Abstract
Telomeres are an unusual component of the genome because they do not encode genes, but their structure and cellular maintenance machinery (which we define as “telotype”) are essential for chromosome stability. Cells can switch between different phenotypic states. One such example is when they switch from maintenance mediated by telomerase (TERT telotype) to one of the two alternative mechanisms of telomere preservation (ALT I and ALT II telotype). The nature of this switch is largely unknown. Reintroduction of telomerase into ALT II, but not ALT I, yeast led to the loss of their ability to survive a second round of telomerase withdrawal. Mating-based genetic analysis of ALT I and II revealed that both types of telomerase-independent telomere maintenance are inherited as a non-Mendelian trait dominant over senescence (SEN telotype). Additionally, inheritance of ALT I and ALT II did not depend on either the mitochondrial genome or a prion-based mechanism. Type I, but not type II, survivor cells exhibited impaired gene silencing, potentially connecting the switch to the ALT telotype epigenetic changes. These data provide evidence that nonprion epigenetic-like mechanisms confer flexibility on cells as a population to adjust to the life-threatening situation of telomerase loss, allowing cells to switch from TERT to ALT telotypes that can sustain viable populations.
CELLS have evolved multiple mechanisms for propagating both reversible and irreversible phenotypic switches to optimize cell growth and division in the face of environmental changes or other selective pressures. One example of an internal change that creates intense selective pressure is telomere shortening. Telomeres, the termini of chromosomes, are incompletely replicated at each cell division by the conventional replication machinery [the end replication problem (Watson 1972; Olovnikov 1973)]. The ribonucleoprotein telomerase counteracts this shortening by extending telomeric DNA at chromosome ends. When lacking either the core protein or the RNA component of telomerase (Est2 and Tlc1, respectively, in yeast), telomeres gradually shorten until they reach a critically short state and prevent cell division (Lundblad and Blackburn 1993). Once most of a population of telomerase-deficient cells has lost viability (called senescence), survivors arise that can continue cell divisions (Lundblad and Blackburn 1993). These surviving cells represent a subpopulation of cells that have undergone a phenotypic switch to a telomerase-independent mode of telomere maintenance (Lundblad and Blackburn 1993). Telomerase-independent telomere maintenance has been observed in a wide range of eukaryotic settings that include human telomerase-negative cancers and yeasts genetically deleted for telomerase (Lundblad and Blackburn 1993; McEachern and Blackburn 1996; Nakamura et al. 1998; Reddel 2003). Mammalian cells, called ALT cells, can use telomerase-independent means to sustain telomere function by mechanisms thought to involve recombination, by analogy with yeast, although the genetic dependencies for vertebrate ALT phenotypes have not been established.
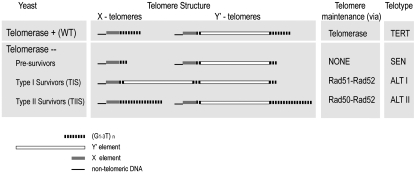
Multiple attributes of cells surviving senescence induced by telomerase deficiency have been reported. The surviving cells of budding yeast Saccharomyces cerevisiae are categorized as either type I or type II on the basis of their telomere composition and mode of maintenance (Lundblad and Blackburn 1993; Teng and Zakian 1999). When telomerase is present normally, all yeast chromosomes are flanked by X elements (one per chromosome end), which, moving out to the chromosome end, are followed directly either by ∼350 bp of (G1–3T)n telomeric DNA repeat tract or by one or more Y′ elements followed by the distal telomeric repeats (Figure 1). The former class are called X telomeres whereas the latter are Y′ telomeres. Before senescence, in Y′ telomeres the terminal G1–3T tract is typically flanked by only a few repeated Y′ elements, each separated from its neighbor by a short internal G1–3T tract (∼50–∼100 bp), as reviewed in Zakian (1996). In type I survivors, all telomeres become Y′-class telomeres: only a very short terminal G1–3T repeat tract (∼100 bp) remains, but it now becomes flanked subtelomerically by an expanded array of tandem repeats of the 5- to 6-kb Y′ element. Hence, type I survivors have amplified the number of subtelomeric Y′ repeat elements. In contrast, type II survivors exhibit little or no Y′ element amplification; instead, telomeres have long but variable-length terminal G1–3T repeat tracts (Lundblad 2002). Not only do the two survivor classes differ in their telomere phenotypes, but also they differ in their genetic requirements for viability. Generation of both survivor types depends on the homologous recombination gene RAD52 (Lundblad and Blackburn 1993; Chen et al. 2001), but type I survivors depend on the RAD51 branch of the recombination pathway while type II survivors rely on RAD50 (Teng et al. 2000; Chen et al. 2001).
Figure 1.—
Yeast telotypes. Telomeres from wild-type (WT) S. cerevisiae have two classes of telomeres: X and Y′, depending on whether they have at least one Y′ element next to the terminal stretch of ∼350 (G1–3T)n telomeric DNA sequence. When more than one Y′ element is present, the Y′'s form an array of direct repeats separated by a short stretch (∼50–100 bp) of (G1–3T)n sequence. Wild-type yeast maintain their telomeres by using TERT and have the TERT telotype. Cells that lose telomerase (presurvivors) show (G1–3T)n tract telomere shortening as a result of incomplete telomere replication. After ∼60–80 generations, they senesce (SEN telotype) and most of them lose viability. A small subpopulation of cells survive. They display changes in telomere architecture and maintain their telomeres through one of the two different branches of recombination: alternative (ALT) modes for telomere replication. Telomeres of type I survivors (ALT I telotype) are all converted to the Y′ class and show amplification of the number of Y′ elements. In telomeres from type II survivors (ALT II telotype), the terminal (G1–3T)n repeat sequence tract is elongated and highly heterogeneous in size.
Yeast lacking telomerase thus change their telomeric properties when they emerge as survivors. Cells can change their properties by means of genetic or epigenetic changes. Epigenetic switches occur through a range of mechanisms: DNA methylation, establishment and maintenance of heterochromatin via histone and other chromatin modifications, and prion-based mechanisms. A defining characteristic of epigenetic changes is the persistence of the cellular phenotype or state through multiple cell divisions even though the original genetic mutation or precipitating event is no longer present in the progeny cells in the dividing population. Telomere states fall into a special class of such phenomena, because changes in the length of the telomeric DNA repeat tract often occur only over the course of many cell divisions. A well-documented example of epigenetic changes relevant to telomeres is gene silencing at telomeres, known as telomere position effect (TPE) (see Perrod and Gasser 2003 for a review). Genes placed close to telomeres are stochastically switched on and off resulting in a bi-stable population. Telomere-length dynamics affect telomeric chromatin and therefore silencing and have been suggested to play a role in TPE (Kyrion et al. 1993). Finally, in telomerase-deleted survivors, the structural state of the telomeres and the ability to maintain them in a telomerase-independent, but recombination-dependent, mode can be propagated indefinitely. Collectively, one can therefore define a particular subclass of epigenetic-like states that we name telotypes. These represent the set of telomere-determined phenotypic characteristics of a cell. Each telotype can be defined by both the structural state of telomeres and the mode of telomere maintenance (Figure 1). Wild-type cells replicate their telomeres through telomerase reverse transcription action and are thus represented as the telomerase reverse transcriptase (TERT) telotype. When telomerase is deleted or not functional in cycling cells, the cells exhibit the senescence (SEN) telotype: telomeres shorten due to the unavailability of any mode of telomere maintenance. The SEN telotype propagation is temporary and limited by the original telomere length and telomere shortening rate, because once telomeres become too short to function, the SEN cells either have to switch to the alternative, recombination-dependent mode of telomere maintenance or lose viability. Therefore, following telomerase loss, cells as a population must eventually switch from the TERT telotype to the alternative lengthening of telomeres (ALT) telotype.
