, CA), 6.25 units MultiScribe reverse transcriptase (Applied Biosystems), 1.25 units RNase inhibitor (Applied Biosystems), 0.2 μm of each primer (al-1F, 5′-TCCAATGTTTCCCCAACTACAAC-3′; al-1R, 5′-CGGTGGTGGGCGAGAA-3′; al-2F, 5′-CGCTATCGCCTACCCCATT-3′; al-2R, 5′-CGACGAGGAAGCCTGTTTG-3′; al-3F, 5′-CATCTCTTCCGCCGGTCTAG-3′; al-3R, 5′-ACCGAGGCCTTGCGTTTAC-3′; tub-2F, 5′-CCCGCGGTCTCAAGATGT-3′; and tub-2R, 5′-CGCTTGAAGAGCTCCTGGAT-3′), and 100 ng of RNA. Quantitative PCR analyses were performed using a 7500 real time PCR system (Applied Biosystems). The reaction included retrotranscription (30 min at 48°), denaturation (10 min at 95°), and 40 PCR cycles (15 sec at 95° and 1 min at 60°). The results for each gene were normalized to the corresponding results obtained with tub-2 to correct for sampling errors. Then, the results obtained with each sample were normalized to the RNA sample from CHZ10 exposed to light for 30 min.
Protein isolation and hybridizations:
Proteins were extracted from mycelia by previously described methods (Garceau et al. 1997) using a modified lysis buffer (50 mm HEPES pH 7.4, 137 mm NaCl, 10% glycerol, 5 mm EDTA, 29.3 μm phenylmethyl-sulphonylfluoride (PMSF), 6.3 μm leupeptin, 4.4 μm pepstatin A) at a ratio of 0.5 ml of buffer to 0.1 g of mycelia. Total protein (200 μg per lane) was subjected to SDS–PAGE on 5% gels and transferred to hybridization membranes. Equal loading was confirmed by staining the hybridization membrane with Pounceau solution. Proteins on membranes were hybridized with a monoclonal antibody against WC-1 (α-WC-1-1m4H4) (Görl et al. 2001). Horseradish peroxidase-conjugated anti-mouse IgG was used as a secondary antibody (Bio-Rad, Hercules, CA). Antibody binding was observed using chemiluminescence (Roche, Indianapolis).
Carotenoid analysis:
Approximately 106–107 viable conidia of CHZ10, SN strains, and the carotenoid overproducer strain vivid, and the wild-type strain as a control were inoculated into 25 ml of Vogel's liquid minimal medium with 0.2% Tween 80 as wetting agent, and placed in sterile petri dishes. Liquid medium was used to prevent conidiation and to allow the growth of N. crassa as submerged vegetative hyphae. The suspensions were grown in the dark for 1 day at 34° and then exposed to light under a set of fluorescent bulbs (10 W/m2 blue light) at 22° for 1 day. After light exposure, mycelia were collected, frozen in liquid nitrogen, and lyophilized. Carotenoids were extracted from 0.1 g dry weight samples as described (Arrach et al. 2002). Total carotenoids were estimated from measurements of the maximal absorption spectra in hexane, assuming an average maximal E (1 mg/liter, 1 cm) = 200.
DNA sequencing:
The DNA sequence of the con-10 segment attached to hph in CHZ10 and the SN mutants was determined after amplification by PCR using primers T7 (5′-GTAATACGACTCACTATAGGGC-3′) and HPH (5′-CGATCAGAAACTTCTCGACA-3′). The primers bound to the sequences in the plasmid polylinker located upstream of con-10 and to the hph ORF (nucleotides 30– 49, GeneBank accession no. K01193). This pairing should ensure that the PCR reactions only amplified con-10 DNA attached to hph and not DNA corresponding to the endogenous con-10 gene. The amplified DNA was cloned into pGEM-T (pGEM-T Easy Vector system I; Promega, Madison, WI) and sequenced by the chain-termination method. The DNA sequence of the gene vivid in CHZ10 and the SN mutants was determined after amplification by PCR using primers VVD-1F (5′-CATCTCGATCGACGGCATTC-3) and VVD-1R (5′-CCCTTGTTTGTCGATGTTGA-3) and sequencing of the 1233-bp PCR product. The amplified DNA included 694 bp of the vivid ORF and two introns, 384 bp upstream of the vivid ORF, and 155 bp downstream of the vivid ORF.
RESULTS
Isolation of mutants altered in con-10 adaptation to photoactivation:
A successful strategy for the isolation of our desired regulatory mutants was based on the use of strains carrying a selectable marker under the control of a light-regulated promoter. N. crassa strains carrying the bacterial gene for hygromycin resistance (hph) driven by the regulatory regions of con-10, or con-6, another conidiation gene, have been used to isolate regulatory mutants that express conidiation genes in vegetative mycelia (Madi et al. 1994). Transcriptional fusions with promoters from light-responsive genes or from genes regulated by the circadian clock have been successfully used in identifying blind mutants and mutants altered in regulation of the circadian clock (Carattoli et al. 1995; Linden et al. 1997b; Vitalini et al. 2004). It was anticipated that mutants with an altered photoadaptation should be capable of producing mRNAs for light-inducible genes when fungal mycelium is grown under continuous light.
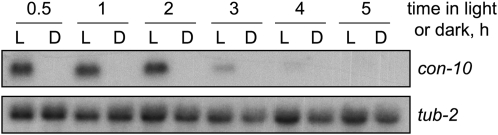
Expression of the gene con-10 of N. crassa is induced by light. However, upon prolonged light exposure, transcription is reduced due to adaptation to light (Lauter and Yanofsky 1993). As shown in Figure 1, the accumulation of con-10 mRNA is high in mycelia exposed to light for 2 hr. Adaptation to light is revealed by the reduced accumulation of con-10 mRNA in mycelia exposed to light for ≥3 hr (Figure 1).
Figure 1.—
Synthesis of con-10 mRNA exhibits photoadaptation. Total RNA was isolated from wild-type mycelia exposed to white light (10 W/m2 blue light) for various periods (L), or kept in the dark (D). RNAs were separated by electrophoresis and hybridized with probes for genes con-10 or tub-2 (tubulin).
