Abstract
In Caenorhabditis elegans embryos, specification of the germ lineage depends on PIE-1, a maternal protein that blocks mRNA transcription in germline blastomeres. Studies in mammalian cell culture have suggested that PIE-1 inhibits P-TEFb, a kinase that phosphorylates serine 2 in the carboxyl-terminal domain (CTD) repeats of RNA polymerase II during transcriptional elongation. We have tested this hypothesis using an in vivo complementation assay for PIE-1 function. Our results support the view that PIE-1 inhibits P-TEFb using the CTD-like motif YAPMAPT. This activity is required to block serine 2 phosphorylation in germline blastomeres, but unexpectedly is not essential for transcriptional repression or specification of the germline. We find that sequences outside of the YAPMAPT are required to inhibit serine 5 phosphorylation, and that this second inhibitory mechanism is essential for transcriptional repression and specification of the germ lineage. Our results suggest that PIE-1 uses partially redundant mechanisms to block transcription by targeting both the initiation and elongation phases of the transcription cycle.
INHIBITION of mRNA transcription is a defining characteristic of the embryonic germ lineage in invertebrates and vertebrates (Seydoux and Braun 2006). In Drosophila and Caenorhabditis elegans, mRNA synthesis appears to be globally, if not completely, inhibited in the embryonic germ lineage from the onset of embryogenesis to gastrulation. Early studies in Drosophila embryos showed that somatic nuclei incorporate radio-labeled UTP at a higher rate compared to germline nuclei (Zalokar 1976). Expression of the transcriptional activator VP16 could turn on a synthetic target gene in somatic cells but not in germ cells (Van Doren et al. 1998). In C. elegans embryos, in situ hybridization experiments using 16 gene-specific probes detected zygotic transcripts in somatic blastomeres, but not in germline blastomeres (Seydoux et al. 1996). The only exceptions were ribosomal rRNAs, which appear to be synthesized in both cell types (Seydoux and Dunn 1997). Further evidence for a lack of transcription specific to mRNAs was obtained using antibodies against the carboxyl-terminal domain (CTD) of RNA polymerase II (Seydoux and Dunn 1997; Martinho et al. 2004).
The CTD is a long extension of the large subunit of RNA polymerase II containing several (42 in C. elegans and 52 in humans) tandem copies of the heptapeptide motif (Y1S2P3T4S5P6S7) (Phatnani and Greenleaf 2006, for review). The phosphorylation status of the serines in these repeats changes as RNA polymerase II proceeds through the transcription cycle. The repeats start out unphosphorylated as RNA polymerase is recruited into the initiation complex at the promoter. During promoter clearance, Ser5 of each repeat becomes phosphorylated by cyclin-dependent kinase in the TFIIH complex (CDK7), and during the elongation phase, Ser2 becomes phosphorylated by cyclin-dependent kinase in the P-TEFb complex (CDK9). These phosphorylations allow the CTD to function as a scaffold to integrate transcription with processing, including capping, splicing, and termination. Phosphorylation of the CTD occurs in competition with CTD phosphatases to allow unphosphorylated RNA polymerase II to recycle back into new initiation complexes. Monoclonal antibodies (H14 and H5) that recognize preferentially P-Ser5 or P-Ser2 (Patturajan et al. 1998) have been used widely to characterize the phosphorylation status of the CTD in vivo. Chromatin immunoprecipitation (ChIP) experiments using H14 and H5 have shown that P-Ser5 predominates at the 5′ end of genes, whereas P-Ser2 predominates near the 3′ end (Phatnani and Greenleaf 2006). Immunofluorescence studies using these same antibodies in Drosophila and C. elegans embryos have shown that somatic nuclei become positive for P-Ser5 and P-Ser2 coincident with the onset of zygotic transcription. In contrast, germ cell nuclei remain negative for P-Ser2 and show only low levels of P-Ser5, until gastrulation. These observations have suggested that mRNA transcription is blocked at a step between initiation and elongation in embryonic germ cells (Seydoux and Braun 2006).
In C. elegans, transcriptional repression in the embryonic lineage requires PIE-1 (Seydoux et al. 1996). PIE-1 is maternal protein that segregates with the embryonic germ lineage and accumulates in the nuclei of each germline blastomere P1–P4 (Mello et al. 1996). PIE-1 contains two predicted RNA-binding domains (CCCH motifs) and does not resemble any known transcriptional repressor. Studies in mammalian tissue culture, however, showed that the C-terminal domain of PIE-1 can inhibit transcription when brought to a promoter via a heterologous DNA-binding domain (Batchelder et al. 1999). This activity depends on a specific sequence near the C terminus of PIE-1. This sequence (YAPMAPT) resembles a nonphosphorylatable version of a CTD repeat, raising the possibility that PIE-1 functions as a competitive inhibitor for a CTD kinase. Subsequent studies, also in mammalian cell culture, found that the C-terminal domain of PIE-1 can inhibit P-TEFb, the complex responsible for phosphorylation of Ser2 (Zhang et al. 2003). P-TEFb is a heterodimer containing the kinase CDK9 and an associated cyclin (typically cyclin T) which binds to the CTD repeats. In vitro, human cyclin T can also bind to alanine-substituted CTD repeats and to C. elegans PIE-1. The cyclin T/PIE-1 interaction was abolished by nonconservative mutations in the YAPMAPT (DAQMEQT). Those same mutations also blocked PIE-1's ability to suppress the stimulatory effect of P-TEFb on transcription in a HeLa cell assay (Zhang et al. 2003). Together these findings have led to a model whereby PIE-1 inhibits transcription by competing P-TEFb away from the CTD, thus blocking transcriptional elongation (Zhang et al. 2003).