Despite considerable study of the molecular genetic features of survivor formation in S. cerevisiae, the origin of this phenotypic switch remains poorly understood. On one hand, frequencies as high as 1 in 104/cell/generation for generating survivors have been reported (Lundblad and Blackburn 1993); this number is considerably higher than would be expected if the survivors arose only through spontaneous mutations, which are expected to occur at the rate of ∼10−8–10−7 (Luria and Delbruck 1943). On the other hand, some senescent small yeast colonies containing 105–106 cells do not form survivors (McEachern and Haber 2006), suggesting that the rate of survivor formation may vary and be rather low. The dynamic growth behavior of type I survivors, reflected in their ability to alternate between healthy and senescent phenotypes, argues against the mutational origin of survivor formation (Lundblad and Blackburn 1993). The authors proposed that survivors are generated “by selection of cells in which successive recombination events have taken place in subtelomeric regions” (p. 357). Consistent with this hypothesis, accumulating experimental data on the inputs that influence the frequency of survivor formation suggest that telomeric DNA structures such as circles or long stretches of telomeric DNA, which are potential template donor molecules for short telomere elongation via recombination, could be the key factors required to switch to ALT (reviewed in McEachern and Haber 2006). However, the hypothesis of the mutational origin of the switch has not been formally ruled out.
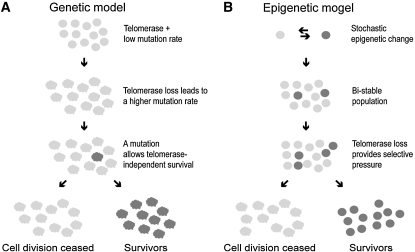
As discussed previously (Lundblad and Blackburn 1993), two formal models could explain survivor formation (Figure 2). In model 1, genomic instability due to the presence of short telomeres produces a mutator phenotype. Consequently, mutation of an appropriate gene for controlling or maintaining telomere length could occur at a relatively high frequency and thus promote survivor formation. In model 2, survivors could arise through an epigenetic-like change. Such a change would occur stochastically and at a low frequency in the cell population and would be positively selected when telomeres became too short. Alternatively, the epigenetic change could also be induced by short telomeres themselves. The changes in models 1 and 2 differ in their predicted modes of inheritance. Chromosomal mutations that generate survivors in model 1 will segregate with a chromosome and thus will be inherited in classic Mendelian fashion. In contrast, epigenetic or epigenetic-like changes (model 2) will be characterized by a dominant, and non-Mendelian, inheritance pattern. Any nonchromosomal determinant such as a protein or nonchromosomal (episomal) DNA will be partitioned potentially equally among all progeny. To distinguish between these two models, we analyzed the patterns of telotype heritability. Here we show that the ALT telotype (telomerase-independent telomere maintenance) of both type I and type II survivors is inherited as a dominant non-Mendelian trait over SEN. This result supports the model of the episomal or epigenetic nature of the TERT-to-ALT telotype switch. We further demonstrate that survivor formation does not depend on the presence of the largest yeast episome, the mitochondrial genome, thereby eliminating one possible mode of non-Mendelian genetic inheritance. Furthermore, elimination of the Hsp104 chaperone, which is required for all known prion-based inheritance in yeast, did not prevent survivor formation or alter the normal survivor formation frequencies. These data provide evidence that a nonprion epigenetic-like change(s) allows the switch from the TERT to the ALT telotype. Finally, ALT I cells had impaired gene silencing, providing a potential connection between epigenetic changes and the switch to this ALT telotype.
Figure 2.—
Hypothetical genetic (A) and epigenetic (B) models of survivor formation upon telomerase loss. Survivors are formed with a frequency of ∼10−4 (Lundblad and Blackburn 1993). In the genetic model (A), loss of telomerase results in a mutator phenotype that would increase the spontaneous mutation rate to ∼10−4. A mutation that allows indefinite viability in the absence of telomerase will allow the host cell to seed a population of survivors. In model B, due to stochastic epigenetic switches, the cell population is not homogeneous, thereby forming a bi-stable population. One of the epigenetic states may confer a growth advantage upon telomerase loss and therefore lead to selective enrichment of cells in the population.
MATERIALS AND METHODS
Yeast strains:
All strains in this study, except for the prion-related experiments, were in the S. cerevisiae A364a background, which was derived from R. Deshaies strain RDY495 (MATa leu2-3, 112 ura3-52 trp1-289 bar1∷LEU2). S. cerevisiae W303 isogenic HSP104 and hsp104Δ strains were used to address the role of prion inheritance in survivor formation.
Yeast manipulations:
MATΔ derivatives of A364a were obtained by disrupting MATa with Kanr using a PCR-generated disruption cassette (Longtine et al. 1998) and by subsequent selection on medium containing G418. Telomerase deletion derivatives of A364a were obtained by similarly disrupting either EST2 or TLC1 with either the TRP1 or URA3 gene cassettes. Disruption of the appropriate gene was confirmed by colony PCR. At this point, telomerase-deficient colonies were still actively growing on the first selection plate (media lacking either tryptophan or uracil) and were called presurvivors (SEN). Survivors (type I and II) were isolated by serially streaking the est2Δ and tlc1Δ strains on plates until cells underwent viability crisis followed by the appearance of healthier-growing survivor cells. To classify survivors as either type I or type II, individual surviving colonies were streaked for single colonies, and the telomere composition of these single colonies was analyzed by Southern hybridization. To mate a presurvivor with a survivor or with another presurvivor, cells from a colony of freshly made presurvivors were mixed with cells of the appropriate mating type on a rich media plate and then incubated for 6–8 hr. Diploids were selected on double drop-out plates.
Halo test:
Approximately 106 MATa bar1 cells were plated on YPD plates using glass beads and ∼107 MATα cells were patched in the center of each plate. Plates were incubated at 30° for 16–36 hr and photographed.
