Abstract
The mouse Mitf gene encodes a transcription factor that is regulated by serine phosphorylation and is critical for the development of melanin-containing pigment cells. To test the role of phosphorylation at a particular serine, S73 in exon 2 of Mitf, we used a standard targeting strategy in mouse embryonic stem cells to change the corresponding codon into one encoding an alanine. By chance, we generated an allele in which 85,222 bp of wild-type Mitf sequence are duplicated and inserted into an otherwise correctly targeted Mitf gene. Depending on the presence or absence of a neomycin resistance cassette, this genomic rearrangement leads to animals with a white coat with or without pigmented spots or a gray coat with obligatory white and black spots. Several independent, genetically stable germline revertants that lacked the duplicated wild-type sequence but retained the targeted codon were then derived. These animals were normally pigmented, indicating that the serine-to-alanine mutation is not deleterious to melanocyte development. The fact that mosaic coat reversions occur in all mice lacking the neo-cassette and that ∼1% of these transmit a reverted allele to their offspring places this mutation among those with the highest spontaneous reversion rates in mammals.
THE molecular properties of nucleic acids and their intricate mechanisms of replication render genes and genomes inherently liable to mutations. These mutations encompass both single nucleotide substitutions and a variety of sequence rearrangements, including insertions, deletions, inversions, and duplications. Among them, gene duplications have long been recognized as an important molecular substrate from which evolutionary changes emerge. In fact, in a variety of species, including mice and humans, gene duplications arise at a high frequency (Lynch and Conery 2000, 2003; Demuth et al. 2006; Conrad and Antonarakis 2007). The evolutionary fates of such duplicates are manifold. For instance, one of the two copies may degenerate by mutations or may acquire novel, beneficial functions while the other copy maintains its original function. Alternatively, both copies may become compromised, leaving them dependent on each other to perform the full function previously associated with the single original gene (Taylor and Raes 2004). Indeed, recent analyses of whole-genome sequences provide many examples for such outcomes (Conrad and Antonarakis 2007).
When duplicates by chance form tandem repeats, however, they may face an altogether different fate: they may simply disappear by either intrastrand homologous recombination or homologous but unequal crossing over. It is conceivable that in such cases sequence alterations such as deletions or inversions associated with the original gene duplication may persist, potentially attesting to the preceding event and affecting the performance of the postrecombination gene. It is equally conceivable, however, that the duplications may vanish without a trace, leaving behind nothing but a single unaltered gene. Although such duplications/disappearances are difficult to track, they need not necessarily go unnoted if the duplication produces a visible phenotype, as we demonstrate here using an intragenic duplication that occurred following targeting of a gene that regulates coat pigmentation. In fact, coat pigmentation is ideal for such studies as it provides a very sensitive and easily visible readout of gene function (Bennett and Lamoreux 2003).
The pigmentation gene in question is Mitf, which resides at the microphthalmia locus. This locus was first described in mice in 1942 with a single allele, mi, that causes the combination of a small-eye and an albino phenotype because of alterations in the retinal pigment epithelium and lack of neural-crest-derived melanocytes (Hertwig 1942). Since its original discovery, >30 distinct forward mutations have been found, each associated with characteristic phenotypes. These phenotypes range from severe microphthalmia, lack of eye and coat pigmentation, deafness, osteopetrosis, and an assortment of other abnormalities to phenotypes so mild that they are invisible to the naked eye even in homozygotes and can be revealed only in compound heterozygotes (reviewed in Steingrimsson et al. 2004; Arnheiter et al. 2006; our unpublished observations). The corresponding gene, Mitf, was first cloned in mice in 1993 and in humans in 1994 (Hodgkinson et al. 1993; Hughes et al. 1993; Tachibana et al. 1994). The studies revealed that Mitf encodes a transcription factor of the basic helix-loop-helix-leucine zipper class that is expressed during development primarily in neural-crest-derived melanocytes and in neuroepithelium-derived pigment cells (Hodgkinson et al. 1993; Nakayama et al. 1998). The analysis of the mutants has established that Mitf cell-autonomously regulates the development of retinal pigment cells and melanocytes in many if not all vertebrates. In fact, Mitf exerts its function at multiple levels, which include the regulation of cell specification, proliferation, and differentiation (reviewed in Steingrimsson et al. 2004; Arnheiter et al. 2006), and it recently has been invoked to play a role as well as a “lineage-addiction oncogene” during melanoma formation and maintenance (Garraway et al. 2005).
An important mechanism controlling MITF activity is post-translational modification induced by extracellular signaling. It has been found, for example, that the activation of the MAP kinase pathway by the KIT ligand, which is required for melanocyte development, leads to phosphorylation of serines 73 and 409 and increases MITF's transcriptional activities while also decreasing its stability (Hemesath et al. 1994; Wu et al. 2000). This finding prompted us to initiate a systematic genetic analysis of mice with mutations in Mitf that affect post-translational modification sites. In the course of analyzing targeted mutants in which Mitf has been specifically changed to encode a nonphosphorylatable alanine instead of a serine at position 73, we came across a targeted allele characterized by a partial gene duplication. This duplication is resolved with high frequency and its resolution is associated with intricate reversions of coat pigmentation. Similar reversions of coat pigmentation have been described with other mutations (De Sepulveda et al. 1995 and references therein), and in the case of two spontaneous alleles of pigmentation genes, dilute-viral (dv) and pink-eyed-unstable (pun), are associated with the resolution of sequence duplications. The allele dv is characterized by the insertion of an ecotropic murine leukemia provirus into the dilute gene, and germline reversion is characterized by intrastrand recombination between the proviral LTRs and excision of the provirus, which occurs at a frequency of 4.5 × 10−6/gamete. Somatic reversion rates of coat pigmentation, however, are less than one in a million mice (Seperack et al. 1988). The allele pun is characterized by the duplication of ∼70 kb of sequence, and reversions are considerably more frequent and primarily somatic (Brilliant et al. 1991; Gondo et al. 1993). The unstable Mitf allele described here is different in that it is a targeted allele and in one of its forms leads to mosaic somatic reversions with a frequency of 100% and a germline reversion rate per gamete of at least 0.77%. Hence, it appears that this targeted allele displays one of the highest reversion rates in mammals and may serve as an excellent model for future studies of somatic recombination.