The model predicts that the C-terminal domain of PIE-1, and the YAPMAPT in particular, should be essential for PIE-1's ability to repress transcription in germline blastomeres. Characterization of the pie-1(zu154) allele, which truncates the last 93 amino acids of PIE-1, including the YAPMAPT, confirmed that this region is essential for transcriptional repression in vivo (Tenenhaus et al. 2001). A direct test of the importance of the YAPMAPT, however, was complicated by (1) the unavailability of pie-1 alleles that affect this motif specifically, and (2) the lack of a reliable transformation system to express transgenic proteins maternally. (PIE-1 is a maternal protein that must be synthesized during oogenesis). Using transient transformants, Batchelder et al. (1999) found that a pie-1 transgene, where YAPMAPT was replaced by DAQMEQT, could still complement a pie-1(null) mutant albeit at a reduced frequency compared to wild type. The transient nature of the transformants precluded any direct assessment of the expression level of the transgenic proteins, further complicating the interpretation of these results (Batchelder et al. 1999).
Fortunately, since these studies, a new transformation technology has been developed for C. elegans (Wilm et al. 1999; Praitis et al. 2001). Ballistic transformation yields transgenes that are integrated singly, or in low copy, at random sites in the genome (Praitis et al. 2001). When driven by the pie-1 promoter, these transgenes express reliably during oogenesis and can be used for structure–function studies of maternal proteins (Hao et al. 2006). We have used the new technology to perform a structure–function study of the PIE-1 C-terminal domain. As predicted by the model, we find that the YAPMAPT is required for inhibition of Ser2 phosphorylation, but surprisingly we also find that this activity is not essential for transcriptional repression in vivo.
MATERIALS AND METHODS
Nematode strains and transgenics:
C. elegans strains were derived from the wild-type Bristol strain N2 using standard procedures (Brenner 1974), except that transgenic strains were kept at 24°.
PIE-1 transgenes were constructed in pID3.01, a GATEWAY destination vector containing the pie-1 promoter, GFP, GATEWAY recombination sequences, and the pie-1 3′-UTR (D'Agostino et al. 2006). Mutations in pie-1 were created in GATEWAY entry clones using the QuickChange site-directed and multisite-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by DNA sequencing.
All transgenes were introduced into worms by ballistic transformation (Praitis et al. 2001). Two independent lines or more were generated for each transgene. In all cases, lines with the same transgene exhibited the same GFP pattern. Transgenic lines were crossed to dpy-18(e364)pie-1(zu127)/qc1 males, balanced, and made homozygous for the transgene. Two independent lines were tested in the rescue assay except for GFP:PIE-1(1–335), GFP:PIE-1(1–299), and GFP:PIE-1(1–259), for which only one line was characterized.
Transgenic rescue assay:
For each line tested, five transgenic hermaphrodites were allowed to lay eggs for 24 hr. The embryos were counted, and 2 days later the number of viable larvae was counted. This experiment was repeated three times for each line. Percentage of lethality was derived from the number of viable larvae/total number of embryos laid.
Immunofluorescence microscopy:
Embryos were permeabilized by freeze cracking and fixed for 30 sec in −20° MeOH and 25 min in formaldehyde fix [1× PBS, 1.6 mm MgSO4, 0.8 mm EDTA, 3.7% formaldehyde]. Slides were washed three times in PBT (1× PBS, 0.1% Triton, 0.1% BSA), blocked for 30 min in PBT, and incubated with primary antibodies overnight at 4°. Secondary antibodies were applied for 1 hr at 4°.
Primary antibodies used were mouse monoclonal antibodies mAb H14 (anti P-Ser5 at 1:2 dilution) and mAb H5 (anti P-Ser2 at 1:5 dilution) (Patturajan et al. 1998). Secondary antibodies used were ALEXA 568-conjugated goat anti-mouse (Molecular Probes, Eugene, OR). DAPI (0.5 μg/ml) was used to visualize nuclei. Samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and examined with a Zeiss-Axioplan2 microscope equipped with a Photometrics coolsnap digital camera.
In situ hybridization:
In situ hybridization was performed as described in (Seydoux and Fire 1995) using an antisense GFP probe to detect pes-10:gfp mRNA as in (Wallenfang and Seydoux 2002).
Confocal microscopy:
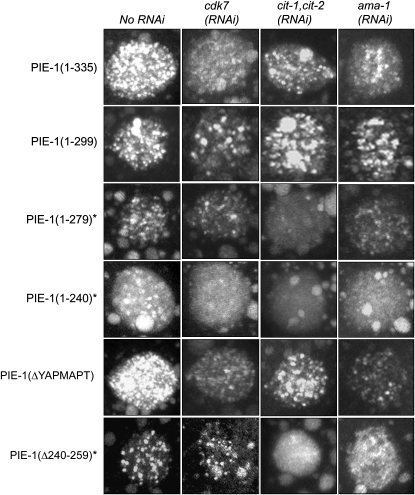
Subcellular localization of GFP:PIE-1 was examined in the germline blastomere P2 using a confocal laser-scanning microscope (Zeiss-LSM 510) and a Krypton-Argon laser (Omnichore, series 43) to generate excitation wavelength of 488 nm. z-Axis images were collected at 0.5-μm intervals through the P2 cell. Figure 5 shows the complete z-series. Both fixed and live samples were examined and no differences in GFP:PIE-1 distribution were observed between the two (data not shown).