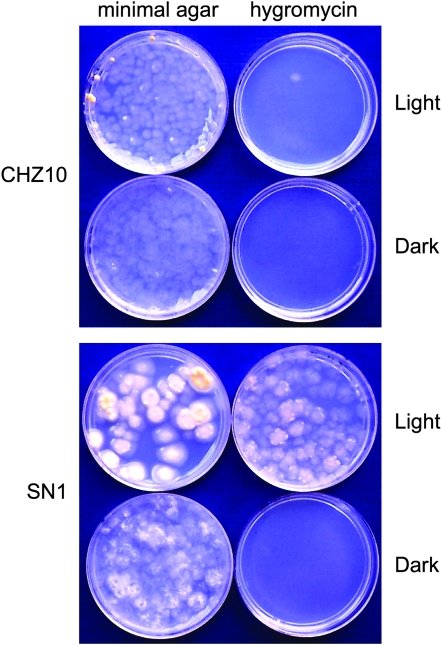
Strain CHZ10 contains a single copy of the con-10∷hph fusion integrated following the his-3 locus. The con-10-associated sequence in this translational fusion begins 517 bp upstream of the transcription initiation site and extends up to codon 40 of the con-10 ORF. That the con-10 regulatory region included in the con-10∷hph fusion should allow regulation of expression of the hph gene by light has been shown in experiments with con-10∷lacZ fusions (Corrochano et al. 1995; Lee and Ebbole 1998a). As expected, strain CHZ10 is sensitive to hygromycin when it is grown under continuous light, presumably due to photoadaptation of the con-10∷hph construct (Figure 2). CHZ10 is also sensitive to hygromycin in the dark, since the con-10 promoter is not active under these conditions (Figure 2). Mutants resistant to hygromycin under continuous light were selected after UV mutagenesis of CHZ10 conidia. Mutagenized conidia were submerged in hygromycin agar to ensure that germinating conidia were not exposed to air, an environmental signal that promotes conidiation and constitutive expression of con-10 (Springer 1993). Hygromycin agar plates with mutagenized conidia were incubated under continuous light for 7 days. Then, all colonies that reached the agar surface were isolated for further screening. From the initial set of hygromycin-resistant colonies we selected strains that grew in hygromycin agar plates under continuous light, but did not grow when the hygromycin agar plates were incubated in the dark. The selected strains were purified by three or more vegetative cycles of growth in minimal agar slants, by conidiation, and colony selection in hygromycin agar plates under continuous light. During strain purification we observed the color of colonies of the mutant strains in X-gal plates that had been incubated in the dark or under continuous light. Strain CHZ10 carried a con-10∷lacZ fusion in addition to the con-10∷hph fusion and the blue color of the mutant colonies on X-gal agar plates served as an indicator of whether expression of the con-10∷lacZ fusion was also increased, suggesting whether or not the mutations that had occurred had acted in cis or in trans. Measurements of light-dependent con-10 or con-10∷hph mRNA accumulation confirmed the cis/trans nature of the mutations. Five mutant strains (SN1–SN5) were finally selected that were resistant to hygromycin under continuous light and were not resistant in the dark. The light response of one of these strains, SN1, is shown in Figure 2. The hygromycin-resistant strains had additional developmental alterations. Strain SN1 produced 100-fold fewer conidia than CHZ10, while strains SN2–SN4 produced ∼10 times less conidia than CHZ10. In addition, SN1 and SN4 grew more slowly than CHZ10 in minimal agar, and SN4 did not develop aerial hyphae. SN1 formed large colonies in sorbose agar, suggesting that it was less sensitive to the restriction on mycelial growth promoted by the components of the sorbose agar. Strains SN2, SN4, and SN5 formed protoperithecia, but no ascospores were observed following attempted mating using these strains as female or male partners. In some instances we isolated a few ascospores, but these failed to germinate, thus preventing further genetic characterization. We amplified and sequenced the con-10 DNA attached to hph in CHZ10 and the five SN strains using primers that specifically prevented amplification of the endogenous con-10 DNA. None of the hygromycin-resistant mutants had mutations in the con-10 DNA sequence attached to hph, an indication that they had mutations in regulatory genes. DNA from the hygromycin-resistant strains, when transformed into CHZ10, did not confer resistance to hygromycin, suggesting that the new mutations are recessive.
Figure 2.—
Properties of SN1, a hygromycin-resistant mutant strain of N. crassa. Strain CHZ10 has an integrated translational fusion containing the regulatory region and part of the coding region of con-10 fused to the coding region of the gene conferring resistance to hygromycin (hph). CHZ10 is sensitive to hygromycin under continuous exposure to light because the con-10 promoter is not active due to photoadaptation. Strain SN1 was derived from CHZ10 after UV mutagenesis. It grows in hygromycin-containing agar under constant light due to the increased accumulation of the con-10∷hph mRNA after light exposure. Conidia from CHZ10 or SN1 were grown in sorbose minimal agar or sorbose agar containing hygromycin (2 mg/ml) at 22° in the dark or under constant white light.
The hygromycin-resistant strains might have suffered mutations in gene vivid since vivid mutants had an alteration in photoadaptation that allowed the accumulation of light-dependent mRNAs after long-light exposures (Heintzen et al. 2001; Schwerdtfeger and Linden 2001, 2003; Shrode et al. 2001). We sequenced the vivid gene, including 694 bp of the ORF and introns, 384 bp of upstream DNA, and 155 bp of downstream DNA, in all the hygromycin-resistant mutants and did not find any nucleotide changes, confirming that none of the mutants carried novel vivid alleles.