MATERIALS AND METHODS
Targeting construct, electroporation, and generation of knock-in mouse:
To generate the Ser73-to-Ala knock-in construct, three nonoverlapping genomic fragments, a 5.3-kb HindIII–BamHI fragment (fragment 1), a 3.9-kb BamHI–HindIII fragment (fragment 2), and a 1.6-kb HindIII–BamHI fragment (fragment 3), were isolated from mouse BAC clone 369-C11, which contains the Mitf gene. Mutations changing the Ser73 codon in exon 2B (in fragment 2) to one encoding an alanine were introduced by PCR-mediated mutagenesis. In addition to the Ser73-to-Ala change, a silent ApaLI restriction site was introduced into exon 2B to facilitate genotyping of the mutant offspring. A 1.85-kb neomycin cassette was inserted into a BglII site in the mutated fragment 2 before fragments 2 and 3 were ligated to form fragment 2/3/neo. Subsequently, fragments 1 and 2/3/neo were ligated at a BamHI site to generate a 12.6-kb fragment. Finally, a herpes simplex virus thymidine kinase (TK) gene under the control of the phosphoglycerate kinase promotor was introduced for negative selection in embryonic stem (ES) cells (Tybulewicz et al. 1991). CJ7 ES cells (strain 129S1/Sv) were electroporated with 20 μg of purified NotI-linearized plasmid, and G418/FIAU selection was applied according to standard protocols, except for experiment 1 where FIAU was started 5 days, rather than the standard 1 day, after electroporation. Targeted colonies were expanded and cells were injected into C57BL/6 blastocysts according to standard protocols.
Breeding:
Two male chimeric mice were first bred with C57BL/6 mice to test for germline transmission of the targeted allele, and positive offspring of each line were interbred. In subsequent breedings, mice either homozygous or heterozygous at the targeted locus were crossed with other Mitf-mutant mice, including Mitfmi-ew (background: NAW) (Minor 1968), Mitfmi-vga-9 (background: mixed C57BL/6;C3H/HeJ) (Tachibana et al. 1992), and Mitfmi (background: C57BL/6) (Hertwig 1942). To remove the floxed neo-cassette, the targeted mice were bred with Meox2tm1(cre)Sor/+ females carrying a Meox2-Cre knock-in allele (background: mixed 129S4/SvJaeSor;C3H/HeJ;C57BL/6) known to lead to efficient Cre-mediated germline recombination (Tallquist and Soriano 2000). The details of further breedings are described in the results.
Genomic Southern blot and PCR analyses:
Genomic DNA from ES cells or mouse liver was digested with either XbaI or EcoRI and probed with PCR-generated probes corresponding to regions lying outside the targeted sequence on either the 5′ or 3′ side (for primer sequences, see supplemental Table S1 at http://www.genetics.org/supplemental/). Similar Southern analyses with appropriate probes (see text and supplemental Table S1) were also performed to detect RFLPs associated with the genomic rearrangement. After resolution of the duplication, genomic DNA was also analyzed by PCR, using primers also listed in supplemental Table S1.
Fluorescent in situ hybridization:
The 12.6-kb targeting construct and an ∼130-kb BAC (L18-25955) covering the 5′ portion of the mouse Mitf gene were used as probes. Metaphase preparations from ES cells or spleen cells were generated by standard air-drying technique and fluorescent in situ hybridization (FISH) was performed with labeled DNA prepared by nick translation using spectrum orange-dUTP (red) or spectrum green-dUTP (green) (Vysis, Downers Grove, IL) essentially as described (Dutra et al. 1996). On each slide, 100 ng of labeled probe were applied. Ten microliters of a hybridization mixture containing the labeled DNA in 50% formamide, 2× SSC, and 10% dextran sulfate were denatured at 75° for 10 min and then incubated at 37° for 30 min for preannealing. Slides were denatured and hybridized for at least 18 hr and counterstained with DAPI–Antifade before viewing.
Comparative genome hybridization:
A fine-tiling, custom, comparative genome hybridization (CGH) array covering the Mitf gene and flanking regions on mouse chromosome 6 (position 97,672,647–98,074,171 according to sequence release NCBIM:36) was prepared by NimbleGen (Madison, WI). The custom array consisted of probes with a length of 50 bases and a median spacing of 10 bp, with appropriate masking of repetitive sequences. Genomic DNA from homozygous S73A-Δneo mice (test sample) was labeled with Cy5 and DNA from S73A2-Δneo mice (reference sample) was labeled with Cy3, and the two samples were cohybridized with the array by NimbleGen, using their methods of hybridization and data analysis.
Northern, RT–PCR, and real-time PCR analyses:
For Northern analyses, total heart RNA was isolated with an RNeasy mini kit (QIAGEN, Valencia, CA), blotted, and probed with a 32P-labeled full-length Mitf cDNA. For RT–PCR analyses, random-primed cDNAs from heart and skin RNA were prepared using superscript III reverse transcriptase (Invitrogen, San Diego) and amplified with primers indicated in supplemental Table S1 at http://www.genetics.org/supplemental/. Real-time PCR was performed using an ABI Prism 7000 real-time PCR machine (Applied Biosystems, Foster City, CA).
Skin sections and immunofluorescence:
Skin patches corresponding to white, gray, and black areas of 3-day-old pups were harvested, fixed in 4% paraformaldehyde in PBS overnight at 4°, transferred to PBS/20% sucrose for overnight incubation at 4°, embedded in tissue-freezing medium (TBS), and cryosectioned at 10 μm thickness. The sections were postfixed in PBS/4% paraformaldehyde for 20 min and permeabilized with PBS/0.1% triton-X-100 for 5 min. MITF protein was detected microscopically using a polyclonal rabbit anti-MITF primary antibody (Opdecamp et al. 1997) and a goat anti-rabbit FITC-coupled secondary antibody (Sigma, St. Louis).
RESULTS
A serine73-to-alanine knock-in allele of Mitf:
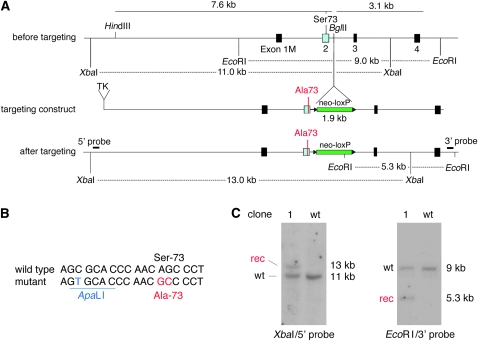
We used a standard targeting strategy in ES cells to generate a mouse with a Ser73-to-Ala (S73A) mutation in the pigment cell transcription factor gene Mitf. The targeting construct, suitable for positive/negative selection, consisted (in the 5′–3′ direction) of a TK cassette, 7.6 kb of 5′ flanking sequence, a loxP-flanked neomycin resistance cassette (neo-loxP) inserted into intron 2, and 3.1 kb of 3′ flanking sequence (Figure 1A). Exon 2, which was part of the 5′ flank, included two alterations compared to wild type: the codon for serine-73 (AGC) was changed to that of an alanine (GCC), and a silent base-pair change was introduced 10 nucleotides upstream of this codon to generate a diagnostic ApaLI site (Figure 1, A and B). CJ7 ES cells were electroporated in two separate experiments, with two targeted clones recovered from a small-scale first experiment (targeting efficiency 10%) and 15 targeted clones recovered from a second experiment (targeting efficiency 20%). Southern analyses of genomic DNA from these clones showed the expected wild type and recombinant bands after appropriate restriction cuts, using 5′ and 3′ probes representing sequences lying outside the targeting construct. Results (Figure 1C) are shown for clone 1 from the first experiment, chosen here because it gave rise to mice with an unusually unstable allele. These mice were obtained from two germline-transmitting high-degree chimeric mice generated by standard injection of clone 1 ES cells into C57BL/6 blastocysts.