Figure 5.—
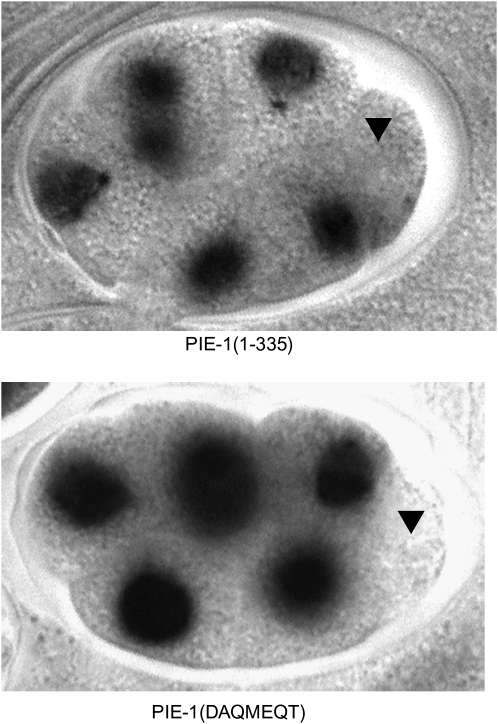
GFP:PIE-1 localization in nuclei. Collapsed confocal Z-stacks through the P2 germline blastomere show accumulation of GFP:PIE-1 in nuclei. Wild-type PIE-1 accumulates in numerous fine nuclear foci. This localization is disrupted in PIE-1 mutants that do not suppress P-Ser5 efficiently (marked with asterisk). Reductions in CDK-7, CIT-1.1/1.2, and AMA-1 (RNA polymerase II) levels also affect this localization. Note that PIE-1 also accumulates in larger cytoplasmic granules (P granules), which are visible around the nuclei in the micrographs.
RNA interference assays:
RNA interference was used to knockout gene function using the bacterial feeding method described by Timmons and Fire (1998). cit1.1 and cit1.2 ORFs were amplified from cDNA and cloned into the vector pCD1.1/L440 to create plasmids pDD71 and pDD72. Ampicillin-resistant transformants in Escherichia coli HT115 were grown in LB with 75 μg/ml ampicillin for 6–8 hr. Cultures were spread on new nematode growth medium plates containing 75 μg/ml ampicillin and 0.3 mm isopropyl β-d-thiogalactoside and incubated overnight at room temperature. L4 hermaphrodites were placed on the bacterial lawn to feed for 19–22 hr at 25°. For cit1.1;cit1.2 (RNAi) equal volumes of pDD72 and pDD73 cultures were mixed before spreading on plates.
Western blotting:
C. elegans embryos of mixed stages were harvested from hermaphrodites by bleaching and suspended in three volumes of 15 mm HEPES (pH 7.6), 10 mm KCl, 1.5 mm MgCl2, 0.1 mm EDTA, 0.5 mm EGTA, 44 mm sucrose, protease inhibitor cocktail (Roche, Indianapolis). The embryo suspension was frozen in liquid nitrogen and stored at −80°. Protein extracts were run on a 4–12% SDS polyacrylamide gel (Invitrogen, Carlsbad, CA). The GFP fusions were visualized by Western blotting using rabbit polyclonal anti-GFP antibody 6455 (1:2000; BD Biosciences, San Jose, CA). Anti-tubulin antibody E7 (1:10,000; Developmental Studies Hybridoma Bank, Iowa City, IA) was used as a loading control. Horseradish peroxidase-conjugated sheep anti-mouse antibody (1:10,000; Amersham Pharmacia, Piscataway, NJ) was used as a secondary antibody. Protein bands were detected using enhanced chemiluminiscence (Amersham Pharmacia).
In vitro binding assay:
cit 1.1, cit 1.2, and par-5 ORFs were cloned from cDNAs into the GATEWAY destination vector pJP1.09 (Pellettieri et al. 2003) to create N-terminal maltose-binding fusion proteins (MBPs). MBPs were grown in E. coli strain CAG456 and induced with 300 μm isopropyl β-d-thiogalactoside. Bacterial pellets were resuspended in 10 ml ice-cold column buffer (20 mm Tris-HCl, 500 mm NaCl, 1 mm EDTA, 1 mm 1,4-dithio-dl-threitol [DTT], 10% glycerol, passed twice through a French press, and centrifuged (SW41 rotor at 36,000 RPM or equivalent for 30 min). One hundred microliters MBP:CIT1.1 fusion protein lysates were bound to amylose resin (New England Biolabs, Beverly, MA) at 4° for 1 hr in 1 ml column buffer. After binding, the beads were washed three times with column buffer.