Generation of ρ° yeast:
Strains lacking mitochondrial DNA (ρ°) were generated by temporarily converting the endogenous PIF1 gene to a version specifically defective in mitochondrial DNA replication: the pif1-m1 mutation (Schulz and Zakian 1994). The pif1-m1 mutation eliminates the first Met codon and generates an ApaI restriction site by changing the endogenous sequence TTAC ATG CCA to TTGG GCC CCA. Eliminating the first Met codon in this way prevents export of the Pif1 protein to mitochondria. To introduce the pif1-m1 mutation, yeast cells were transformed with the integrating plasmid pYT118, which was designed to create one full-length pif1-m1 gene and one truncated pif1 gene when integrated at the endogenous PIF1 locus. pYT118 carries the PIF1 promoter followed by the 5′ coding sequence up to the first Acc65I restriction site and contains the pif1-m1 mutation. pYT118 was generated by ligating pRS406 digested with EagI and Acc65I to two PIF1 fragments; one contained 500 bp of the PIF1 promoter region flanked by EagI and ApaI sites, and the other contained the PIF1 coding region flanked by Acc65I and ApaI sites (the ApaI sites replaced the first Met codon in both fragments). The PIF1 fragments were amplified from genomic DNA using standard PCR techniques and the appropriate primers. Cells were transformed with pYT118 linearized with PmeI, and plasmid integration was selected by growth on plates lacking uracil. Loss of mitochondrial DNA was confirmed in two ways: (1) inability of petite colonies to grow on plates containing glycerol as the only carbon source and (2) loss of the Southern hybridization signal when total genomic DNA was probed with 32P-labeled total mitochondrial DNA. The pif1-m1 genotype of each ρ° derivative was returned to PIF1+ by selecting for plasmid loop-out events on plates containing 5-FOA. Return to PIF1+ was confirmed by digesting PCR products spanning the PIF1 5′ coding end with ApaI. ρ° colonies containing PIF1 (PCR products were resistant to ApaI digestion) were analyzed for survivor formation.
Southern hybridization:
Genomic DNA was isolated according to standard protocols using cells scraped from a plate. For analysis of telomeric restriction fragment patterns (“teloblot”), this DNA was then digested with XhoI, separated on a 0.7% agarose gel, and transferred to a MagnaGraph nylon membrane (Osmonics). Then membrane was probed for the telomeric TG1–3 sequence with the 32P-end-labeled oligonucleotide (CCCACA)4.
RESULTS
Resenescence of type I and type II survivors:
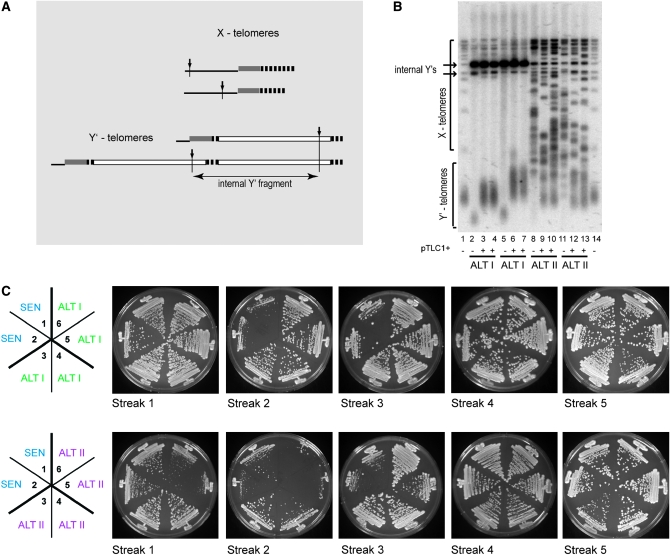
The simplest test for whether genetic (irreversible) or epigenetic (reversible) changes are responsible for the switch from TERT to the ALT telotypes would be to reverse ALT cells to telomerase-dependent telomere maintenance, i.e., to switch them back to TERT, and then remove telomerase once again and measure the frequency of resenescence. If the change in the course of the original senescence was genetic, the cells would not be expected to undergo a viability crisis the second time they lose telomerase, because all the cells in the population would already have the “survivorship” mutation from the first round of senescence. If, however, the TERT-to-ALT switch results from an epigenetic change and the ALT cells fully switch their epigenetic state back to TERT upon regaining telomerase, then they will resenesce after telomerase is lost once again. We reintroduced TLC1 on a CEN/ARS plasmid to tlc1 type I (ALT I) and type II (ALT II) survivors and propagated them for ∼300 generations (15 restreaks). As reported (Lundblad and Blackburn 1993; Teng and Zakian 1999), upon reintroduction of telomerase the length of the telomeric TG tracts in ALT cells reequilibrated close to that of the wild-type cells (Figure 3B). However, the transition from ALT to TERT was not complete for either ALT I or ALT II cells: although the telomeres of ALT I cells rapidly returned to wild-type or longer lengths, the Y′ telomeres did not revert back to X, even after ∼300 cell generations. The ALT II cells shortened their telomeres after telomerase was recovered and some of the telomeres were in the range of the wild-type cells (Figure 3B, lanes 8–13). However, a full return to the Southern blot band pattern characteristic of the wild-type telomeres was not observed even after ∼300 generations (Figure 3B, lanes 1 and 14). Therefore, complete reversion from ALT I or II to TERT did not occur.
Figure 3.—
Resenescence of ALT I and ALT II cells. (A) Schematic of the yeast telomeric terminal XhoI fragment analyzed by Southern blot hybridization in B. Since terminal (G1–3T)n tracts are variably sized, all telomeric bands are characteristically smeared. Telomere-proximal XhoI sites are shown by vertical arrows. Since neither X elements nor (G1–3T)n tracts have XhoI sites, the cleavage occurs within the neighboring unique chromosomal sequence. As a result, each X-telomeric XhoI fragment runs as a smeared band of a certain size range. The Y′ element contains a unique site and therefore the XhoI terminal fragments from the multiple different Y′ telomeres run together as a group. Internal Y′ fragments, resulting from XhoI digestion of tandem Y′ elements in an array, run as ∼5.2- or 6.8-kb bands. A telomeric sequence oligonucleotide (C3ACA)4 was used as a probe to detect the telomeric XhoI fragments. (B) Southern blot hybridization analysis of telomeres of tlc1Δ ALT I and ALT II cells before and after reintroduction of functional telomerase using pTLC1+ (TLC1 cloned in a CEN/ARS vector). Independently isolated ALT I and ALT II survivors (two each) were transformed with the pTLC1+ plasmid. Two clones from each transformation were propagated for ∼300 generations (15 plate passages). Genomic DNA from the original survivors (lanes 2, 5, 8, and 11) and their telomerase-positive derivatives propagated for ∼300 generations was purified and analyzed by Southern hybridization. DNA from wild-type TLC1 cells was run as a reference control in lanes 1 and 14. Brackets specifying the X and Y′ telomeres on the left of the blot are relevant only to wild-type and ALT I samples (lanes 1–7 and 14). The observed change in mean telomere length in one of the two ALT I survivors (lanes 6 and 7) may suggest the possibility of a mutation gained during the original senescence. In ALT II cells, Y′ telomeres may contain very long stretches of TG tracts, resulting in a much increased length of the terminal XhoI fragment. (C) Cell viability of ALT I and ALT II cells after secondary telomerase loss. Approximately 300 generations after reintroduction of TLC1 on a plasmid into ALT I and ALT II yeast, cells that underwent spontaneous plasmid loss were isolated and their growth was observed during the next ∼100 generations (streaks 1–5). Two freshly made tlc1Δ strains were used as positive controls for senescence shown in sectors 1 and 2 (top and bottom). Cells grown in sectors 3–6 (top) were generated from the strains analyzed in B, lanes 3, 4, 6, and 7, respectively. Strains in sectors 3–6 (bottom) were derived from the parents in B, lanes 9, 10, 12, and 13, respectively.