Figure 1.—
Targeting construct and analysis of a targeted ES clone. (A) Schematic of a portion of the mouse Mitf gene and targeting construct. (Top) The genomic region before targeting around exon 1M–4, the position of S73 in exon 2, the position of the BglII site where the neo-cassette will be inserted, and the position of the EcoRI and XbaI restriction sites used to identify legitimate recombination events. (Middle) The targeting construct, consisting (in the 5′–3′ direction) of a TK cassette, 7.6 kb of 5′ flanking sequence containing a modified codon, the neocassette (green) flanked by loxP sites (triangles), and 3.1 kb of 3′ flanking sequence. (Bottom) The genomic arrangement after targeting, showing the position of the diagnostic restriction fragments and the position of the probes used for Southern hybridization. The XbaI fragment in wild type is 11 kb but after targeting is increased to 13 kb because of the insertion of the neo-cassette. The EcoRI fragment in wild type is 9 kb and, after targeting, a novel EcoRI fragment of 5.3 kb is generated because of an EcoRI site in the neo-cassette. (B) Wild-type and modified sequence around S73. The mutant contains a silent ApaLI site and an AGC-to-GCC codon change, leading to a Ser-73-to-Ala change. (C) Analysis of wild-type and clone 1 ES cells by Southern hybridization. Note the expected recombinant bands after indicated restriction cuts and hybridization with the indicated probes.
Homozygous targeted mice are white but retain wild-type Mitf sequences:
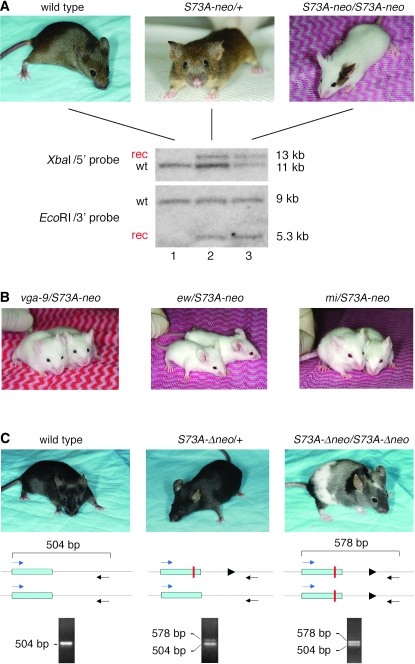
From both clone 1 chimeric mice, we eventually derived offspring that were completely white. In 15–20% of them, however, a small pigmented spot on the head or at the base of the tail was present (see example in Figure 2A, labeled S73A-neo/S73A-neo). The white offspring occurred with a frequency corresponding to the expected Mendelian ratios for homozygosity at the targeted allele, and further breeding tests confirmed that they were indeed homozygous for S73A-neo while heterozygous carriers were normally pigmented (Figure 2A, labeled S73A-neo/+). The pigmented spots, however, were not stably inherited since they occurred randomly in offspring, regardless of whether or not the corresponding parents carried spots. Interestingly, the targeted allele did not cause microphthalmia, in contrast to other Mitf alleles that lead to white coats.
Figure 2.—
Breeding of targeted mice and color change after removal of the neo-cassette. (A) Coat appearance of wild type, heterozygous, and homozygous S73A-neo mice and corresponding genomic Southern analyses. Both wild-type and heterozygous mice are normally pigmented while homozygotes are largely white except for pigmented spots seen in 15–20% of the animals as shown in the example. Genomic DNA was digested and probed as described in Figure 1 for ES cells. The targeted heterozygous mouse shows the expected Southern pattern (although with different intensities for the wild-type and recombinant bands, particularly clear with XbaI and the 5′ probe) and the homozygous mouse shows wild-type and recombinant bands of equal intensities. (B) Compound heterozygotes of the indicated genotypes are white mice. (C). After Cre-mediated recombination to remove the neo-cassette, homozygous mice (labeled S73A-Δneo/S73A-Δneo) are largely gray but have extensive white spotting and at least one darkly pigmented area. Also, they retain a wild-type PCR band as indicated at the bottom of the figure. The solid triangles represent the loxP site. For details, see text.
Genomic tail DNA from wild-type, heterozygous, and homozygous mice was then digested and probed as shown for ES DNA. Wild-type DNA (Figure 2A, lane 1) gave a single band and heterozygous DNA (Figure 2A, lane 2) the expected recombinant and wild-type bands, whereby the wild-type band was slightly more intense than the corresponding recombinant band. In homozygous DNA (Figure 2A, lane 3) probed with either one of the two probes, a wild-type band was retained, and its intensity was similar to that of the recombinant band. Reprobing with a neo-specific probe exclusively labeled the recombinant band (not shown). These results suggested that, within the genomic distance bracketed by the 5′ and 3′ probes, neither the wild-type nor the recombinant band was grossly rearranged and that the unexpected wild-type band of homozygotes lay outside the boundaries of the recombinant band. It also suggested that the targeted allele, despite the presence of a wild-type sequence, was not able to provide functional MITF protein, except perhaps in cells giving rise to the occasional pigmented spots and in the retinal pigmented epithelium where absence of Mitf would normally lead to microphthalmia. In further support of this notion, the targeted allele was unable to complement the coat pigmentation of three distinct Mitf alleles: Mitfmi-vga-9 (an insertional null allele, abbreviated as vga-9), Mitfmi (encoding a nonfunctional protein with a DNA-binding domain mutation), and Mitfmi-ew (encoding a similarly nonfunctional protein with a DNA-binding domain mutation, abbreviated as ew) (Figure 2B). In fact, these compound heterozygotes displayed pigmented spots more rarely than S73A-neo homozygotes, but their eyes were still of normal size. The data indicated that a wild-type sequence was retained in the homozygous knock-ins and likely represented a contiguous stretch of DNA with a minimal size corresponding to the distance between the left-most XbaI site and the right-most EcoRI site (14.2 kb). The presence of this wild-type sequence could be the result of a gene duplication (partial or full) that preceded or followed the targeting event, or it could represent a hitherto unrecognized pseudogene linked to the targeted gene.