Wild-type and mutant pie-1 ORFs, and elongin C (DeRenzo et al. 2003), were cloned into pDD91, a GATEWAY destination vector for making in vitro translated proteins. The pie-1 clones were transcribed and translated (IVT) in the presence of 35S-labeled proteins in vitro using the TNT SP6-coupled rabbit reticulocyte system according to the manufacturer's instructions (Promega, Madison, WI). Five to 25 μl of in vitro translated PIE-1 proteins (the exact amount of this was determined for each mutant; equal amounts of proteins were used for each mutant) were added to MBP:CIT1.1, MBP:PAR5, or MBP protein bound to beads in 1 ml interaction buffer (20 mm HEPES, 1.0 mm EDTA, 200 mm NaCl, 0.1% NP-40, 6.0% glycerol, 1 mm DTT). The proteins were allowed to bind for 2 hr at 4° with gentle agitation. After the binding reaction the beads were washed five times with the interaction buffer, and the bound proteins were eluted by boiling in the SDS sample buffer and resolved by SDS–PAGE on a 4–12% gel, which was analyzed for autoradiography. One-twentieth of the radioactive proteins used for binding was loaded for input. After exposure, the gel was stained with Coomassie Blue to make sure that equal amounts of MBP and MBP-fusion proteins had been loaded. Bands were quantified using the Imagequant software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
PIE-1 binds C. elegans cyclin T in vitro:
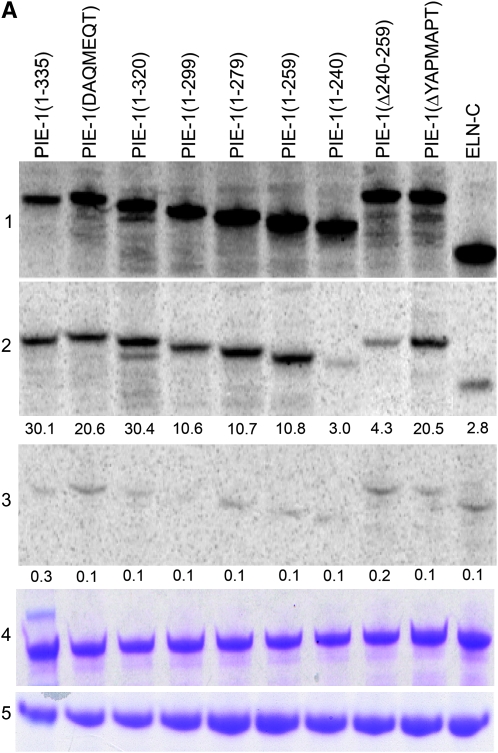
Zhang et al. (2003) reported that the C-terminal domain of PIE-1 can bind to human CycT1. C. elegans has two closely related cyclin T1 homologs, cit-1.1 and cit-1.2 (Shim et al. 2002). To test for binding with PIE-1, we synthesized CIT-1.1 and CIT-1.2 fused to MBP in E. coli (materials and methods). MBP:CIT-1.1 and MBP:CIT-1.2 were immobilized on amylose resin and incubated with 35S-labeled, in vitro translated full-length PIE-1(aa 1–335). Bound proteins were resolved by SDS–PAGE (materials and methods). We found that MBP:CIT-1.1 and MBP:CIT-1.2 both bound to PIE-1 (Figure 1A and data not shown). Control proteins (MBP:PAR-5 and in vitro translated elongin C) bound only weakly, confirming the specificity of the assay.
Figure 1.—
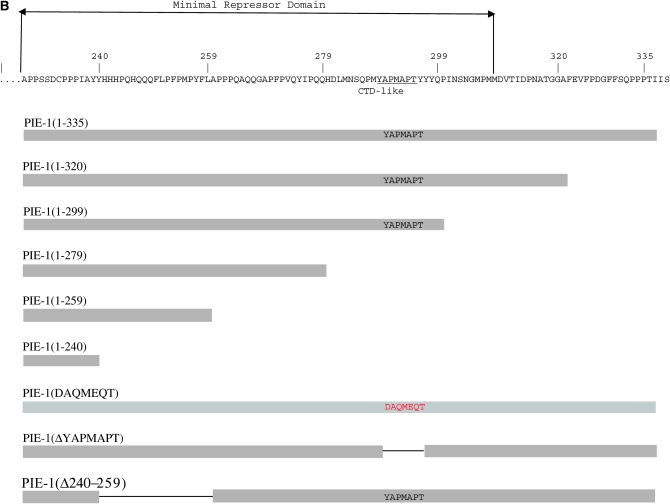
PIE-1 binds to CIT-1.1 in vitro. (A) In vitro translated and 35S-labeled full-length PIE-1 and mutant derivatives (section 1—input) were incubated with immobilized MBP:CIT-1.1 (section 2) or negative control MBP:PAR-5 (section 3) and bound proteins were resolved by SDS–PAGE. Sections 4 and 5 show Coomassie staining of MBP:CIT-1.1 (section 4) and MBP:PAR-5 (section 5) to control for loading. ELN-C is elongin C (DeRenzo et al. 2003) used here as a negative control. Numbers below sections 2 and 3 indicate percentage bound (bound/input × 100%), as calculated by measuring band intensities using Imagequant software (Molecular Dynamics). (B) Diagram showing the sequence of the C-terminal domain of PIE-1 and the mutant derivatives used in this study. The minimal repressor domain is the minimal PIE-1 fragment that can inhibit transcription when artificially brought to a promoter in HeLa cells (Batchelder et al. 1999).
To determine which domain in PIE-1 interacts with cyclin T, we constructed five truncation derivatives spanning the last 95 amino acids of PIE-1 (Figure 1B). We showed previously that deletion of this region blocks PIE-1's ability to repress transcription in vivo, but does not affect other aspects of PIE-1 function (Tenenhaus et al. 2001). This region partially overlaps the domain [PIE-1(204–335)] sufficient for binding to human CycT1 in vitro (Zhang et al. 2003) and PIE-1's “minimal repressor domain” [PIE-1(223–304)] defined in mammalian cell culture (Batchelder et al. 1999). We also made two additional PIE-1 mutants, targeting specifically the YAPMAPT motif (aa 285–291): PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT), where the YAPMAPT has been precisely deleted.
All fusions bound MBP:CIT-1.1 above background except for PIE-1(1–240), suggesting that aa 240–259 are essential for binding (Figure 1A). To test this requirement directly, we constructed one additional mutant lacking only this region (Figure 1B). As expected, PIE-1(Δ240–259) did not bind MBP:CIT-1.1 above background (Figure 1A). We conclude that PIE-1 binds CIT1.1 in vitro and that this interaction requires amino acids 240–259 (“CIT-1.1 binding domain”).