Two different groups of hygromycin-resistant mutants are defined by the photoactivation of the con-10∷hph fusion:
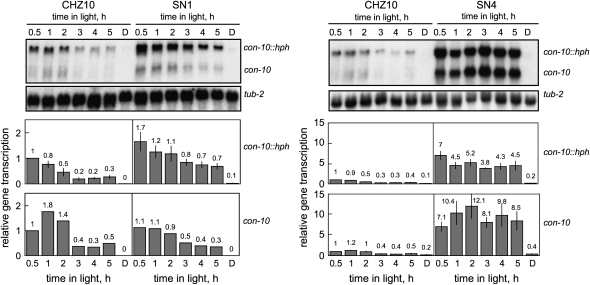
To assay the status of the mechanism of photoadaptation in our isolated hygromycin-resistant mutants we exposed mycelia from CHZ10 and the five mutant strains to light for different periods. Total RNA was extracted from each mycelium. The RNAs were then separated by electrophoresis, and hybridized with gene-specific probes to detect the presence of specific mRNAs. As expected, the mRNAs corresponding to the con-10∷hph fusion and the endogenous con-10 gene were observed in CHZ10 mycelia after 30 min of light, but the amounts of these mRNAs were reduced after exposure to light for ≥2 hr (Figure 3). Similar experiments were performed with hygromycin-resistant strains, and the results suggested that the strains could be grouped by the transcriptional behavior of their con-10∷hph fusion after light exposure, as observed for the two representative strains examined in Figure 3. Mycelia from strains SN1 and SN3 had elevated levels of con-10∷hph mRNA after 30 min of light exposure compared to the con-10∷hph mRNA level in CHZ10. However, the amount of con-10∷hph mRNA accumulated in mycelia was reduced after several hours of light, suggesting that the mechanism of photoadaptation was still functional, although it was weaker. The reduction in the amount of con-10∷hph mRNA in SN1 and SN3 after light exposures lasting several hours was not as pronounced as in CHZ10, and allowed the growth of mutant mycelia in hygromycin agar plates under continuous light (Figure 2). The amount of the endogenous con-10 mRNA was not significantly increased in these strains after light exposure.
Figure 3.—
Photoactivation of the con-10∷hph fusion and the endogenous con-10 gene in CHZ10 and two representative hygromycin-resistant strains. RNA was isolated from mycelia exposed to white light (10 W/m2 blue light) for various periods, or kept in the dark (D). RNAs were separated by electrophoresis and hybridized with probes for con-10 (hybridizing to the mRNAs of the con-10∷hph fusion and the endogenous con-10) or tub-2 (tubulin) genes. The plots show the average and standard error of the mean of the relative photoactivation, in two independent experiments (a single experiment was performed measuring con-10 mRNA in SN1). Each hybridization signal was normalized to the corresponding tub-2 hybridization signal to correct for loading errors. Then, the hybridization signal was normalized to the signal obtained for CHZ10 after 30 min of light. Note the different scales in the relative transcription plots for SN1 and SN4. See also supplemental Figure 1 at http://www.genetics.org/supplemental/ for the light-dependent accumulation of the con-10 and con-10∷hph mRNAs in the five hygromycin-resistant strains.
Interestingly, the transcriptional response to light was clearly different in the other hygromycin-resistant mutant strains. After light exposure, strains SN2, SN4, and SN5 accumulated large amounts of con-10∷hph mRNA and the endogenous con-10 mRNA. The high mRNA accumulation remained constant even after exposures to light lasting up to 5 hr (SN1 and SN4 data are shown in Figure 3). These results suggest that these strains, SN2, SN4, and SN5, had suffered mutations in proteins repressing con-10 photoactivation, thus disrupting the mechanism of photoadaptation. Alternatively, it is possible that the mutations have prevented the synthesis of the light-dependent repressor after changes in the cellular environment. These adaptation mutant strains were selected for further characterization.
Mutations in strains SN2, SN4, and SN5 modify the mechanism of photoadaptation of some, but not all, of the light-regulated genes of N. crassa:
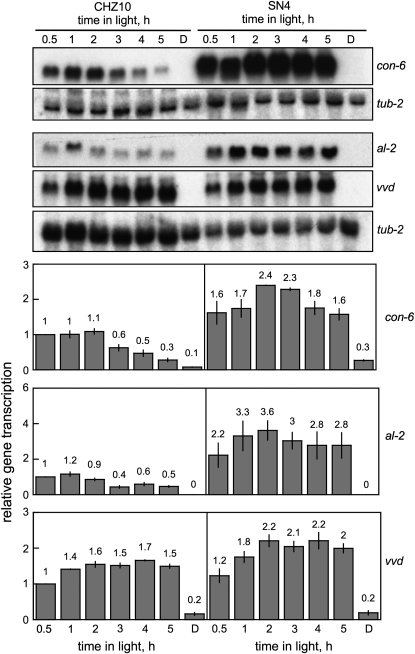
We next examined light regulation of other light-regulated N. crassa genes. These include another conidiation gene, con-6 (White and Yanofsky 1993), the gene for carotenoid biosynthesis, al-2 (Schmidhauser et al. 1994), and the photoreceptor genes, wc-1 (Ballario et al. 1996) and vivid (Heintzen et al. 2001). In addition, we assayed the mRNA levels for the wc-2 gene after light exposure, since the WC-2 protein forms a complex with WC-1 (Linden and Macino 1997) (Figure 4 and supplemental Table 1 at http://www.genetics.org/supplemental/). Regulation of con-10 and con-6 by light was similarly altered in strains SN2, SN4, and SN5 (Figure 4 and supplemental Table 1). Following light exposure these three strains accumulated large amounts of con-10 and con-6 mRNAs, and the accumulation remained constant even after exposures to light lasting up to five hours (Figure 4 and supplemental Table 1). In addition, mycelia from strain SN4 exposed to light accumulated more al-2 mRNA than CHZ10 (Figure 4 and supplemental Table 1). To the contrary, photoactivation of other well characterized light-inducible genes like vivid and wc-1 were not significantly altered in these or the other hygromycin-resistant mutants (Figure 4 and supplemental Table 1). This suggests that the altered proteins of the presumed regulatory genes are specific in which genes they activate. We did not observe any change in the accumulation of wc-2 mRNA in our adaptation mutant strains (supplemental Table 1).