Removal of the neomycin resistance cassette in intron 2 enhances pigmentation:
It was conceivable that the white coats of the homozygotes were due to the S73A mutation, to gene rearrangement, or simply to the presence of the neo-cassette in intron 2 that may reduce mRNA levels. To exclude the latter possibility, we crossed the mice with a germline deleter strain (Meox2-Cre) (Tallquist and Soriano 2000) and eventually obtained homozygous mice, identified by breeding tests, that lacked the neo-cassette and consistently displayed a patchwork coat with white, gray, and darkly pigmented areas (Figure 2C). Their targeted allele was termed S73A-Δneo. Using genomic PCR with primers in exon 2 and intron 2 as indicated in Figure 2C, we obtained a single 504-bp band with wild-type DNA and a 504-bp band as well as a 578-bp band with both heterozygous and homozygous DNA. The wild-type band in heterozygotes was more intense compared to the 578-bp band but was of an intensity similar to that of the 578-bp band in homozygotes. This result was reminiscent of that obtained by Southern analysis before crossing with the deleter strain. Restriction with ApaLI, for which there was a diagnostic site associated with the S73A mutation, selectively cut the 578-bp band (not shown), suggesting that this band represented the targeted sequence. Direct sequencing confirmed that the smaller band corresponded to the wild-type Mitf sequence between the two primers whereas the larger band represented the same sequence except that it contained the ApaLI site, the S73A mutation, and a single loxP site flanked by linker sequences (amounting to a total of 74 bp) inserted into intron 2 (supplemental Figure S1 at http://www.genetics.org/supplemental/). Hence, the results confirmed the presence of a wild-type band in homozygous mice even after the removal of the neo-cassette and suggested that the more extensive pigmentation in S73A-Δneo homozygotes was due to the removal of the neo-cassette and not to the removal of an extraneous wild-type sequence.
Frequent reversion of the coat-color phenotype:
Upon subsequent breeding of mice carrying the S73A-neo or the S73A-Δneo allele, we obtained and analyzed three germline revertants of coat pigmentation that occurred spontaneously in offspring from different parents, involved either sex, and behaved as phenotypically dominant. Each was associated with the retention of the targeted mutation and the loss of the extra wild-type band. They were identified by visual inspection during a year of breeding among 93 live pups carrying S73A-neo and 100 pups carrying S73A-Δneo. The analysis included homozygotes and compound heterozygotes but excluded heterozygotes carrying a wild-type allele since, in such animals, normal pigmentation would mask phenotypic reversion of the targeted allele. Given that the reversions are phenotypically dominant in homozygotes and can occur in either gamete, the frequency per gamete is 3/386 or 0.77%. Although the data set is small, this frequency appears to be extremely high. Since the appearance of these three revertants, we have observed additional revertants at similar frequencies that also lost the wild type and retained the targeted sequence, but they were not analyzed further.
Event 1:
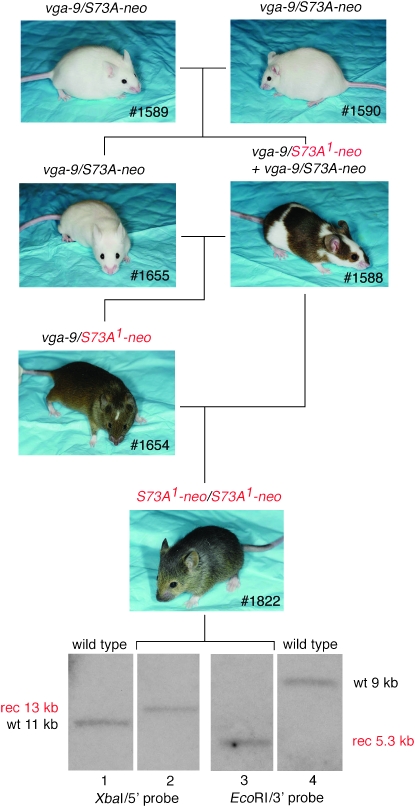
The first event was observed in an offspring from the intercross of two vga-9/S73A-neo compound heterozygotes (Figure 3, #1589 and #1590). Such crosses normally produce only white mice of genotypes vga-9/vga-9, which are microphthalmic; vga-9/S73A-neo, which are normophthalmic (similar to the one depicted as #1655 in Figure 3); and S73A-neo/S73A-neo, which are normophthalmic, the latter two sometimes with pigmented spots. Figure 3 shows the exceptional mouse, #1588, which was extensively pigmented, and which, when crossed to its vga-9/S73A-neo sibling (#1655), gave rise to a pigmented offspring (1 of 3 pups) with a small white head blaze, white feet, and a white belly spot (1654). Genotyping indicated that #1654 contained a vga-9 allele and an S73A-neo allele. Because this mouse was different in pigmentation compared to vga-9/S73A-neo mice previously observed, it is labeled vga-9/S73A1-neo, the superscript reflecting the assumption that the S73A-neo allele had undergone a further mutation. When #1654 was backcrossed to #1588, we obtained pigmented offspring (6 of 12 pups). Two of these lacked a head blaze although they still had white feet and a small belly spot. An example is shown as #1822. Southern analysis of this mouse's DNA showed that it retained the recombinant band but lacked the extraneous wild-type band, regardless of which of the two probes was used. Subsequent breeding showed that #1822 and similar offspring were indeed homozygous for the S73A1-neo allele and that the pigmentation phenotype remained stable through multiple generations. These mice were also crossed with the Meox2-Cre deleter strain, resulting in offspring carrying a S73A1-Δneo allele. Homozygosity for this allele was associated with a wild-type pigmentary phenotype (no head blaze, no belly spot, normally pigmented feet), which also remained stable through subsequent breedings. We interpret these results to indicate that (1) #1588 has selectively lost the wild-type sequence that initially was associated with its targeted gene; (2) the loss happened early in development, perhaps at a preimplantation state and in a single cell that then served as a common precursor to melanocytes and germ cells; and (3) the rearranged gene now produced functional MITF protein in sufficient quantities to lead to near-normal pigmentation even in combination with a null allele and to fully normal pigmentation after removal of the neo-cassette. Although this scenario is plausible, a direct proof cannot easily be provided.