The YAPMAPT motif and CIT-1.1 binding region are required to inhibit Ser2 phosphorylation:
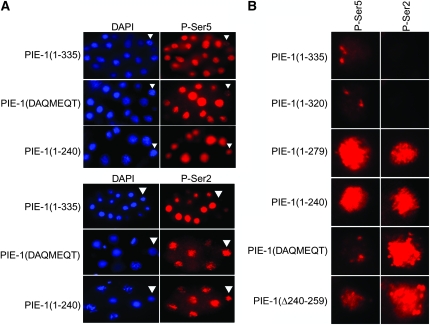
To test for activity in vivo, we introduced the PIE-1 mutants in the pie-1 expression vector pID3.01 to create amino terminal GFP fusion under the control of the pie-1 promoter and 3′-UTR. These constructs were transformed into worms by microparticle bombardment and crossed into the pie-1 null mutant pie-1(zu127) (see materials and methods). Embryos derived from pie-1(zu127) hermaphrodites [hereafter referred to as pie-1(zu127) embryos] lack endogenous PIE-1 and activate mRNA transcription in the P lineage starting in the four-cell stage (Seydoux et al. 1996). pie-1(zu127) embryos have high levels of P-Ser5 and P-Ser2 in 100% of P2, P3, and P4 (hereafter collectively referred to as P blastomeres) (Table 1 and Seydoux and Dunn 1997). The wild-type pie-1 transgene rescues this defect, with 0 and 13% of pie-1(zu127);GFP:PIE-1(1–335) showing high Ser-5P and Ser-2P, respectively, in P blastomeres (Table 1 and Figure 2). GFP:PIE-1(1–320) and GFP:PIE-1(1–299) also rescue. The longest truncation that did not rescue was GFP:PIE-1(1–279). This construct retains the CIT-1.1 binding domain but lacks the YAPMAPT. GFP:PIE-1(1–279) failed to suppress both P-Ser5 and P-Ser2. Interestingly, the two mutants affecting only the YAPMAPT [PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT)] suppressed P-Ser5 but not P-Ser2 (Table 1 and Figure 2). Similarly, the mutant construct lacking CIT-1.1 binding domain PIE-1(Δ240–259) partially suppressed P-Ser5 but completely failed to suppress P-Ser2. We conclude that the CIT-1.1 domain and YAPMAPT are both required to inhibit Ser2 phosphorylation, and that neither is sufficient on its own. Furthermore, additional sequences around the YAPMAPT are required to inhibit Ser5 phosphorylation.
TABLE 1.
| Transgene | % lethality | n | % P-Ser5 (high) | n | % P-Ser2 | n |
|---|---|---|---|---|---|---|
| No transgene | 100 | >50 | 100 | >50 | 100 | >50 |
| PIE-1(1–335) | 9 | 701 | 0 | 15 | 13 | 105 |
| PIE-1(1–320) | 3 | 1002 | 0 | 25 | 7 | 92 |
| PIE-1(1–299) | 11 | 298 | 0 | 25 | 5 | 50 |
| PIE-1(1–279) | 100 | 623 | 100 | 25 | 97 | 67 |
| PIE-1(1–259) | 100 | 346 | ND | ND | 100 | 25 |
| PIE-1(1–240) | 100 | 1038 | 100 | 25 | 100 | 151 |
| PIE-1(DAQMEQT) | 8 | 829 | 0 | 30 | 82 | 165 |
| PIE-1(ΔYAPMAPT) | 8 | 934 | 0 | 39 | 89 | 54 |
| PIE-1(Δ240–259) | 62 | 634 | 52 | 25 | 94 | 74 |
Percentage of pie-1(zu127) embryos expressing the indicated transgene that did not survive embryogenesis (% lethality), that exhibited a germline blastomere with high P-Ser5 levels (equivalent to somatic blastomeres) (% P-Ser5), or that exhibited a germline blastomere positive for P-Ser2 (% P-Ser2) is shown. Italics indicate no rescue of the pie-1(zu127) phenotypes, underlining indicates partial rescue, and no italics or underlining indicates rescue comparable to that obtained with the wild-type transgene PIE-1(1–335). Note that rescued embryos no longer have high P-Ser5 levels (equivalent to somatic blastomeres), but maintain the low intermediate levels observed in wild-type germline blastomeres (Figure 2). Examination of digital images revealed that, for the majority of pie-1(zu127) embryos expressing PIE-1(DAQMEQT) (16/20) and PIE-1(ΔYAPMAPT) (18/21), P-Ser2 levels in germline blastomeres were equivalent to P-Ser2 levels in somatic blastomeres (also see Figure 2).
Figure 2.—
Inhibition of P-Ser5 and P-Ser2 by the PIE-1 transgenes. (A) Eight- to 15-cell pie-1(zu127) embryos expressing the indicated PIE-1 transgenes and stained for DAPI and P-Ser2 or P-Ser5. Arrow points to the germline blastomeres. (B) Close-up of germline blastomere nuclei stained for P-Ser2 or P-Ser5 in pie-1(zu127) embryos expressing the indicating PIE-1 transgenes.
The affinity of anti-P-Ser5 (H14) and anti-P-Ser2 (H5) antibodies may be influenced by phosphorylation at nearby sites (Palancade and Bensaude 2003), raising the concern that, under certain conditions, these antibodies may not report directly on the activity of the initiation-specific kinase CDK-7, and the elongation-specific kinase CDK-9 (P-TEFb), respectively. To investigate this possibility, we probed for P-Ser5 and P-Ser2 in embryos depleted for cdk-7 or cit-1.1/cit-1.2. As expected (Shim et al. 2002; Wallenfang and Seydoux 2002), we found that the residual levels of P-Ser5 in the P blastomeres of pie-1(zu127);GFP:PIE-1(1–335) embryos depend on cdk-7 but not cit-1.1/cit-1.2 (Table 2). Similarly, the high levels of P-Ser5 present in pie-1(zu127);GFP:PIE-1(1–240) were eliminated by cdk-7(RNAi) but not cit-1.1/cit-1.2(RNAi). P-Ser2 present in pie-1(zu127);PIE-1(1–240) was eliminated by both cdk-7(RNAi) and cit-1.1/cit-1.2(RNAi). All signals were eliminated by depletion of RNA polymerase II [ama-1(RNAi)] (Table 2). We conclude that our detection methods are likely to accurately discriminate between CDK-7 (initiation-) and P-TEFb (elongation-) dependent phosphorylation events.