Despite these complications, we exposed cells to a second round of telomerase loss. The ALT I cells survived it with an efficiency so high that no sign of senescence was detected in the population (Figure 3C, top). In contrast, clones originating from one ALT II survivor underwent a strong viability crisis (Figure 3C, bottom, streak 2, sectors 5 and 6). Thus, a mutation as a cause of the TERT-to-ALTII switch could be ruled out for this ALT II strain. The resenescing clones generated from the other ALT II survivor (Figure 3C, bottom, sectors 3 and 4) showed little if any viability crisis. It remains possible that the original TERT-to-ALT conversion of this strain occurred through a mutation, which then allowed cells to avoid the viability crisis during resenescence. Thus, to gain a further insight into the mechanism of TERT-to-ALT switching, we performed a mating-based genetic analysis of the ALT phenotypes.
Design of suitable genetic crosses to detect telotype inheritance:
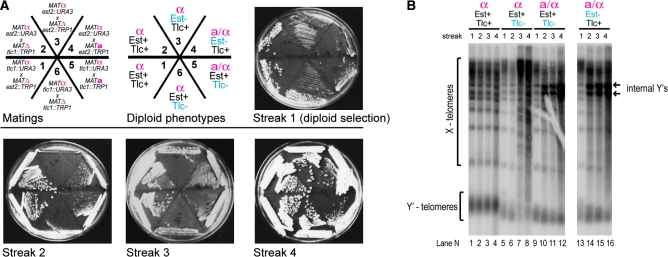
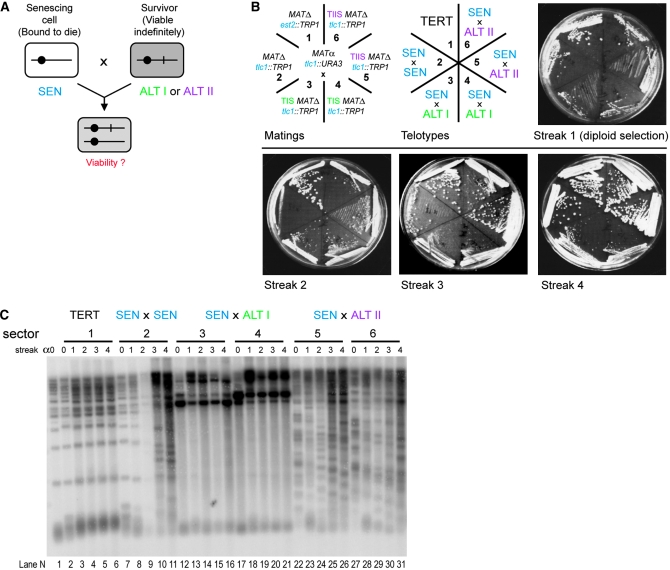
The procedure of making crosses among strains of different mating type and various genotypes underpins genetic studies in yeast. To test whether the senescence phenotype, normally observed in haploid cells, is affected through the mating process, we mated cells with recently inactivated telomerase (presurvivors, SEN telotype; see Figure 1) and followed the senescence of the resulting diploids. Crosses between tlc1 and est2 presurvivors were used as positive controls for survival, as they generate telomerase proficient EST2/est2 TLC1/tlc1 diploids that restore the TERT telotype. As expected, no senescence for these cells was observed (Figure 4A, sectors 1 and 2). Matings between two presurvivors lacking the same telomerase component (Est2 or Tlc1) result in telomerase-deficient zygotes that were expected to senesce. However, clear cell viability crisis could not be observed in telomerase-deficient MATa/α diploids (Figure 4A, sectors 4 and 5). As previously described (Lowell et al. 2003), the severity of the cell viability crisis is suppressed by the MATa/α genotype. To overcome this problem, we replaced MATa cells with MATΔ; MATΔ cells phenocopy MATa and therefore mate with MATα, but the crosses result in MATα diploids. The senescent phenotype in a diploid was now clearly observed when we mated presenescent MATα to MATΔ cells (Figure 4A, sectors 3 and 6). This obviated the problem of viability crisis alleviation in a/α diploids.
Figure 4.—
Determination of mating-type background suitable for analysis of telotype inheritance in diploids lacking telomerase. (A) Growth senescence in diploids is most clearly observed in MATα cells. Cell growth over ∼80 generations (∼20/plate) was followed by serially restreaking. Each EST2 or TLC1 allele was disrupted in haploid ura3 trp1 cells with either TRP1 or URA3. The matings were designed so that Ura+ Trp− cells were crossed with Ura− Trp+ cells (top left), allowing for diploid selection on media lacking both uracil and tryptophan (streak 1) and then passaged on YPD plates (streaks 2–4). (B) Southern blot of telomeres from diploid strains in A. Cells from sectors 1, 6, 5, and 4 (from left to right) were used for telomere analysis.
In addition to growth phenotype, telomere length and composition were analyzed by telomere-specific Southern blotting (teloblot). In telomerase-positive diploids recovered from crosses 1 and 2, the average length and a well-defined size range of the terminal TG repeat tracts was stably maintained over many generations at both Y′ and X telomeres (Figure 4B, lanes 1–4). Telomerase-deficient MATa/α diploids showed amplification of the Y′-element-specific bands at early generations after mating (by streak 2; see Figure 4B, lanes 10–12 and 14–16), consistent with the previously reported higher frequency of type I survivor formation (Lowell et al. 2003). In contrast, in MATα diploids lacking telomerase, telomeres shortened to a critical length and at that point most cells subsequently ceased to divide. As the number of cells surviving this crisis increased in the population, a more heterogeneous set of telomere lengths arose and the pattern of bands became less distinct (Figure 4B, lanes 7 and 8). This telomere pattern is indicative of type II survivors, in which the terminal TG repeat tract is considerably but variably lengthened and subtelomeric Y′ elements are not amplified. A predominance of type II telomeres upon senescence of our haploid S. cerevisiae A364a was common. Therefore, MATα diploids resemble haploids in their senescence features. Hence, all the following genetic analyses of survivors were carried out through generation of MATΔ/MATα diploids.
The survivor phenotype is dominant to senescence:
To determine the type of genetic or cellular change(s) that allow cells to survive a telomere-length-related crisis, we subjected survivors to genetic analysis by mating them to cells with recently inactivated telomerase (presurvivors, SEN) and followed the viability/senescence of the resulting diploids.