Figure 4.—
Photoactivation of the light-inducible genes in CHZ10 and the light adaptation mutant SN4. The RNAs analyzed were from mycelia exposed to white light (10 W/m2 blue light) for various periods, or kept in the dark (D). RNAs were separated by electrophoresis and hybridized with probes for the con-6, al-2, vvd, or tub-2 (tubulin) genes. The plots show the average and standard error of the mean of the relative photoactivation in two independent experiments. Each hybridization signal was normalized to the corresponding tub-2 hybridization signal to correct for loading errors. Then, the hybridization signal was normalized to the signal obtained with CHZ10 after exposure to 30 min of light. See also supplemental Figure 1 at http://www.genetics.org/supplemental/ for the light-dependent accumulation of the con-6 mRNA in the five hygromycin-resistant strains.
The light-adaptive mutations in strains SN2, SN4, and SN5 do not change the threshold of these strains for photoactivation:
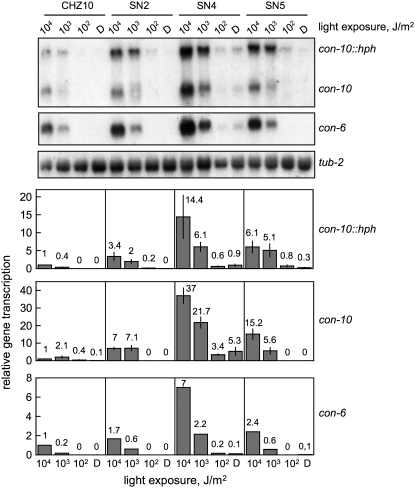
The high levels of con-10 and con-6 mRNAs observed in strains SN2, SN4, and SN5 after light exposure may have been caused by a reduction in the light threshold that can be perceived by the fungus. Therefore we measured the threshold of gene photoactivation for the con-10∷hph fusion and the endogenous con-10 and con-6 genes by exposing mycelia from CHZ10 and the adaptation mutants to light of different intensities (Figure 5). Gene photoactivation was clearly observed using 3 × 104 J/m2 in CHZ10 and the mutant strains, while light exposures of less intensity reduced gene photoactivation. A light exposure of 3 × 102 J/m2 applied to CHZ10 or strains SN2, SN4, and SN5 did not increase the mRNA accumulation observed for con-10, con-10∷hph, or con-6 over the values observed in dark cultures (Figure 5). We therefore concluded that the threshold for photoactivation of con-10 and con-6 is about 102 J/m2 in CHZ10, and it was the same in strains SN2, SN4, and SN5, suggesting that a large change in the light threshold is not responsible for the high and constant gene photoactivation observed in these adaptation mutants.
Figure 5.—
Threshold of gene photoactivation in CHZ10 and the light adaptation mutants. RNA was extracted from mycelia exposed to white light of various intensities during 30 min, or kept in the dark (D). The light-exposures used were: 3.4 × 104, 3.4 × 103, and 3.4 × 102 J/m2. RNAs were separated by electrophoresis and hybridized with probes for con-10 (hybridizing to the mRNAs of the con-10∷hph fusion and the endogenous con-10), con-6, or tub-2 (tubulin) genes. The plots show the average and standard error of the mean of the relative photoactivation values in two independent experiments (a single experiment was performed to detect con-6 mRNA). Each hybridization signal was normalized to the corresponding tub-2 hybridization signal, to correct for loading errors. Then, the hybridization signal was normalized to the signal obtained with CHZ10 after exposure to 30 min of light of the highest intensity.
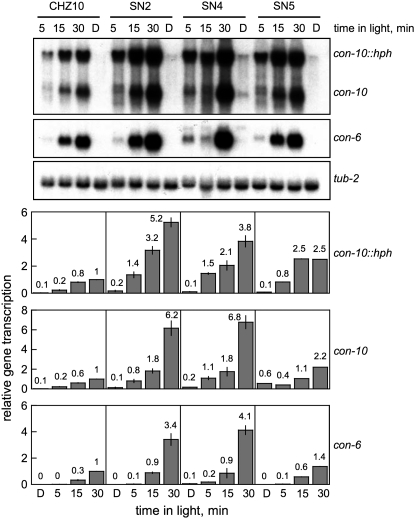
The pattern of mRNA synthesis and degradation after light induction is not appreciably altered in the adaptation mutants:
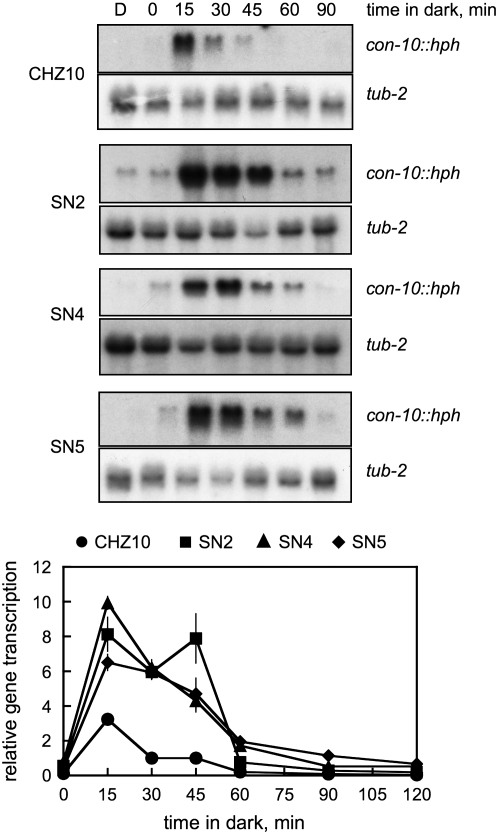
The accumulation of mRNAs for the con-10∷hph fusion, and the endogenous con-10 and con-6 genes may have been caused by a quick transcriptional response after light exposure or to the synthesis of unusually stable mRNAs after light induction. However, short illumination times (5–15 min) resulted in a similar pattern of mRNA accumulation in CHZ10 and the adaptation mutant strains (Figure 6). The mRNAs for the con-10∷hph fusion and for the endogenous con-10 and con-6 genes were detected after 5 min of light exposure, while longer light exposures resulted in higher mRNA accumulation in CHZ10 and mutant mycelia (Figure 6). The adaptation mutant strains always accumulated more con-10, con-10∷hph, and con-6 mRNAs than CHZ10, but the mRNA accumulation in all the strains increased as illumination time increased from 5 to 30 min. After light exposure, maximum con-10∷hph mRNA accumulation occurred after 15 min in the dark in CHZ10, and after 15–45 min in the dark in the adaptation mutants (Figure 7). Messenger RNA from con-10∷hph was not detectable in CHZ10 or the adaptation mutant mycelia after 90 min in the dark (Figure 7), suggesting that the adaptation mutants were not producing unusually stable con-10∷hph mRNAs. These results suggest that the mutations in strains SN2, SN4, and SN5 have specifically altered Neurospora's photoadaptation, resulting in high, sustained mRNA accumulation after exposure to light.