TABLE 2.
| % P-Ser5
|
% P-Ser2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Transgene | % lethality | N | None | Low | High | N | None | Positive | N |
| PIE-1(1–335) | 9 | 701 | 0 | 100 | 0 | 15 | 87 | 13 | 105 |
| PIE-1(1–355) cdk-7(RNAi) | 96 | 422 | 100 | 0 | 0 | 42 | 100 | 0 | 42 |
| PIE-1(1–335) cit-1.1/1.2(RNAi) | 91 | 408 | 4 | 96 | 0 | 51 | 100 | 0 | 40 |
| PIE-1(1–335) ama-1(RNAi) | 97 | 445 | 100 | 0 | 0 | 51 | 100 | 0 | 45 |
| PIE-1(1–240) | 100 | 1038 | 0 | 0 | 100 | 25 | 0 | 100 | 151 |
| PIE-1(1–240) cdk-7(RNAi) | 100 | 441 | 100 | 0 | 0 | 46 | 100 | 0 | 45 |
| PIE-1(1–240) cit-1.1/1.2(RNAi) | 100 | 446 | 0 | 0 | 100 | 44 | 100 | 0 | 29 |
| PIE-1(1–240) ama-1(RNAi) | 100 | 364 | 100 | 0 | 0 | 45 | 100 | 0 | 43 |
Control RNAi experiments to evaluate the specificity of the P-Ser5 and P-Ser2 signals in pie-1(zu127) embryos carrying the indicated transgenes. P-Ser5 levels are divided into three categories depending on whether P-Ser5 was detected at levels equivalent to that seen in somatic blastomeres (high), at the intermediate level typical of wild-type germline blastomeres (low, with two prominent foci), or absent (none) (also see Figure 2). As expected for a modification linked to initiation, P-Ser5 signals (both in the high and low categories) are eliminated upon depletion of cdk-7 and ama-1, but remain unaffected by depletion cit-1.1/1.2. In contrast, as expected for a modification linked to elongation, P-Ser2 signals are eliminated upon depletion of all three genes.
Inhibition of Ser2 phosphorylation is not essential for embryonic viability or to inhibit synthesis of a zygotic mRNA in P blastomeres:
Next, we investigated the ability of each transgene to rescue the embryonic lethality of pie-1(zu127) embryos. In these embryos, descendents of the P2 blastomere adopt the fates of descendents of the EMS blastomere, resulting in excess pharyngeal and intestinal cell types and embryonic lethality. This cell-fate transformation is thought to be due to activation of transcription in P2, which causes the SKN-1 transcription factor to activate an EMS-specific transcription program in the P2 lineage (Mello et al. 1996; Tenenhaus et al. 2001).
As expected, full-length PIE-1(1–335) and the two truncations able to block Ser2 and Ser5 phosphorylation [PIE-1(1–320) and PIE-1(1–299)] rescued the pie-1(zu127) lethality efficiently (<15% lethality) (Table 1). In contrast, truncations that fail to suppress Ser2 and Ser5 [PIE-1(1–279), PIE-1(1–259), and PIE-1(1–240)] did not rescue (Table 1). Surprisingly, PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT), which suppressed P-Ser5 but not P-Ser2, rescued as efficiently as wild type. Similarly PIE-1(Δ240–259), which partially suppresses P-Ser5 but not P-Ser2, partially rescued the embryonic lethality (<65% lethality) (Table 1). We conclude that, whereas suppression of P-Ser5 correlates with viability, suppression of P-Ser2 is not essential.
The ability of PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT) to restore viability suggests that these mutants are able to inhibit transcription in P blastomeres. To test this hypothesis directly, we crossed into pie-1(zu127);GFP:PIE-1(DAQMEQT), an integrated array containing multiple copies of the pes-10:gfp transgene. pes-10 encodes an early zygotic transcript that can be detected in somatic blastomeres as early as the four-cell stage (Seydoux and Fire 1994). Transcripts derived from the multicopy pes-10:gfp array are readily visualized by in situ hybridization in somatic blastomeres in wild-type embryos, and in both somatic and germline blastomeres in pie-1(zu127) embryos (Seydoux et al. 1996; Wallenfang and Seydoux 2002). We found that, as in wild type, pes-10:lacZ transcripts were present only in somatic blastomeres in pie-1(zu127);PIE-1(DAQMEQT) embryos (Figure 3). We conclude that PIE-1(DAQMEQT) retains the ability to inhibit the transcription of at least one zygotic transcript (pes-10), even though it has lost the ability to inhibit Ser2 phosphorylation.
Figure 3.—
Wild-type PIE-1 and PIE-1(DAQMEQT) inhibit transcription of a pes-10:gfp transgene. In situ hybridization shows nuclear accumulation of zygotic pes-10:gfp RNA in pie-1(zu127) embryos expressing the indicated transgenes. Arrows point to germline blastomeres, which do not accumulate pes-10:gfp RNA. Fourteen of 14 embryos expressing wild-type PIE-1 and 28/28 embryos expressing PIE-1(DAQMEQT) showed this pattern.