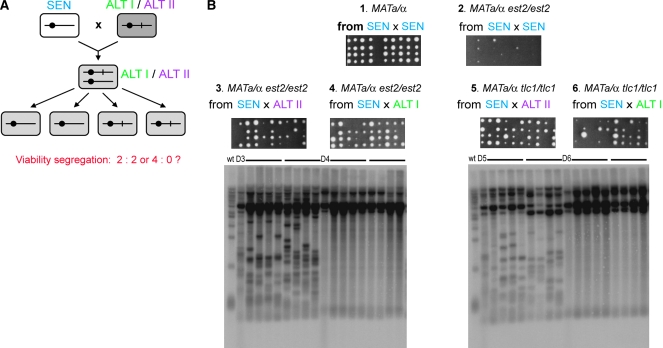
To test whether the survivor growth phenotype is dominant or recessive to senescence, type I or type II haploid survivors (ALT I and ALT II telotypes, respectively) were mated to presurvivors (SEN telotype) as described above (Figure 5A). If survivors were originally generated due to a recessive loss-of-function (LOF) mutation, then the resultant diploid would senesce, as the intact copy of the normal gene from the presurvivor would complement the LOF mutation in the survivor. If survivors were generated by some other mechanism, including a dominant gain-of-function mutation, then the resultant diploid would not senesce. Inspection of the growth phenotypes showed that the diploids, generated from crosses between ALT and SEN, grew well regardless of whether the SEN cells were mated to ALT I or ALT II (Figure 5B, sectors 3–6). In contrast, very poor growth was observed at streak 2 in diploids resulting from mating two SEN populations as these cells senesced and underwent crisis (Figure 5B, sector 2). Colonies were generally small in diploid strains derived from ALT I crosses, reflecting the characteristic slow-growth phenotype of type I survivors (Figure 5B, sectors 3 and 4). Colonies from crosses involving ALT II cells were more heterogeneous and grew similarly to ALT II haploids. Thus, both the type I and type II survivor growth phenotype was dominant over presurvivor senescence.
Figure 5.—
ALT I and ALT II telotypes are dominant over SEN. (A) Schematic of the genetic analysis experiment shown in B. Each cell is shown as a rectangle; a chromosome is indicated by a solid line and a centromere by a circle. SEN cells and ALT cells could differ by a hypothetical chromosomal mutation (shown as vertical line on chromosome), expected to be inherited in a classic Mendelian fashion. Alternatively, ALT cells have a different change (episomal DNA, epigenetic switch, prion, etc.), predicted to show non-Mendelian inheritance. The hypothetical changed state is indicated by shading. (B) Cell growth over ∼80 generations of diploids derived from crosses between SEN tlc1 cells and SEN est2 (sector 1, negative control for senescence), SEN tlc1 (sector 2, positive control for senescence), two independently isolated ALT I tlc1 strains (sectors 3 and 4), and two independently isolated ALT II tlc1 derivatives (sectors 5 and 6). Detailed genotypes of the strains involved in the crosses are shown on the diagram (top left). The experiment was performed as described in Figure 4. (C) Southern blot of telomeres from the haploids prior to matings and the resultant diploid cells in B. The DNA from MATα presurvivor common to all six crosses is in lane 1 (streak α0). The DNA samples from the six MATΔ strains prior to matings are shown as streak 0 lanes. Streaks 1–4 represent serial passages of the diploids obtained in crosses between the haploids from the streak 0 to the MATα presurvivor. The six horizontal bar numbers on the top of the gel and the telotypes above them correspond to the sector numbers and telotypes in B (top middle).
The ability of each survivor type to confer the corresponding telotype on the presurvivor telomeres was then assessed by teloblot. Telomerase-positive diploids derived from telomerase-deficient SEN haploids regained the TERT telotype: telomeres returned to their equilibrium length within the first diploid selection streak and were then maintained at this length (Figure 5C; compare lanes 2–6). When ALT I cells were mated to presurvivors, DNA restriction fragments indicative of the presurvivor X telomeres were lost within the first 20 generations of growth, suggesting early conversion of telomeres from SEN to ALT I (Figure 5C; compare lanes 1 and 8 to lanes 13 and 18). When ALT II yeast were mated to a presurvivor, telomeres appeared heterogeneous in length in the characteristic ALT II pattern. This heterogeneity made it difficult to track the X telomeres from the presurvivor genome. However, the TG repeats of Y′ telomeres gradually shortened at the rate normally observed in senescing cells and were then lengthened in a type II pattern (Figure 5C; compare bottom broad band in lanes 23–26 and 28–31 to lanes 8–11). These results suggest that, in the SEN × ALT II diploids, telomeres originating from the presurvivor parent are lengthened through addition of TG repeat sequence only once they approach or have reached a critical minimum length.
These results indicate that a recessive mutation in survivors cannot account for the switch to ALT telotype. Had there been such a mutation, the diploids generated by crossing a haploid presurvivor to a haploid survivor would have acquired a complementing wild-type allele from presurvivors and senesced rapidly. Rather, both ALT telotypes act dominantly in the diploid products of the cross, but in different fashions: the ALT I telotype spreads to presurvivor telomeres quickly regardless of the TG repeat length, whereas the presurvivor telomeres are converted to ALT II only after the terminal stretches of TG sequence have become short.
The ability to switch to the ALT telotype is inherited in a non-Mendelian fashion:
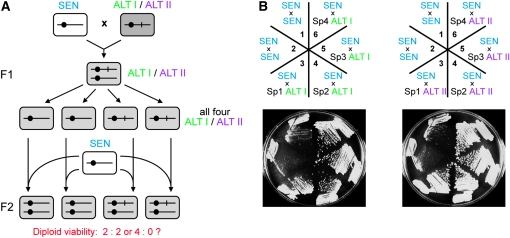
To determine whether the dominant ALT telotypes observed above were due to a classical mutation in a single genetic locus, MATa/α diploids derived from ALT × SEN crosses were sporulated. The growth and telomere morphology of the spore progeny were then analyzed. Diploids derived from ALT × SEN matings should be heterozygous for any hypothetical genes conferring the survivor phenotype. If a single genetic locus were required to maintain the survivor phenotype, then senescence:viability should segregate 2:2 upon sporulation (see Figure 6A). If the viability of the original ALT cells (haploid survivors) does not come from a genetic change in a single chromosomal locus, then 4:0 segregation is expected. Examination of the growth phenotype of tetrads derived from four independently obtained ALT × SEN diploids revealed that from both type I and type II survivor diploids a large fraction of tetrads contained three or four live spores, although tetrads containing fewer live spores were also observed (Figure 6B, diploids 3–6). The growth of these spores—two complete tetrads from each of the diploids—was further examined over many generations. None of the spore progeny of all eight tetrads showed any sign of senescence for at least 120 generations (data not shown). In contrast, when a telomerase-deficient presurvivor diploid was sporulated, for most tetrads in all spores loss of growth was observed immediately; thus, as expected, these spores senesced early because telomeres had continued to shorten in the diploid state (Figure 6B, diploid 2).
Figure 6.—
Non-Mendelian inheritance of the dominant survivor phenotype. (A) Schematic of the genetic analysis experiment (shown in B) addressing possible inheritance patterns for a hypothetical change that confers ALT telotypes. If the change is a chromosomal mutation, then upon diploid sporulation it is expected to segregate with the chromosome bearing it, thereby being inherited by only two of the four spores (the two on the bottom right). The other two spore progeny are expected to senesce. If, however, the change in ALT is of a different nature (see Figure 5 legend), it will be inherited by all four spores. (B) Viability segregation and telomere composition analysis of diploids derived by mating a presurvivor (SEN) with either ALT I or ALT II haploids. SEN cells were mated ∼20 generations after telomerase deletion and the diploids were sporulated immediately after being selected (prior to senescence). Cross 1 is from mating tlc1 and est2 cells to recover TERT telotype diploids as a negative control for senescence. Cross 2 is a positive control for senescence. Two full-tetrad progeny from each of the four SEN × ALT matings (see spore growth for crosses 3–6) were assayed for telomere composition by teloblot, shown under the corresponding tetrad growth panels. Samples from the parental diploids are in lanes D3–D6. Horizontal bar indicates the lanes in which DNA from spores of the same tetrad was analyzed. wt, wild-type DNA.