Figure 3.—
Phenotypic reversion, event 1. The crossing of two vga-9/S73A-neo compound heterozygotes led to an extensively pigmented mouse (#1588) that apparently was mosaic for a novel Mitf allele, S73A1-neo, and that gave rise to phenotypically and genotypically different offspring. Using the indicated breeding scheme, a mouse (#1822) homozygous for S73A1-neo, was generated. Genomic Southern hybridization as described in Figure 1 indicated that this mouse lost the extraneous wild-type band but retained the targeted recombinant band.
Event 2:
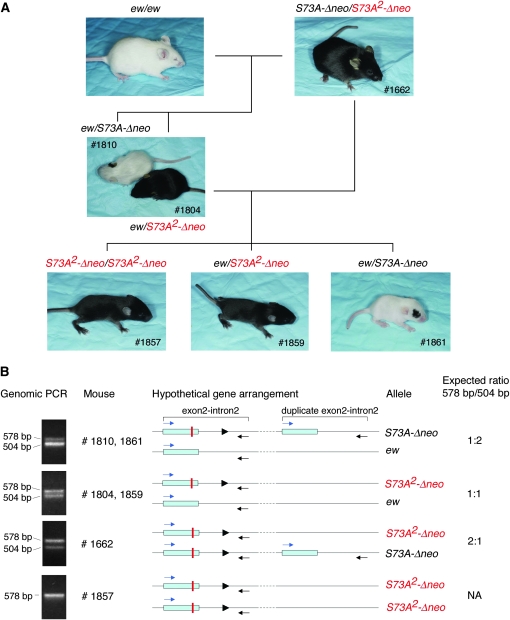
This event occurred in a pedigree that segregated ew and S73A-Δneo, i.e., after removal of the neo-cassette. Among the offspring between two ew/S73A-Δneo compound heterozygotes, we normally obtain white mice of genotypes ew/ew (microphthalmic) and ew/S73A-Δneo (normophthalmic)—the former never with pigmented spots, the latter nearly always with at least one black spot—and white/gray/black, normophthalmic S73A-Δneo/S73A-Δneo homozygotes resembling the one depicted in Figure 2C. One such cross, however, produced an offspring that was fully pigmented as shown in Figure 4A (#1662). When this mouse was crossed with ew homozygotes, we obtained normally pigmented offspring (two of nine pups) as exemplified by #1804, along with white compound heterozygotes (ew/S73A-Δneo) (seven of nine pups), usually sporting a black spot as exemplified by #1810. Backcrossing of #1804 to #1662 gave two types of pigmentation: fully pigmented pups (six of eight pups, examples #1857 and #1859) and white pups with pigment spots (two of eight pups, example #1861). As shown in Figure 4B, genomic PCR using the primers depicted in Figure 2C showed that the fully pigmented #1857 mouse lacked the wild-type band of 504 bp but retained the recombinant band of 578 bp. All other offspring shown in Figure 4A had both bands. The relative intensities of these two bands, however, differed between the mice. In #1810 and #1861, for example, the wild-type band was more intense, in #1662 the mutant band was more intense, and in #1804 or #1859 the two bands were of equal intensity. These results are consistent with a model of gene rearrangement, depicted in Figure 4B, that assumes an association in cis of the mutant and extra wild-type sequences in S73A-Δneo and the loss of the wild-type sequence in S73A2-Δneo. As in event 1, the loss of the wild-type band was probably the result of a somatic rearrangement that happened early in development, affected the germline, and resulted in functional MITF protein.
Figure 4.—
Phenotypic reversion, event 2. (A) The crossing of two ew/S73A-Δneo compound heterozygotes (not shown) generated a mouse (#1662), which was fully pigmented. This mouse carried a novel Mitf allele, S73A2-Δneo, that could be bred to homozygosity using the indicated breeding scheme. The presence of S73A2-Δneo renders mice fully pigmented even if their second Mitf allele is ew, a strong Mitf allele. (B) Genomic PCR of the indicated mice, using PCR primers as in Figure 2. The analysis indicates that #1857 contains only the targeted band of 578 bp, while the other mice all contain both the wild-type 504-bp band and the targeted 578-bp band, although at different relative intensities. A hypothetical gene rearrangement and the expected ratios of the two bands, consistent with the PCR results, are shown on the right. The solid triangles represent the loxP site.
Event 3:
The third phenotypic reversion happened in a line homozygous for S73A-Δneo and hence among offspring of two similarly white/gray/black pigmented parents. There was one pigmented offspring with a gray head blaze (#1790, not shown), which, upon crossing with vga-9 homozygotes, produced a fully pigmented offspring (1 of 13 pups, #1895, not shown). From this mouse, we eventually derived mice homozygous for S73A3-Δneo. Again, these homozygous mice lacked a wild-type sequence and retained only the recombinant sequence, and both genotype and phenotype remained stable through subsequent breedings.
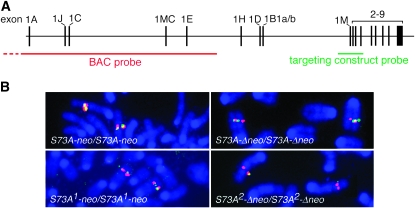
Analysis by fluorescent in situ hybridization:
To gain further insights into the underlying gene rearrangement and reversions, we then performed FISH analyses on metaphase spreads prepared from ES cells and spleen cells of select representatives of the various mutant and revertant mice. Two probes, depicted schematically in Figure 5A, were used. One corresponded to the 12.6-kb targeting construct (containing the neo-cassette) and was used to detect small genomic rearrangements. The other corresponded to a 130-kb BAC covering the 5′-end of the gene without, however, overlapping with the targeting construct.
Figure 5.—
Two-color FISH. (A) Schematic of the entire Mitf gene containing 17 exons and spanning ∼214 kb. The position of the BAC probe (red) covering the 5′ portion of the gene is shown, as is the position of the 12.6-kb targeting construct probe (green), covering exons 1M–4. (B) Metaphase spreads were prepared from spleen cells harvested from homozygous mice of the indicated genotypes. The red signal is of equal strength regardless of genotype. Also, compared to cells still containing the extra wild-type sequence (S73A-neo and S73A-Δneo), the green signal is weaker in cells from mice that have lost the wild-type band (S73A1-neo and S73A2-Δneo). Chromosome banding (not shown) indicates that the signals are on chromosome 6 at a location where Mitf is expected.