Localization of PIE-1 to nuclear foci correlates with inhibition of Ser5 phosphorylation:
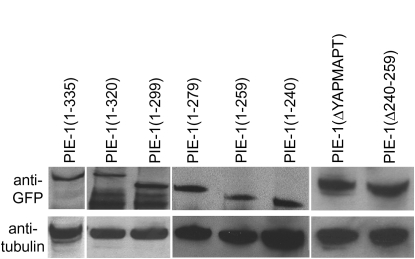
An important control when comparing the activity of transgenes is to verify that the transgenic proteins are expressed at similar levels. Using Western blotting, we found that all GFP:PIE-1 fusions were expressed at comparable levels (Figure 4). We also examined the localization of each fusion in embryos by confocal microscopy. We found that every fusion localized to the P blastomeres and was enriched on cytoplasmic P granules, as is the case for wild-type PIE-1. We noticed, however, that not all fusions showed the same distribution pattern in nuclei. Wild-type GFP:PIE-1 concentrates in numerous, fine nuclear foci of unknown origin (Figure 5). Similar nuclear foci are also observed when endogenous PIE-1 is visualized by immunofluorescence (Tenenhaus et al. 1998). We found that GFP:PIE-1 mutants that retained the ability to inhibit P-Ser5 accumulated in numerous nuclear foci similar to wild type. This was even true for PIE-1(ΔYAPMAPT), which inhibits P-Ser5 but not P-Ser2. In contrast, GFP:PIE-1 mutants that failed to suppress P-Ser5 (identified with asterisks in Figure 5) localized to fewer nuclear foci. The most dramatic phenotype was seen with GFP:PIE-1(1–240), which accumulated only on a few nuclear foci and was primarily diffuse throughout the nucleoplasm.
Figure 4.—
Western blot of GFP:PIE-1 fusions. Total worm extracts from strains expressing the indicated PIE-1 fusions were immunoblotted with anti-GFP antibody. Anti-tubulin is used as the loading control.
To determine whether the PIE-1 nuclear foci are dependent on CDK-7 or P-TEFb activity, we examined the distribution of GFP:PIE-1 in embryos where cdk-7 or cit-1.1/cit-1.2 was depleted by RNAi. We found that in all cases depletion of these factors reduced the apparent number of nuclear foci (Figure 5). This was true even for fusions such as GFP:PIE-1(1–240) whose localization to foci was already severely compromised in wild-type embryos. We conclude that the PIE-1 localization to nuclear foci is sensitive to reduction in the levels of CDK-7, P-TEFb, and RNA polymerase II, and that all PIE-1 mutants tested in this study remain sensitive to changes in these factors.
DISCUSSION
We have performed an in vivo structure–function analysis of the PIE-1 transcriptional repression domain. Our findings support the view that the YAPMAPT motif in this domain is essential for inhibition of P-TEFb activity by PIE-1. Our results also indicate, however, that this activity is not essential to suppress transcription in vivo and that PIE-1 also uses sequences outside the YAPMAPT to inhibit RNA polymerase II.
PIE-1 as an inhibitor of P-TEFb:
The model put forth by Zhang et al. (2003) predicts that the YAPMAPT motif in PIE-1 should be essential (1) to bind to cyclin T, (2) to inhibit Ser2 phosphorylation, and (3) to inhibit transcriptional elongation. We were not able to demonstrate a requirement for the YAPMAPT in our cyclin T binding assay, although we identified a sequence 5′ to the YAPMAPT essential for binding (“cyclin T binding domain”). In their GST pull-down experiments, Zhang et al. (2003) used human CycT1 and the C-terminal domain of PIE-1, whereas we used C. elegans CIT-1.1 and full-length PIE-1, which could account for the different results. It will be important to test directly whether PIE-1 and CIT-1.1 interact in vivo and what specific sequences are required for this interaction.
Consistent with the model of Zhang et al. (2003), however, we found that the YAPMAPT (and the cyclin T binding region) are essential for inhibition of Ser2 phosphorylation in germline blastomeres. This finding supports the view that PIE-1 functions, at least in part (see below), by binding and inhibiting P-TEFb. The earlier finding that high levels of PIE-1 are required for complete suppression of P-Ser2 in germline blastomeres (Tenenhaus et al. 2001) is also consistent with PIE-1 functioning as a competitive inhibitor.
Inhibition of Ser2 phosphorylation, however, is unlikely to be the only mechanism used by PIE-1 to repress transcription. PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT) were able to rescue the embryonic lethality of pie-1(zu127) as efficiently as wild-type PIE-1. Because the embryonic lethality of pie-1(zu127) is thought to be a direct consequence of transcriptional activation in germline blastomeres (Tenenhaus et al. 2001), we infer that PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT) are able to suppress transcription. Consistent with this interpretation, we showed that PIE-1(DAQMEQT) suppresses the transcription of one zygotically expressed transgene pes-10:gfp. If transcription is suppressed, why then is P-Ser2 activated? One possibility is that PIE-1 inhibits transcription using two partially redundant mechanisms: one mechanism targeting pTEFb to prevent elongation and another mechanism targeting a different component, with the net effect of reducing the efficiency of initiation. The latter could function directly by targeting a component of the initiation complex, or indirectly by preventing recycling of the polymerase, or by reversing P-Ser5 phosphorylation. Loss of P-TEFb inhibition, as with the PIE-1(DAQMEQT) and PIE-1(ΔYAPMAPT) transgenes, would cause loci to become transcribed throughout their length, leading to increased P-Ser2 levels. However if initiation and/or recycling of the polymerase were still inefficient, P-Ser5 levels would remain low. Although this interpretation fits the available data, it will remain speculative until we can determine what types of transcripts are made in germline blastomeres.
PIE-1 as an inhibitor of Ser5 phosphorylation:
Our structure–function analyses indicate that (1) PIE-1 can inhibit P-Ser5 (or block its accumulation) independently of PIE-1's effect on P-Ser2 and (2) inhibition of P-Ser5, not P-Ser2, correlates best with PIE-1's ability to inhibit transcription and promote germ cell fate. Low levels of P-Ser5 are a conserved characteristic of embryonic germ cells in C. elegans and Drosophila (Seydoux and Dunn 1997), yet studies in other organisms have focused primarily on monitoring P-Ser2 (Knaut et al. 2000; Tomioka et al. 2002; Deshpande et al. 2004; Martinho et al. 2004). In ascidian embryos, in situ hybridization experiments have suggested that transcription is inhibited in germ cell precursors, but P-Ser2 was detected in those cells (Tomioka et al. 2002). The authors concluded that a different mechanism of repression might operate in ascidians, but it would be interesting to also evaluate P-Ser5 levels. Recent studies in mice have shown that migrating primordial germ cells lack both P-Ser2 and P-Ser5 (Seki et al. 2007).