Southern blotting analysis showed that the telotype of the survivor parent was also propagated in the telomeres of all surviving spores. This segregation analysis had required the use of MATa/α diploids. Therefore, as expected, Y′ amplification was observed in all spores, even from type II survivor diploids (Figure 6B). Telomeres were examined using the same four diploids and the eight full tetrads (two from each) as described above for the senescence phenotype. All spores from all the ALT × SEN diploids inherited the telomere composition characteristic of the original ALT strains (Figure 6B; see teloblots).
These findings on both the growth and the telomere phenotypes of the tetrads from the ALT × SEN crosses suggest that maintenance of the ALT I and II telotypes does not rely on a mutation at a single genetic locus in survivors; otherwise, the haploid tetrad progeny would have become senescent in a 2:2 pattern. Two possible explanations exist for the results observed. First, once the switch to the ALT telotype has occurred, its maintenance no longer requires the switch-inducing factor to be present. Rather, it is only the establishment of the ALT telotype (i.e., conversion of the telomeres to those of the survivor type), which occurred during the diploid stage, that requires a hypothetical mutation to be present (e.g., of a single genetic locus). Second, both the ability to switch to and to maintain the ALT telotype is inherited in a non-Mendelian fashion.
We ruled out the first hypothesis, i.e., that a single genetic locus in the survivor is required to transduce the ALT telotype onto presurvivor telomeres. Tetrad spores from both ALT I and ALT II heterozygous diploids were each mated with a SEN tester strain (see Figure 7A). If mutation of a single genetic locus is required to convert presurvivor telomeres in the diploid to the corresponding ALT type and thereby prevent senescence, then only two of the four spores would inherit the competence to confer survival when mated to SEN cells. Since, as described above, telomerase-deficient MATa/α diploids do not display a clear viability crisis, these crosses were performed by first deleting the MATa locus from the appropriate spores to generate MATΔ derivatives, which were then crossed to MATα SEN cells. MATα spores were mated to MATΔ SEN yeast. As expected, in control SEN × SEN crosses, telomerase-deficient diploids clearly underwent a viability crisis ∼40 generations after mating (Figure 7, sectors 1 and 2). In contrast, diploids derived by mating the same SEN haploids to each of the four tetrad spores grew robustly and did not senesce (Figure 7, sectors 3–6). These same results were observed whether the tetrads analyzed were from ALT I or ALT II diploids. Thus, the ability of a survivor to rescue a presurvivor is inherited and passed on to progeny in a manner inconsistent with mutation of a single genetic locus; rather, this ability exhibits non-Mendelian inheritance.
Figure 7.—
A non-Mendelian heritable change acts dominantly both to establish and to maintain the survivor state. (A) Schematic of the genetic analysis experiment (shown in B) addressing possible inheritance patterns for a hypothetical change that confers ALT telotypes. Because all four spores inherited the ALT telotype (see Figure 6), either the change in the original ALT cells is not a chromosomal mutation and therefore inherited in a non-Mendelian fashion (hypothesis 1) or it is a chromosomal mutation, which might be required not for the maintenance but for the establishment of the ALT telotype (hypothesis 2). The crosses of the spore progeny from F1 to SEN cells should distinguish between these two possibilities. If hypothesis 1 is correct, then all four spore progeny are expected to confer the ALT telotype to the diploids in F2. If the hypothetical change is a mutation that is required for the establishment of ALT, then two of the spore progeny that do not have the mutation will not rescue the senescence phenotype of SEN cells. (B, top) Diagram indicating the crosses examined. Cells in sectors 1 and 2 are positive controls for senescence obtained as described in Figure 5. Sectors 3–6 represent mating presurvivors (SEN) with four progeny spores from the same tetrad (from Figure 6). Each spore (Sp) colony was subsequently mated to another presurvivor, as shown. Plates from the second restreak are shown (∼40 generations after mating). Diploids were passaged for ∼80 generations, as in Figure 5. No viability crisis was observed for the diploids from sectors 3–6. One example each for tetrads derived from type I and type II survivors are shown; duplicate experiments gave similar results.
A mitochondrial DNA mutation or a prion-based mechanism are not responsible for survivor formation or maintenance:
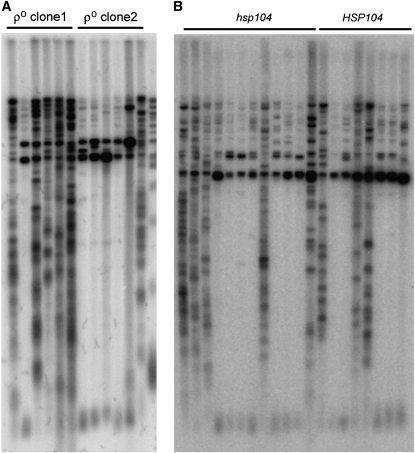
The analysis thus far indicates that the ability of cells to survive a telomere-length-induced crisis is due to a dominant change that is inherited in a non-Mendelian fashion. Known examples of non-Mendelian modes of inheritance include genetic changes encoded by episomal (extrachromosomal) DNA, epigenetic changes, and prions. The mitochondrial genome represents the largest yeast episomal DNA molecule. Although each yeast cell contains only one highly branched mitochondrion that partitions into daughter cells, the copy number of mitochondrial DNA molecules is not tightly controlled so that heteroplasmy can result in different progeny cells with the same nuclear genomes (Piskur 1994). To test whether the switch to ALT telotype occurred due to a dominant mutation in the mitochondrial genome, we determined whether survivors appeared in cells lacking mitochondrial DNA (ρ° cells). For these experiments, either the protein or the RNA component of telomerase was inactivated in ρ° haploids. These strains senesced as expected after ∼80–100 generations and formed survivors at a frequency roughly similar to that of the equivalent ρ+ haploid strains (data not shown). Telomere-specific Southern analysis of survivors from two independently derived ρ° strains showed that both type I and type II survivors occurred (Figure 8A). Thus, establishment and maintenance of the survivor state did not require the mitochondrial genome.
Figure 8.—
Neither mitochondrial DNA nor prion propagation is required for survivor formation. (A) ρ° yeast form survivors with telomere structure similar to ρ+ cells. Teloblot of 12 independently obtained est2Δ and tlc1Δ survivors (lanes 1–12). DNA of the parental telomerase-proficient strain (wt) is in the far right lane. (B) Teloblot of survivors obtained from hsp104 and HSP104 isogenic strains.
Prion inheritance over many cell divisions can occur in the absence of a nuclear gene mutation. The heat-shock protein Hsp104 is required for all known prion-based inheritance in yeast (Tuite and Lindquist 1996). Therefore, we determined whether survivor formation, maintenance, and telomere phenotypes were altered when cells were deleted for Hsp104 as well as telomerase. We found that Hsp104 does not notably affect the survivor formation or survivor telotype frequencies (Figure 8B). This independence of survivor formation on the heat-shock protein Hsp104 implies that survivorship is not prion based.