Figure 6.—
Photoactivation of gene expression in CHZ10 and the light adaptation mutants, during short exposure times. RNA was isolated from mycelia exposed to white light (10 W/m2 blue light) for various periods, or kept in the dark (D). RNAs were separated by electrophoresis and hybridized with probes for con-10 (hybridizing to the mRNAs of the con-10∷hph fusion and the endogenous con-10), con-6, or tub-2 (tubulin) genes. The plots show the average and standard error of the mean of the relative photoactivation values in two independent experiments. Each hybridization signal was normalized to the corresponding tub-2 hybridization signal to correct for loading errors. Then, the hybridization signal was normalized to the signal obtained with CHZ10 after exposure to 30 min of light.
Figure 7.—
Messenger RNA accumulation after photoactivation in strain CHZ10 and the light adaptation mutants. RNAs were obtained from mycelia kept in the dark (D) or exposed to 1 min of white light (10 W/m2 blue light) and then kept in the dark for various durations from 0 min (no incubation in the dark) to 120 min. RNAs were separated by electrophoresis and hybridized with probes for the con-10 or tub-2 (tubulin) genes. The plots show the average and standard error of the mean for the relative photoactivation, in two independent experiments. Each hybridization signal was normalized to the corresponding tub-2 hybridization signal to correct for loading errors. Then, the hybridization signal was normalized to the signal obtained with CHZ10 RNA following 1 min in light and after 30 min in the dark. Dark controls were not included in the plots. The results of the experiments with dark incubations lasting 120 min are only shown in the relative photoactivation plots.
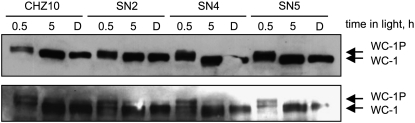
The light-dependent phosphorylation of WC-1 is not altered in our adaptation mutants:
Light exposure triggers the phosphorylation of the WC-1 photoreceptor and the exclusion of the White Collar complex (WCC) from the promoters of light-activated genes (Talora et al. 1999; He and Liu 2005). Since the phosphorylation status of WC-1 may play a role in the mechanism of photoadaptation, we investigated the presence of phosphorylated forms of WC-1 after light exposure by determining the extent of phosphorylation by gel electrophoresis and staining with a specific antibody. Light-dependent phosphorylation of WC-1 is observed by the presence of forms of WC-1 with low mobility after gel electrophoresis that disappear after a treatment with phosphatase (Talora et al. 1999; He and Liu 2005). As expected, a WC-1 form with reduced electrophoretic mobility was detected in mycelia of CHZ10 after 30 min of light, presumably due to the light-dependent phosphorylation of WC-1 (Figure 8). Phosphorylation of WC-1 was greatly reduced in mycelia grown in the dark or after 5 hr in the light (Figure 8). The pattern of light-dependent phosphorylation of WC-1 was not grossly altered in the adaptation mutants SN2, SN4, and SN5 (Figure 8), suggesting that the alteration in the mechanism of photoadaptation in these mutant strains is not due to a major alteration in the light-dependent phosphorylation of WC-1.
Figure 8.—
Light-dependent phosphorylation of WC-1 in CHZ10 and in light adaptation mutants. Total proteins from mycelia exposed to white light (10 W/m2 blue light) during 30 min or five hours, or kept in the dark (D) were separated by electrophoresis and their presence determined with an antibody specific for WC-1. The phosphorylated form of WC-1, which has a slightly higher migration position on the gel, was preferentially observed after 30 min of light (WC-1P). The results obtained in two independent experiments are shown.
Adaptation mutants accumulate more mycelial carotenoids than the wild type, after light exposure:
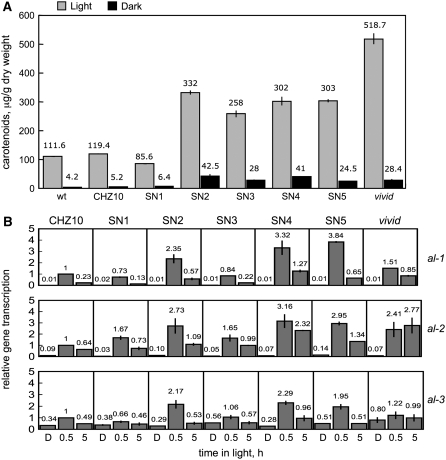
The most conspicuous photoresponse in vegetative mycelia of N. crassa is the induction of carotenoid biosynthesis (Linden et al. 1997a). To determine if our novel mutations had altered carotenoid accumulation, we measured the total amount of carotenoids in mycelia after 1 day of light exposure compared to the levels observed in mycelia cultured in the dark (Figure 9). The increased amount of al-2 mRNA observed in the adaptation mutant SN4 (Figure 4) suggested that the presence of extra AL-2 protein would increase the amount of carotenoids in cultures exposed to light. Accordingly, we assayed photocarotenogenesis in the wild type, the CHZ10 strain, and all of our hygromycin-resistant strains, including the adaptation mutants SN2, SN4, and SN5 (Figure 9A). As a control we compared carotenoid production in a vivid mutant, since the alteration in photoadaptation in this strain results in high levels of mycelial carotenoids after light exposure (Shrode et al. 2001). Most of the hygromycin-resistant strains, with the exception of SN1, exhibited a threefold increase in the accumulation of carotenoids after 1 day of light exposure compared to the amount accumulated by the wild type or CHZ10, and less than the fivefold increase observed in the vivid mutant (Figure 9). Dark-grown mycelia of SN2–SN5 also contained more carotenoids than the wild type, CHZ10, or SN1. These findings suggest that most of the new mutations, including those that appeared to have disrupted the mechanism of photoadaptation, also altered regulation of carotenoid biosynthesis in N. crassa mycelia. The increased accumulation of carotenoids in most of the hygromycin-resistant strains prompted us to characterize in detail the light-dependent transcription of the albino genes al-1 (Schmidhauser et al. 1990), al-2 (Schmidhauser et al. 1994), and al-3 (Nelson et al. 1989) that are required for carotenoid biosynthesis (Figure 9B). Strains SN2, SN4, and SN5 showed a two- to threefold increase in the amount of al-1, al-2, and al-3 mRNAs after 30 min of light, but light exposures of 5 hr clearly reduced the corresponding mRNA levels, suggesting that photoadaptation was not grossly altered. The mRNA reduction after 5 hr of light was less pronounced for al-2 mRNA in strain SN4, as we had shown previously (Figure 4). As expected, photoadaptation of albino gene transcription was clearly altered in the vivid strain: the accumulation of mRNAs for al-1, al-2, and al-3 remained elevated after 5 hr of light. The increase in the light-dependent accumulation of the albino genes in strains SN2, SN4, and SN5 may explain the increased accumulation of carotenoids that we observed in these strains.