The ability of PIE-1 to limit P-Ser5 levels in germline blastomeres does not require the YAPMAPT and only partially requires the cyclin T binding motif, suggesting that this activity does not involve a direct interaction with P-TEFb. We propose that PIE-1 also interacts with other component(s) of the transcriptional machinery, and that these interactions prevent RNA polymerase II from successfully initiating at most loci. Consistent with PIE-1 associating with transcription complexes, we have found that PIE-1 localizes to numerous nuclear foci and that this localization is disrupted by depletion of CIT1.1/CIT1.2, CDK-7, and RNA polymerase II. Furthermore, all the PIE-1 mutants that fail to inhibit P-Ser5 are defective in this localization, suggesting that recruitment of PIE-1 to nuclear foci is central to PIE-1 function. Identification of the proteins that interact with the PIE-1 C-terminal domain will be critical to further our understanding of PIE-1's remarkable ability to globally repress transcription. PIE-1 homologs with similar activities have not been identified in other organisms. In Drosophila, several partially redundant factors have been implicated in inhibiting transcription in germ cells (Leatherman et al. 2002; Deshpande et al. 2004; Martinho et al. 2004; Deshpande et al. 2005). It will be interesting to determine whether these factors also act by targeting different phases of the transcription cycle.
Acknowledgments
We thank Jeff Corden and Keith Blackwell for insightful comments on the manuscript. This work was supported by National Institutes of Health grant HD-37047. G.S. is an investigator of the Howard Hughes Medical Institute.
References
- Batchelder, C., M. A. Dunn, B. Choy, Y. Suh, C. Cassie et al., 1999. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 13: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, I., C. Merritt, P. L. Chen, G. Seydoux and K. Subramaniam, 2006. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 292: 244–252. [DOI] [PubMed] [Google Scholar]
- DeRenzo, C., K. J. Reese and G. Seydoux, 2003. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. Schedl, 2004. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development 131: 1247–1257. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun, T. M. Jinks, A. D. Polydorides and P. Schedl, 2005. Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mech. Dev. 122: 645–657. [DOI] [PubMed] [Google Scholar]
- Hao, Y., L. Boyd and G. Seydoux, 2006. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell 10: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut, H., F. Pelegri, K. Bohmann, H. Schwarz and C. Nusslein-Volhard, 2000. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 149: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman, J. L., L. Levin, J. Boero and T. A. Jongens, 2002. Germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr. Biol. 12: 1681–1685. [DOI] [PubMed] [Google Scholar]
- Martinho, R. G., P. S. Kunwar, J. Casanova and R. Lehmann, 2004. A noncoding RNA is required for the repression of RNA polII-dependent transcription in primordial germ cells. Curr. Biol. 14: 159–165. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., C. Schubert, B. Draper, W. Zhang, R. Lobel et al., 1996. The PIE-1 protein and germline specification in C. elegans embryos. Nature 382: 710–712. [DOI] [PubMed] [Google Scholar]
- Palancade, B., and O. Bensaude, 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270: 3859–3870. [DOI] [PubMed] [Google Scholar]
- Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent et al., 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273: 4689–4694. [DOI] [PubMed] [Google Scholar]
- Pellettieri, J., V. Reinke, S. K. Kim and G. Seydoux, 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell 5: 451–462. [DOI] [PubMed] [Google Scholar]
- Phatnani, H. P., and A. L. Greenleaf, 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20: 2922–2936. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, Y., M. Yamaji, Y. Yabuta, M. Sano, M. Shigeta et al., 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and R. E. Braun, 2006. Pathway to totipotency: lessons from germ cells. Cell 127: 891–904. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and M. A. Dunn, 1997. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124: 2191–2201. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and A. Fire, 1994. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120: 2823–2834. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and A. Fire, 1995. Whole-mount in situ hybridization for the detection of RNA in Caenorhabditis elegans embryos. Methods Cell Biol. 48: 323–337. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., C. C. Mello, J. Pettitt, W. B. Wood, J. R. Priess et al., 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382: 713–716. [DOI] [PubMed] [Google Scholar]
- Shim, E. Y., A. K. Walker, Y. Shi and T. K. Blackwell, 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus, C., C. Schubert and G. Seydoux, 1998. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev. Biol. 200: 212–224. [DOI] [PubMed] [Google Scholar]
- Tenenhaus, C., K. Subramaniam, M. A. Dunn and G. Seydoux, 2001. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 15: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tomioka, M., T. Miya and H. Nishida, 2002. Repression of zygotic gene expression in the putative germline cells in ascidian embryos. Zool. Sci. 19: 49–55. [DOI] [PubMed] [Google Scholar]
- Van Doren, M., A. L. Williamson and R. Lehmann, 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246. [DOI] [PubMed] [Google Scholar]
- Wallenfang, M. R., and G. Seydoux, 2002. cdk-7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99: 5527–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm, T., P. Demel, H. U. Koop, H. Schnabel and R. Schnabel, 1999. Ballistic transformation of Caenorhabditis elegans. Gene 229: 31–35. [DOI] [PubMed] [Google Scholar]
- Zalokar, M., 1976. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev. Biol. 49: 425–437. [DOI] [PubMed] [Google Scholar]
- Zhang, F., M. Barboric, T. K. Blackwell and B. M. Peterlin, 2003. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 17: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]