ALT I but not ALT II cells have decreased silencing at the silent mating-type locus HML:
The best-characterized example of epigenetic switching in yeast is gene silencing at telomeres (Gottschling et al. 1990) and at the silent mating-type loci HML and HMR (Klar et al. 1981; Nasmyth et al. 1981). Transcription of a gene placed at a telomere stochastically switches on and off and the switched states are inherited through multiple generations. Furthermore, telomeres and silencing at internal chromosomal regions interact, as longer telomeres attenuate silencing at HMR (Buck and Shore 1995). Since silencing factors, such as Sir2/3/4, bind both telomeres and HML/HMR, telomere elongation is thought to enrich the Sir proteins at telomeres, thus depleting them from HMR (Buck and Shore 1995). We hypothesized that telomerase loss may affect silencing too.
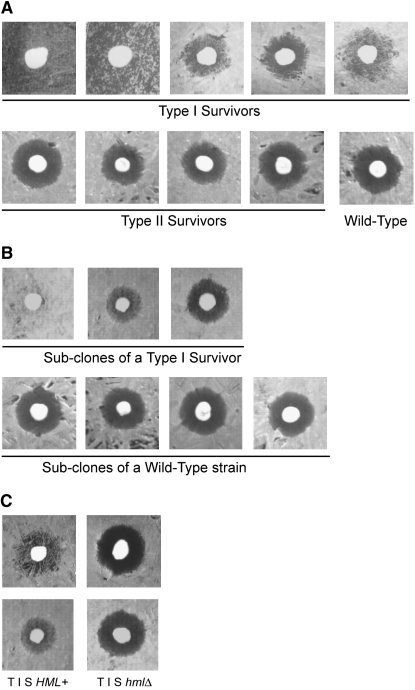
MATa, but not MATα or MATa/α, cells are sensitive to the mating pheromone α-factor, secreted by MATα cells. Therefore, the growth of MATa cells on plates is inhibited around a patch of MATα cells, resulting in a “halo” that represents the zone of growth inhibition (see Figure 9). When the silencing at HML, which contains the α1 and α2 genes defining the MATα cell type, is perturbed, MATa cells become phenotypically MATa/α. Hence they are no longer sensitive to α-factor and can grow in close proximity to MATα cells. Depending on the frequency of desilencing in the population of the MATa cells, the halo becomes turbid to different degrees. We tested for any evidence of epigenetic changes upon the switch to ALT telotype by assaying silencing at HML using this “halo” test. All independently isolated type II survivors behaved similarly to the telomerase-positive (wild-type) control, showing clear, nonturbid halos (Figure 9A, bottom). However, independently isolated type I survivors showed heterogeneous behavior: some failed to form a visible halo, and others produce halos but with variable degrees of turbidity, indicating that many cells were unresponsive to the pheromone (Figure 9A, top). To check that the difference in halo response was not specific to this particular survivor subclone, we assayed different subclones obtained from the same survivor and compared them to subclones from the wild-type telomerase-positive strain (Figure 9B). The subclonal variation in halo test response of type I survivors persisted, suggesting an epigenetic nature of the decreased HML silencing phenotype. When the HML locus was deleted, only clear halos were formed (Figure 9C), confirming that decreased silencing was responsible for the observed halo phenotype of the type I survivors. We therefore tested whether stochastic desilencing of HML was required for the TERT-to-ALT I switch and ALT I viability. However, the ALT I hmlΔ cells stayed viable over multiple generations and the HML locus also was not required for survivor formation (data not shown). Hence ALT I cells have altered epigenetic properties with respect to HML silencing.
Figure 9.—
Type I, but not type II, survivors have decreased silencing at HML. (A) Silencing at HML assayed by halo test in MATa bar1Δ type I and type II survivors. Cells with silenced HML are phenotypically MATa and do not grow in close proximity to MATα cells (patched in the center), thereby forming a halo of no growth. When HML silencing is abrogated, MATa cells become phenotypically MATa/α and their growth is no longer inhibited by the α-factor secreted by the central MATα cells. Hence the more cell growth within the halo (“turbid halo”), the lower the HML silencing. (B) Clonal variation of silencing at HML. Subclones of a type I survivor (top) and of a telomerase-positive wild-type control strain (bottom) assayed for halo formation. (C) Deletion of the HML locus suppresses the turbid halo phenotype of type I survivors. Halo assays for two independently generated type I survivors and their hmlΔ derivatives are shown.
DISCUSSION
The inheritable stochastic changes that lead to survivor formation in theory could have been irreversible on the basis of the alterations of DNA sequence (genetic), such as spontaneous mutations, or by analogy to the programmed cell-to-cell diversity resulting from, for example, VDJ recombination in B-cells. In contrast, we have shown that the reversible, mitotically inheritable changes defined by the survivorship state are not caused by genetic mutations. The two specific telotypes, the ALT I and II telotypes, found in yeast cells that survive loss of telomerase, can be passed on by a dominant and non-Mendelian inheritance mode to a presurvivor cell that otherwise would be destined to die. Therefore, a genomic mutation in its classical definition can be ruled out as a cause of the survival, at least for some of the ALT II cells. However, it remains formally possible that the original TERT-to-ALT switch does require a genomic mutation, but the resulting alterations in the telomere state confer the ALT phenotype in an epigenetic manner on the future progeny. Alternatively, such a mode of inheritance can be a characteristic of episomal and epigenetic change, as well as of prions—self-propagating switches in protein conformation. No evidence for a prion-like mechanism was found on the basis of the insensitivity of survivor formation, propagation, and telotypes to deleting Hsp104, a chaperone required for the prion propagation. The yeast mitochondrial genome, which can be considered an episome, was also not required for survivor formation. We cannot exclude a possible contribution from other episomes such as rDNA circles or 2μ plasmids that could be responsible for survival, although no evidence for any such mechanism has emerged to date. The telotype could, however, determine the abundance of episomal DNA molecules consisting of the circles of Y′ repeats and (G1–3T)n telomeric DNA found in yeasts, which have been hypothesized to be critical for survivor formation (Tomaska et al. 2004; Larrivee and Wellinger 2006). Notably, the prevalence of Y′ repeat circles is much higher in both type I and type II survivors than in control yeast (Larrivee and Wellinger 2006; S. Makovets and E. Blackburn, unpublished observations). However, the vast majority of Y′ repeats are still part of linear chromosomes, as analyzed by two-dimensional native–native gel electrophoresis (Larrivee and Wellinger 2006; S. Makovets and E. Blackburn, unpublished observations), and because of their numerical dominance these Y′ elements could be the main donor of the template sequence for telomere replication in ALT I cells. The same could be true for ALT II yeast, where the long stretches, rather than circles, of (G1–3T)n DNA on telomeres can be used for telomere maintenance. Such chromosomally located, highly repeated DNA sequences could be inherited in a dominant non-Mendelian fashion if a substantially large number of the 32 telomeres have them and only a few are required for the inheritance of the ALT telotypes.