Figure 9.—
Biosynthesis of carotenoids in N. crassa wild-type and mutant mycelia. (A) Light-dependent accumulation of carotenoids in mycelia of CHZ10 and hygromycin-resistant strains. Total carotenoids were extracted from mycelia that had been exposed to white light (10 W/m2 blue light) for 1 day (24 hr) or kept in the dark. Values obtained with wild type and a carotenoid overproducing strain (vivid) are included for comparison. The plot shows the average and standard error of the mean, for carotenoid accumulation in two or three independent experiments. (B) Photoactivation of the albino genes al-1, al-2, and al-3 in CHZ10, and hygromycin-resistant strains. Quantitative PCR experiments were performed to measure the relative accumulation of mRNA in mycelia exposed to white light (10 W/m2 blue light) for 30 min or five hours, or kept in the dark (D). The plots show the average and standard error of the mean of the relative mRNA accumulation in two independent experiments. The results from each PCR for each albino gene were normalized to the corresponding PCR for tub-2 to correct for sampling errors. Then, the results were normalized to those obtained with CHZ10 after exposure to 30 min of light.
DISCUSSION
In previous searches for blind mutants of N. crassa, alterations in only two genes, wc-1 and wc-2, were detected, indicative of the major role(s) played by the WC-1 and WC-2 proteins in photoreception (Linden et al. 1997a). An interesting feature of regulation of gene transcription by light is the transiency of the photoresponse, since mRNA accumulation is greatly reduced when the illumination period is extended (Lauter and Yanofsky 1993; Arpaia et al. 1999; Schwerdtfeger and Linden 2001, 2003). This phenomenon, dubbed photoadaptation, is presumably regulated by a mechanism involving a novel regulatory protein or proteins. To identify the regulatory genes required for photoadaptation we designed a novel genetic selection procedure using a N. crassa strain with the bacterial gene for resistance to hygromycin, hph, under the control of the regulatory region of con-10, a gene regulated by photoadaptation (Lauter and Yanofsky 1993). Growth of the strain containing this fusion, CHZ10, was sensitive to hygromycin in the dark, or upon exposure to continuous light, since the con-10 promoter was not active due to photoadaptation. Using the strain with this construct allowed the isolation of hygromycin-resistant mutants following culture under continuous light. These mutants presumably had suffered mutations that disrupted the shut-down mechanism of photoadaptation, allowing continued light activation of the con-10 promoter that regulates con-10∷hph gene expression.
We isolated five mutant strains that grew in hygromycin agar plates exposed to continuous light. Three of these hygromycin-resistant strains, SN2, SN4, and SN5, accumulated high amounts of the con-10∷hph mRNA after light exposure, and this mRNA accumulation remained constant after exposure to light for extended periods of up to 5 hr. These findings suggest that the mutational changes in these strains have inactivated a repression mechanism, or activated an activation mechanism, that is involved in photoadaptation. The mutations also altered photoadaptation of the endogenous con-10 gene and the conidiation gene con-6, however other light-regulated genes (al-2, wc-1, and vivid) were not affected (with the exception of al-2 in SN4), suggesting the existence of rather specific mechanisms causing photoadaptation in N. crassa. In addition, we observed a two- to threefold increase in the light-dependent accumulation of al-1, al-2, and al-3 mRNAs in strains SN2, SN4, and SN5, suggesting that the mutations can also modify the initial transcriptional response to light. In two other hygromycin-resistant mutants, SN1 and SN3, we observed that con-10∷hph mRNA accumulation was reduced after long exposures to light, an indication that photoadaptation in these strains remained functional. However, after long light exposures, con-10∷hph mRNA was still detected in these strains, explaining their hygromycin-resistance phenotype. The absence of a clear change in the photoregulation of the endogenous con-10 gene in these mutant strains may indicate that they have suffered mutations in the con-10 regulatory sequence attached to hph. However since we did not detect any change in the con-10 regulatory sequence fused to hph in any of our hygromycin-resistant mutants we concluded that the genetic changes in mutants SN1 and SN3 were not present in this region.
Our characterization of light-dependent gene transcription in the three adaptation mutants SN2, SN4, and SN5 showed that they did not suffer major changes in the mechanism governing activation of transcription by light exposure. The threshold for light activation of transcription, and mRNA production and stability did not appear to be grossly altered in these adaptation mutants. Light-dependent phosphorylation of the photoresponsive White Collar complex (WCC) resulting in its release from light-regulated promoters, has been proposed as a major event leading to photoadaptation (He and Liu 2005). Our characterization of the phosphorylation status of WC-1 has shown that the WCC continues to be transiently phosphorylated after light exposure in our adaptation mutants and in CHZ10, although we cannot rule out minor changes in the light-dependent phosphorylation of the WCC in the adaptation mutants. Our results suggest that major alterations in the light-dependent phosphorylation of the WCC are unlikely to be responsible for the adaptation mutant phenotype.