A 12.6-kb probe is at the limit of sensitivity in this type of assay and on the nontargeted chromosome can hybridize with only 10.7 kb of sequence. Not surprisingly, using this short probe, no signal was found on normal ES cells, but spreads from clone 1 ES cells gave a clear signal on one of the two chromosome 6's where Mitf resides (not shown). This finding is consistent with the notion that the duplicated sequence provided for a larger hybridization target and sits in the vicinity of Mitf. For two-color FISH, the BAC probe (labeled with spectrum orange, giving a red signal) was mixed with the targeting construct probe (labeled with spectrum green, giving a green signal). As shown in Figure 5B, a red Mitf signal of similar strength was seen in spreads of spleen cells from all homozygous mice, regardless of genotype, suggesting that gene amplification did not include, or only partially included, the region of the gene covered by the BAC. The targeting construct probe gave a green signal, colocalized with the red signal, in spreads from cells of the S73A-neo or the S73A-Δneo genotype. Both of these alleles still contain the extra wild-type sequence and hence contain sequences available for hybridization that are at least double the size of the probe. In contrast, metaphase spreads from homozygous S73A1-neo cells (event 1), which lack the duplicated sequence and have a hybridizable target of only 12.6 kb, gave a weaker green signal, and spreads from homozyogus S73A2-Δneo cells (event 2), whose hybridizable sequence is only 10.7 kb, gave no green signal or only occasionally a barely visible one (Figure 5B). Taken together, these results suggested that the targeting in ES cells led to a local partial gene duplication that was resolved through subsequent recombinations.
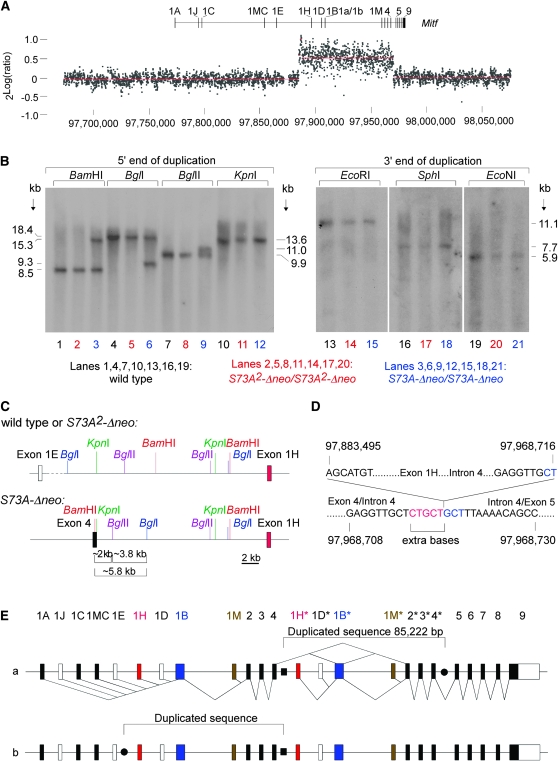
Comparative genome hybridization:
Semiquantitative Southern analyses using a variety of cDNA probes suggested that exons 1B and 2–4 were duplicated but exons 1A and 5–9 were not (not shown). To determine more precisely the extent of the duplication, we performed CGH analysis, using a fine-tiling, custom oligonucleotide array covering 401,524 bp of sequence that included the entire 214-kb Mitf gene. Preparations of genomic DNA from homozygous S73A-Δneo and S73A2-Δneo mice were labeled with Cy5 and Cy3, respectively, mixed, and hybridized with the array. The normalized results, shown in Figure 6A and expressed as 2Log (ratio of signal intensities), indicated that a sequence spanning exon 1H–exon 4 (corresponding approximately to position 97,883,500–97,968,700 according to sequence release NCBIM:36) showed a copy number increase in S73A-Δneo mice.
Figure 6.—
Molecular analysis of the duplication. (A) A fine-tiling custom CGH array spanning the indicated region of chromosome 6 (based on sequence release NCBIM:36) was cohybridized with Cy3-labeled DNA from homozygous S73A2-Δneo mice and with Cy5-labeled DNA from S73A-Δneo mice. A copy number difference is seen for a sequence approximately covering positions 97,883,500–97,968,700, extending from upstream of exon 1H to a position between exon 4 and 5. (B) Southern analysis using probes corresponding to the 5′- and 3′-end of the duplication (left and right, respectively). Probing with the 5′ probe reveals RFLPs in DNA from S73A-Δneo for BamHI, BglI, and BglII, but not for KpnI. Probing with the 3′ probe shows no RFLPs between the different DNAs. (C) Restriction map for wild type or S73A2-Δneo (top) or for S73A-Δneo (bottom) on the basis of the above Southern analyses. The novel restriction fragments indicate the presence of a BamHI–KpnI–BglII–BglI constellation that happens to be present in intron 4 of Mitf. (D) Junction sequences of the inserted partial gene duplication in S73A-Δneo. Five extra bases (red) are found between the intron 4/upstream exon 1H junction. The downstream junction in intron 4 reads exactly like the wild-type sequence. Interestingly, the sequence marked in blue is a direct repeat of the extra sequence marked in red. (E) Two possible arrangements of the insertions of the duplicate. The novel intron 4/upstream exon 1H junction is marked by a solid square, and the potential other junctions by solid circles. In a, the duplicate spanning exons 1H–4 (marked with asterisks) is inserted into intron 4. In b, the duplicate is inserted between exons 1E and 1H. The two possible arrangements, however, are sequence identical. Splice patterns (known and predicted following the insertion of the duplicate) are indicated only for version a.
On the basis of the reversion events and the FISH results, it was likely that this sequence represented a contiguous stretch inserted in the vicinity of, or within, the Mitf gene. As shown in Figure 6B, Southern analysis of genomic DNA from wild-type, S73A2-Δneo, and S73A-Δneo mice, digested with three different enzymes and hybridized with a probe covering the 5′-end of the duplication, revealed RFLPs in S73A-Δneo DNA compared to the other two DNAs. With a fourth enzyme, KpnI, however, only one band of wild-type size was seen, likely because the wild type and the novel junction band were of similar length. Using a probe covering the 3′-end of the duplication and three restriction enzymes, only bands of wild-type size were found, suggesting that the 3′ junction of the duplication corresponded to the wild-type sequence. Figure 6C compares the restriction maps of wild type/S73A2-Δneo DNA and S73A-Δneo DNA on the basis of the above findings with the 5′ probe. This analysis revealed a novel BamHI–KpnI–BgllI–BglI constellation in the S73A-Δneo DNA. Such a constellation is in fact found in exon 4/intron 4 of Mitf, suggesting that the duplicated sequence is inserted at this location. Using appropriate primers, we amplified the predicted junction and confirmed by sequencing the link between position 97,968,716 (in intron 4) and position 97,883,495 (upstream of exon 1H), whereby 5 extra bases were inserted upstream of the insertion point (Figure 6D). As expected, the same primers were unable to amplify a product in wild-type or S73A2-Δneo DNA. They were also unable to amplify the corresponding product in untargeted control ES cells from the first electroporation (the one that gave rise to the mice described in this article) or in targeted ES cells from the second electroporation, suggesting that the observed duplication did not preexist in the ES cells used for targeting (data not shown). The sequence lying 84,868–85,955 bp downstream of this junction (position 97,986,363–97,969,450, covering the end of the duplication as determined by CGH) was as in wild type. As shown in Figure 6E(a), these findings are consistent with a tandem duplication of 85,222 bp placed 6784 bp downstream of exon 4 into intron 4 of Mitf. As shown in Figure 6E(b), however, it is equally plausible that the duplicated sequence is inserted 20,542 bp downstream of exon 1E. Nevertheless, like the potential junction sequence between exons 4 and 5, the potential junction sequence between exons 1E and 1H did not show an alteration compared to wild type, and so the question of which portion of the rearranged gene is the “duplicate” has to be left open (for a determination of which of the two exon 2's contains the targeted mutation, see below). In any event, whatever led to the duplication in the first place, the revertant allele S73A2-Δneo was indistinguishable from wild type, except for the mutations intended by the targeting.