The inheritance mode of survival vs. senescence also can be explained by an epigenetic switch. Other epigenetic changes in various situations are known to involve heritable transcriptional switches, which occur through DNA methylation, histone modifications, or gene expression from positive feedback loop modules. Analysis of transcription profile dynamics during yeast replicative senescence identified multiple genes whose expression was turned on and that stayed on in type II survivors (Nautiyal et al. 2002). One or more of these genes could be regulated in a positive feedback loop fashion, such that their upregulated state would be inherited in a dominant non-Mendelian fashion. Such populations of yeast will exist as a bi-stable system where a small subpopulation is able to survive telomerase loss due to a stochastic upregulation of a gene; the actual loss of telomerase will lead to selective reproduction of these cells that have undergone the switch.
The switch to either ALT telotype results in changes in both telomere state and the mode of telomere maintenance. It is not clear which of these events is primary. One possibility is that the telomere can stochastically switch to a state that allows it to be maintained through recombination (ALT). Such a switch for an X telomere could be an acquisition of a Y′ repeat, which provides the telomere with an extensive stretch of homology to a relatively large pool of Y′ telomeres, thereby allowing it to be involved in intratelomeric recombination processes. This hypothesis would explain the observed lack of any viability crisis during the resenescence of ALT I cells.
Heterochromatic switches are known to occur due to changes in the amounts and modifications of bound telomeric chromatin factors (Perrod and Gasser 2003). Normally, they affect transcription of telomere-positioned genes, but in the absence of telomerase, they may also influence the availability of telomeres for recombination. It is not known whether such switches can be spread from telomere to telomere within a single cell; conceivably, mechanisms that cluster multiple telomeres could be involved. Alternatively, the stochastic change could affect the recombination machinery so that it becomes more active on telomeres. In either scenario, the switch is expected to work in trans, rather than being specific to a single telomere. Survivor formation, which involves conversion of a set of 32 yeast telomeres, occurs with a relatively low frequency. However, when we challenged survivors with another 32-piece set of “unconverted” telomeres by crossing them to SEN cells, the second round did not lead to any senescence-like viability crisis (see Figure 5). These results suggest that the telomere conversion per se is not a stochastic event; but rather that the stochastic event is acquisition of the ability to convert, whether via changes in recombination machinery or in telomere states. The latter could be connected to accumulation of Y′ repeats and/or total amount of telomeric TG sequences in ALT I cells and long continuous TG stretches in ALT II cells (Topcu et al. 2005).
In the case of ALT I, the telomeres/subtelomeres from presurvivors were processed by the recombination machinery well before they reached critical shortness. The ability to subject telomeres to recombination early in senescence, before they are critically short, could be the key to the formation of the type I survivors: more time would be available to allow conversion of all the telomeres to the Y′ class before they become short enough to cause cell-cycle arrest.
We found that type I survivors had impaired silencing at the chromosomal internal locus, HML. HML desilencing in MATa strains is known to lead to the MATa/α phenotype, which results in a higher frequency of type I survivor formation (Lowell et al. 2003). However, HML was still dispensable for generating survivors, suggesting that it does not have a causative effect. Decreased HML silencing could be explained by titration of the Sir proteins away from HML to the amplified Y′ repeats in ALT I cells, consistent with the previously observed effect of long telomeres on HML silencing (Buck and Shore 1995). However, in type II survivors, in which telomeric G1–3T DNA repeat sequences are amplified even more, HML silencing was intact. Therefore, the decreased silencing at HML specific to ALT I cells may reflect changes in chromatin architecture that could affect either telomere states or gene expression upon the switch from TERT to ALT I.
In summary, adaptation of cell populations to telomerase absence, i.e., survivor formation in yeast, can be considered as an inheritable stochastic change from a telomerase-dependent (TERT telotype) to a telomerase-independent (ALT telotype) mode of telomere maintenance in response to a life-threatening challenge. While the importance of programmed epigenetic changes during development in higher eukaryotes is increasingly well recognized, accumulating examples of phenotypic polymorphism in populations of bacteria and lower eukaryotes highlight the potential biological significance of epigenetic changes in microbial populations as well. It is likely that epigenetic diversity is used by both prokaryotes and eukaryotes to increase their flexibility as a population to adjust to multiple environmental conditions.
Acknowledgments
We are grateful to the three anonymous reviewers for helpful comments and suggestions. We thank members of the Blackburn lab for encouraging discussions and support. We also thank Bradley A. Stohr for critical reading of the manuscript and helpful suggestions. This work was supported by National Institutes of Health grant GM26259.
This article is dedicated to the memory of Ira Herskowitz.
References
- Buck, S. W., and D. Shore, 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370–384. [DOI] [PubMed] [Google Scholar]
- Chen, Q., A. Ijpma and C. W. Greider, 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21: 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., J. N. Strathern, J. R. Broach and J. B. Hicks, 1981. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature 289: 239–244. [DOI] [PubMed] [Google Scholar]
- Kyrion, G., K. Liu, C. Liu and A. J. Lustig, 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7: 1146–1159. [DOI] [PubMed] [Google Scholar]
- Larrivee, M., and R. J. Wellinger, 2006. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 8: 741–747. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lowell, J. E., A. I. Roughton, V. Lundblad and L. Pillus, 2003. Telomerase-independent proliferation is influenced by cell type in Saccharomyces cerevisiae. Genetics 164: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V., 2002. Telomere maintenance without telomerase. Oncogene 21: 522–531. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and E. H. Blackburn, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Luria, S. E., and M. Delbruck, 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10: 1822–1834. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., and J. E. Haber, 2006. Telomerase-independent telomere maintenance in yeast, pp. 199–224 in Telomeres, edited by T. de Lange, V. Lundblad and E. H. Blackburn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nakamura, T. M., J. P. Cooper and T. R. Cech, 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493–496. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. A., K. Tatchell, B. D. Hall, C. Astell and M. Smith, 1981. A position effect in the control of transcription at yeast mating type loci. Nature 289: 244–250. [DOI] [PubMed] [Google Scholar]
- Nautiyal, S., J. L. DeRisi and E. H. Blackburn, 2002. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 9316–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov, A. M., 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41: 181–190. [DOI] [PubMed] [Google Scholar]
- Perrod, S., and S. M. Gasser, 2003. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell. Mol. Life Sci. 60: 2303–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur, J., 1994. Inheritance of the yeast mitochondrial genome. Plasmid 31: 229–241. [DOI] [PubMed] [Google Scholar]
- Reddel, R. R., 2003. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 194: 155–162. [DOI] [PubMed] [Google Scholar]
- Schulz, V. P., and V. A. Zakian, 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155. [DOI] [PubMed] [Google Scholar]
- Teng, S. C., and V. A. Zakian, 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., J. Chang, B. McCowan and V. A. Zakian, 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 6: 947–952. [DOI] [PubMed] [Google Scholar]
- Tomaska, L., M. J. McEachern and J. Nosek, 2004. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 567: 142–146. [DOI] [PubMed] [Google Scholar]
- Topcu, Z., K. Nickles, C. Davis and M. J. McEachern, 2005. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl. Acad. Sci. USA 102: 3348–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M. F., and S. L. Lindquist, 1996. Maintenance and inheritance of yeast prions. Trends Genet. 12: 467–471. [DOI] [PubMed] [Google Scholar]
- Watson, J. D., 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239: 197–201. [DOI] [PubMed] [Google Scholar]
- Zakian, V. A., 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30: 141–172. [DOI] [PubMed] [Google Scholar]