It is conceivable that our adaptation mutants have alterations in genes for proteins that are required for the release of the WCC from the con-10 and con-6 promoters after light-dependent phosphorylation. If the WCC remains bound to promoters throughout light exposure, then a high and sustained accumulation of mRNAs would be produced resulting in the high and constant light-dependent mRNA accumulation that we have observed. Characterization of WCC binding to the promoters of con-10 and con-6 during exposure to light in the adaptation mutants should allow us to examine this possibility.
In the three adaptation mutants, SN2, SN4, and SN5, the amount of carotenoids accumulated after light exposure increased threefold, probably as a result of their two- to threefold increase in the light-dependent accumulation of al-1, al-2, and al-3 mRNAs. Only strain SN3 had a clear increase in the amount of carotenoids accumulated after light exposure without a corresponding increase in the light-dependent accumulation of al-1, al-2, and al-3 mRNAs. These findings suggest that enhanced light-dependent carotenoid accumulation may occur by other regulatory mechanisms. However, in the absence of a detailed genetic characterization of the adaptation mutants we cannot rule out the possibility that the alteration in the biosynthesis of carotenoids in these strains may have been caused by the presence of additional mutations in their genomes in addition to those altering photoadaptation.
Mutants altered in the gene vivid accumulate five times more carotenoids than the wild type after light exposure. This is believed to be a consequence of a defect in the mechanism of photoadaptation, allowing the accumulation of light-dependent mRNAs after long-light exposures, as is seen for con-10, con-6, and al-1 (Heintzen et al. 2001; Schwerdtfeger and Linden 2001, 2003; Shrode et al. 2001). The VIVID protein is a small polypeptide with a flavin-binding domain similar to that found in WC-1, suggesting that VIVID may function as a photoreceptor involved in the mechanism of photoadaptation (Heintzen et al. 2001; Schwerdtfeger and Linden 2003; Zoltowski et al. 2007). None of the hygromycin-resistant strains that we have isolated had the carotenoid accumulation observed in vivid. The absence of nucleotide changes in the vivid gene from each hygromycin-resistant strain allowed us to conclude that we have isolated mutants in novel genes involved in the regulation of photoadaptation.
The increased accumulation of carotenoids after light exposure observed in most of the hygromycin-resistant strains suggests that our genetic selection strategy could be used to identify carotenoid-overproducing strains that may be of interest to the biotechnology industry. A strain carrying a fusion of hph to the regulatory region from any of the al genes may also allow the isolation of novel mutants specifically altered in the photoadaptation of al genes. This may lead to the accumulation of even higher levels of carotenoids after light exposure. These experiments may lead to new N. crassa strains optimized for carotenoid production.
We have described here a strategy for the isolation of novel mutants with defects in the mechanism of photoadaptation. Unfortunately, our attempts to use the photoadaptation mutant strains in genetic crosses failed, thus preventing a detailed genetic characterization of the mutant genes. It is possible that heterokaryons containing a photoadaptation mutant nuclei and a helper wild-type nuclei would be able to participate in crosses, normally. The use of the appropriate genetic markers in each nucleus should allow us to map the photoadaptation mutations and proceed to identify the mutant genes on the Neurospora genetic map. Since proteins similar to WC-1 and WC-2 have been described in ascomycete, basidiomycete, and zygomycete fungi (Purschwitz et al. 2006; Corrochano 2007; Herrera-Estrella and Horwitz 2007), we expect that the same regulatory proteins that participate in the mechanism of photoadaptation in N. crassa will play similar roles in other fungi.
Acknowledgments
The WC-1 antibody was kindly provided by M. Merrow and Till Roenneberg (University of Munich, Germany). The Fungal Genetics Stock Center (FGSC, http://www.fgsc.net) provided N. crassa strains. This work was supported by European funds (European Regional Development Fund, ERDF), the Spanish Ministerio de Educación y Ciencia (BIO2006-14897), the Scientific Program of NATO (CRG 960166), and Junta de Andalucía (CVI 0119).
References
- Arpaia, G., F. Cerri, S. Baima and G. Macino, 1999. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 262: 314–322. [DOI] [PubMed] [Google Scholar]
- Arrach, N., T. J. Schmidhauser and J. Avalos, 2002. Mutants of the carotene cyclase domain of al-2 from Neurospora crassa. Mol. Genet. Genomics 266: 914–921. [DOI] [PubMed] [Google Scholar]
- Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo et al., 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15: 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Belden, W. J., J. J. Loros and J. C. Dunlap, 2007. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25: 587–600. [DOI] [PubMed] [Google Scholar]
- Carattoli, A., E. Kato, M. Rodríguez-Franco, W. D. Stuart and G. Macino, 1995. A chimeric light-regulated amino acid transport system allows the isolation of blue light regulator (blr) mutants of Neurospora crassa. Proc. Natl. Acad. Sci. USA 92: 6612–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett, M. A., N. Garceau, J. C. Dunlap and J. J. Loros, 2002. Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics 160: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano, L. M., 2007. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem. Photobiol. Sci. 6: 725–736. [DOI] [PubMed] [Google Scholar]
- Corrochano, L. M., and P. Galland, 2006. Photomorphogenesis and gravitropism in fungi, pp. 233–259 in The Mycota I. Growth, Differentiation and Sexuality, edited by U. Kües and R. Fischer. Springer-Verlag, Berlin.
- Corrochano, L. M., F. R. Lauter, D. J. Ebbole and C. Yanofsky, 1995. Light and developmental regulation of the gene con-10 of Neurospora crassa. Dev. Biol. 167: 190–200. [DOI] [PubMed] [Google Scholar]
- Crosthwaite, S. K., J. C. Dunlap and J. J. Loros, 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276: 763–769. [DOI] [PubMed] [Google Scholar]
- Davis, R. H., 2000. Neurospora. Contributions of a Model Organism. Oxford University Press, Oxford.
- Dunlap, J. C., and J. J. Loros, 2005. Neurospora photoreceptors, pp. 371–389 in Handbook of Photosensory Receptors, edited by W. R. Briggs and J. L. Spudich. Wiley-VCH, Weinheim, Germany.