As depicted in Figure 6E(a), the sequence rearrangement in S73A-neo and S73A-Δneo mice makes predictions for the composition of the corresponding transcripts. Shown below the sequence line in Figure 6E(a) are the known splicing patterns (Steingrimsson et al. 2004; Hershey and Fisher 2005; Arnheiter et al. 2006) and shown above are the aberrant splicing patterns anticipated on the basis of the presence of the duplication. A detailed analysis of these transcripts is given below.
Transcript analyses:
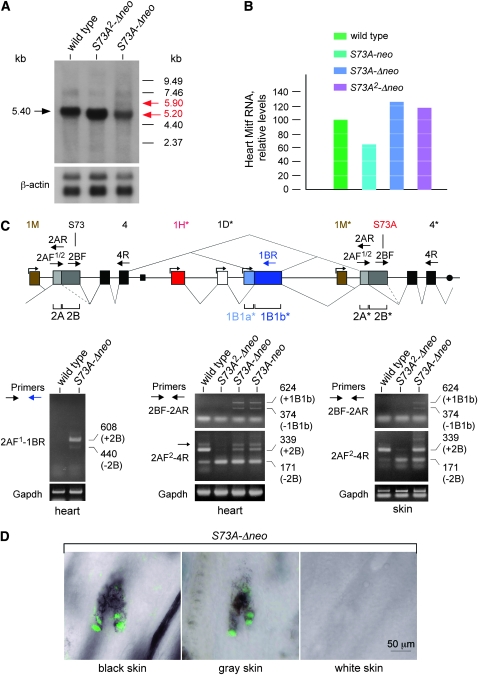
We first analyzed Mitf transcripts by Northern blots, using heart tissue as the source of RNA because the heart is not obviously affected by the mutations. As shown in Figure 7A, a cDNA probe detected a major 5.40-kb band in wild-type heart and a band of ∼5.20 kb in both S73A2-Δneo and S73A-Δneo heart. In S73A-Δneo heart, the major band was weaker by comparison, but there was an additional larger, although weak, band at ∼5.9 kb. The size of this larger band would be consistent with an mRNA representing an internal duplication of exons 2–4. Real-time PCR of heart cDNA based on primers in exons 8 and 9, common to all Mitf transcripts, is shown in Figure 7B. This analysis revealed that, compared to wild type, Mitf RNA levels of S73A-neo hearts were slightly reduced, and those of S73A-Δneo and S73A2-Δneo hearts were slightly increased.
Figure 7.—
Mitf transcript analyses with the mutated gene before and after resolution of the sequence rearrangement. (A) Northern analysis of total RNA harvested from heart of wild-type mice and S73A2-Δneo and S73A-Δneo homozygotes. The blots were probed with a radiolabeled Mitf cDNA. A single major band of 5.4 kb is in wild type and a single major band of 5.2 kb is in S73A2-Δneo and S73A-Δneo RNA. In addition, a barely visible minor band of ∼5.9 kb is seen only in S73A-Δneo RNA, along with a presumably unspecific minor band at ∼7.46 kb present regardless of genotype. (B) Quantitation of RNA levels in heart of the indicated genotypes by real-time PCR, using primers in exon 8 and exon 9 common to all Mitf RNAs. After normalization for Gapdh levels, there is a relative reduction in Mitf RNA levels in the presence of the neo-cassette and a relative increase in Mitf levels in the absence of the cassette. (C, top) Schematic of the splice patterns around the duplication. Both exons 2 and 1B are bipartite, and exons 1M, 1H, 1D, and 1B1a each have their own promoters (indicated by small forward arrows). The relative position of primers (larger forwardand reverse arrows) is indicated on top of exons 2A, 2B, 4, 1B1b*, 2A*, 2B*, and 4*. (Bottom left) Wild type and S73A-Δneo RNA were amplified with the indicated primers to demonstrate that exon 4 splices into exon 1B1b* only in S73A-Δneo as predicted from the genomic rearrangement. Two bands that differ in the presence or absence of the alternatively spliced exon 2B are visible. Furthermore, their sequence is wild type at S73, indicating that the wild-type exon 2B is located upstream of 1B1b*. (Bottom middle) Heart RNA of the indicated genotypes was analyzed with primers 2BF and 2AR to demonstrate splicing from exon 4 to exon 2A*. Two products are seen when the duplicate sequence is present. The 624-bp band represents splicing of exon 4 into 1B1b* and 1B1b* into 2A* and the 374-bp band splicing of exon 4 directly into exon 2A*. Primers 2AF2 and 4R were used to analyze the presence and absence of exon 2B. Wild type mostly generates a 339-bp band containing exon 2B while S73A2-Δneo mostly generates a 171-bp band lacking exon 2B. With the duplicated sequence present, the 339-bp band is more prominent. There is also a larger band in heart RNA, indicated by a small arrow, that likely represents a differentially spliced message that was not further analyzed. (Bottom right) The use of the same primer pairs as in the bottom middle shows similar results on skin RNA. The skin of S73A-neo mice was not analyzed as it is mostly white and hence lacks melanocytes, the major contributors to skin Mitf RNA. (D) Indirect immunofluorescence of MITF protein in skin of homozygous S73A-Δneo pups. Cryosections of skin were labeled with a rabbit serum against mouse MITF protein, and black skin, gray skin, and white skin was analyzed separately. Note the normal nuclear appearance of MITF fluorescence in follicular pigment cells except in white skin. The labeling intensity appears slightly higher in black skin compared to gray skin.