- Franchi, L., V. Fulci and G. Macino, 2005. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 56: 334–345. [DOI] [PubMed] [Google Scholar]
- Froehlich, A. C., Y. Liu, J. J. Loros and J. C. Dunlap, 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819. [DOI] [PubMed] [Google Scholar]
- Galland, P., 1991. Photosensory adaptation in aneural organisms. Photochem. Photobiol. 54: 1119–1134. [Google Scholar]
- Garceau, N. Y., Y. Liu, J. J. Loros and J. C. Dunlap, 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476. [DOI] [PubMed] [Google Scholar]
- Görl, M., M. Merrow, B. Huttner, J. Johnson, T. Roenneberg et al., 2001. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 20: 7074–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., and Y. Liu, 2005. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 19: 2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., P. Cheng, Y. Yang, L. Wang, K. H. Gardner et al., 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297: 840–843. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., J. J. Loros and J. C. Dunlap, 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464. [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella, A., and B. A. Horwitz, 2007. Looking through the eyes of fungi: molecular genetics of photoreception. Mol. Microbiol. 64: 5–15. [DOI] [PubMed] [Google Scholar]
- Lauter, F. R., and V. E. A. Russo, 1991. Blue light induction of conidiation-specific genes in Neurospora crassa. Nucleic Acids Res. 19: 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter, F. R., and C. Yanofsky, 1993. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 90: 8249–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., and D. J. Ebbole, 1998. a Analysis of two transcription activation elements in the promoter of the developmentally regulated con-10 gene of Neurospora crassa. Fungal Genet. Biol. 23: 259–268. [DOI] [PubMed] [Google Scholar]
- Lee, K., and D. J. Ebbole, 1998. b Tissue-specific repression of starvation and stress responses of the Neurospora crassa con-10 gene is mediated by RCO1. Fungal Genet. Biol. 23: 269–278. [DOI] [PubMed] [Google Scholar]
- Lee, K., J. C. Dunlap and J. J. Loros, 2003. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., and G. Macino, 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 16: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., P. Ballario and G. Macino, 1997. a Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 22: 141–150. [DOI] [PubMed] [Google Scholar]
- Linden, H., M. Rodríguez-Franco and G. Macino, 1997. b Mutants of Neurospora crassa defective in regulation of blue light perception. Mol. Gen. Genet. 254: 111–118. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Q. He and P. Cheng, 2003. Photoreception in Neurospora: a tale of two White Collar proteins. Cell. Mol. Life Sci. 60: 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madi, L., D. J. Ebbole, B. T. White and C. Yanofsky, 1994. Mutants of Neurospora crassa that alter gene expression and conidia development. Proc. Natl. Acad. Sci. USA 91: 6226–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, T., Y. Onda, E. Fujiwara, M. Wada and Y. Toyoshima, 2004. Two independent light signals cooperate in the activation of the plastid psbD blue light-responsive promoter in Arabidopsis. FEBS Lett. 571: 26–30. [DOI] [PubMed] [Google Scholar]
- Nelson, M. A., G. Morelli, A. Carattoli, N. Romano and G. Macino, 1989. Molecular cloning of a Neurospora crassa carotenoid biosynthetic gene (albino-3) regulated by blue light and the products of the white collar genes. Mol. Cell. Biol. 9: 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschwitz, J., S. Muller, C. Kastner and R. Fischer, 2006. Seeing the rainbow: light sensing in fungi. Curr. Opin. Microbiol. 9: 566–571. [DOI] [PubMed] [Google Scholar]
- Roberts, A. N., V. Berlin, K. M. Hager and C. Yanofsky, 1988. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol. Cell. Biol. 8: 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Romero, J., and L. M. Corrochano, 2006. Regulation by blue light and heat shock of gene transcription in the fungus Phycomyces: proteins required for photoinduction and mechanism for adaptation to light. Mol. Microbiol. 61: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Sachs, M. S., and C. Yanofsky, 1991. Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev. Biol. 148: 117–128. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmidhauser, T. J., F. R. Lauter, V. E. Russo and C. Yanofsky, 1990. Cloning, sequence, and photoregulation of al-1, a carotenoid biosynthetic gene of Neurospora crassa. Mol. Cell. Biol. 10: 5064–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser, T. J., F. R. Lauter, M. Schumacher, W. Zhou, V. E. A. Russo et al., 1994. Characterization of al-2, the phytoene synthase gene of Neurospora crassa. Cloning, sequence analysis, and photoregulation. J. Biol. Chem. 269: 12060–12066. [PubMed] [Google Scholar]
- Schwerdtfeger, C., and H. Linden, 2001. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol. Microbiol. 39: 1080–1087. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger, C., and H. Linden, 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22: 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrode, L. B., Z. A. Lewis, L. D. White, D. Bell-Pedersen and D. J. Ebbole, 2001. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet. Biol. 32: 169–181. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays 15: 365–374. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., and C. Yanofsky, 1992. Expression of con genes along the three sporulation pathways of Neurospora crassa. Genes Dev. 6: 1052–1057. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., K. M. Hager, C. Garrett-Engele and C. Yanofsky, 1992. Timing of synthesis and cellular localization of two conidiation-specific proteins of Neurospora crassa. Dev. Biol. 152: 255–262. [DOI] [PubMed] [Google Scholar]
- Talora, C., L. Franchi, H. Linden, P. Ballario and G. Macino, 1999. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 18: 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalini, M. W., L. W. Morgan, I. J. March and D. Bell-Pedersen, 2004. A genetic selection for circadian output pathway mutations in Neurospora crassa. Genetics 167: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, B. T., and C. Yanofsky, 1993. Structural characterization and expression analysis of the Neurospora conidiation gene con-6. Dev. Biol. 160: 254–264. [DOI] [PubMed] [Google Scholar]
- Yamashiro, C. T., O. Yarden and C. Yanofsky, 1992. A dominant selectable marker that is meiotically stable in Neurospora crassa: the amdS gene of Aspergillus nidulans. Mol. Gen. Genet. 236: 121–124. [DOI] [PubMed] [Google Scholar]
- Zoltowski, B. D., C. Schwerdtfeger, J. Widom, J. J. Loros, A. M. Bilwes et al., 2007. Conformational switching in the fungal light sensor Vivid. Science 316: 1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]