To analyze which mRNAs are made and why the major bands in S73A2-Δneo and S73A-Δneo are smaller than in wild type, we generated and sequenced a number of RT–PCR products. The choice of primers was dictated by the genomic rearrangement shown at the top of Figure 7C. If transcripts are initiated from the downstream exons 1H*, 1D*, 1B1a*, or 1M*, they are expected to be entirely normal, except for the presence of the codon changes if exon 2*, rather than exon 2, were mutated. In contrast, if transcripts are initiated from the upstream exons 1A–1M, after exon 4 they would have to splice into the next available downstream exon, i.e., either subexon 1B1b* (hereafter referred to as exon 1B1b*) or exon 2*. Transcripts containing exon 1B1b*, however, would include a premature stop codon 90 bases downstream of the splice junction in exon 1B1b* and hence likely be subjected to nonsense-mediated decay. Splicing from exon 4 directly into exon 2* would retain the open reading frame but generate abnormal proteins with internally duplicated exons 2–4, whereby one exon 2 would be wild type and the other one mutated.
Previous results have shown that exon 2 is in fact bipartite, divided into an exon 2A of 20 codons and an exon 2B of 56 codons, and that exon 2B is absent in a small percentage of Mitf mRNAs (Hallsson et al. 2000). Taking this information into account for RT–PCR analyses, we first chose a forward primer at the beginning of exon 2A (2AF1) and a reverse primer corresponding to exon 1B1b (1BR). As shown at the bottom left of Figure 7C, these primers gave no band in wild type but two bands in S73A-Δneo: one of 608 bp that contained a wild-type exon 2B, and a weak one of 440 bp that lacked exon 2B. This finding shows that an RNA that links exon 4 with exon 1B1b* indeed accumulated and clearly places the wild-type exon 2 upstream of the novel intron 4/1H* junction and hence the mutated one downstream of this junction. It also confirms that wild-type exon 2B is eliminated in some mRNAs as mentioned above (Hallsson et al. 2000). We then used a forward primer at the end of exon 2B (2BF) in combination with a reverse primer at the beginning of exon 2A (2AR). As shown at the middle bottom of Figure 7C, this combination did not give a product in the absence of the duplication but again gave two bands in S73A-Δneo and S73A-neo heart: one of 624 bp that contained exon 1B1b* and one of 374 bp that lacked this exon (Figure 7C, middle bottom). This finding indicates that in hearts from mice with the rearrangement, there are RNAs linking exon 4 with exon 2A*, with or without inclusion of exon 1B1b*. Also shown in Figure 7C (middle bottom) are the results obtained with the forward primer 2AF2 and the reverse primer 4R. In wild type, this combination gave the expected 339-bp band containing exon 2B and a less intense 171-bp band lacking this alternatively spliced exon. With S73A2-Δneo (which has only one exon 2A/2B), however, the 171-bp band was prominent while the 339-bp band was barely visible. This finding is consistent with the fact that the Northern analysis showed a major band that was smaller in these mice compared with wild type. It further suggested that the mutation at S73A, or the introduced ApaLI site, or both together, altered an exonic splice enhancer sequence or created an exonic splice silencer sequence that we plan to analyze in detail in the future. Interestingly, in RNA from either S73A-Δneo or S73A-neo hearts (which have the two exons 2A/2B), the 339-bp band was more intense, suggesting that it came from the S73 and not the S73A sequence. Bands that would link the wild-type exon 2B with the mutated exon 2B were not detected in these reactions, likely because they were not amplified efficiently because of their larger size. Using the same primer pairs as for heart, RNA from skin gave similar results, as shown at the bottom right of Figure 7C. We did not test, however, skin from S73A-neo mice because melanocytes, which are the major contributors to Mitf RNA in skin, are missing in such mice.
Protein expression analyzed by immunofluorescence:
The fact that both S73A-Δneo and S73A2-Δneo mice are substantially pigmented predicted that MITF proteins, however aberrant by carrying internal exon duplications or simply lacking exon 2B, might still accumulate in the nucleus and be active. In fact, as shown in Figure 7D, in immunofluorescently stained sections of gray or black skin of S73A-Δneo mice, follicular pigmented cells with a characteristic nuclear MITF fluorescence were seen. Such cells were totally absent in the white parts of the skin. These results suggest that melanocytes accumulate in these mice because Mitf produces enough functional protein despite the internal gene duplication.
DISCUSSION
Coat-color loci have played, and still play, important roles in genetics chiefly because alterations in coat pigmentation can easily be seen in vivo and by themselves have no effects on reproduction or survival. Here, we used changes in coat pigmentation in the mouse to track the reversion of an unstable targeted allele of Mitf, a gene encoding a transcription factor primarily responsible for the early development of pigment cells and their eventual differentiation. That the instability of this allele is due to the resolution of a partial gene duplication by homologous recombination rests on two main arguments. First, homozygous targeted mice, which are white or white spotted, depending on the presence or absence of a neomycin resistance cassette, harbor an intragenic insertion of 85 kb of duplicated wild-type Mitf sequence spanning seven exons. Second, genetically stable revertants, which show minor white spots or are pigmented just as wild-type mice, have lost the duplicated sequence and reconstructed an Mitf gene indistinguishable from wild type except for the specific mutations introduced by targeting. In fact, in all reversions observed to date, the targeted exon 2 was retained and the wild-type exon 2 was lost. This is readily explained by the observation that the S73A mutation sits near one end of the duplication, an arrangement that, upon homologous pairing, favors recombinations within the several-fold larger region between the other end and the mutation.
While the loss of the duplicated sequence by homologous recombination is evident after germline transmission, it is more difficult to prove that the frequent mosaic reversions of coat pigmentation in homozygotes are also due to similar recombinations. Because the reversions are phenotypically dominant, and because a normal wild-type sequence is always present, a loss of the novel exon4/1H junction would be manifested only as copy number changes of the novel junction and the wild-type sequence. Such changes, however, might be masked by the continuous presence of the duplication in nonmelanocytic skin cells that are unlikely to revert in synchrony with melanocytes. Nevertheless, the observation that, in two revertant lines, germline transmission was preceded by unusually extensive pigmentation in one of the parents argues strongly for a scenario in which the same molecular event affected both germ cells and melanocytes, most likely because a common precursor cell was affected. It cannot be decided at this stage, however, whether such somatic reversions were due to mitotic intrastrand recombination or homologous but unequal crossing over and whether meiotic recombinations might occur as well. We favor intrastrand recombination in all observed reversions because we have no indication from the analysis of the offspring of a number of distinct compound heterozygotes that recombinations have occurred between the targeted mutation and the respective mutations in other Mitf alleles. For instance, for mice derived from event 2, which was not foreshadowed by parental pigmentation changes, we can be sure that recombination did not occur between the targeted mutation and the Mitfmi-ew mutation as cDNA cloning from this line of mice did not indicate the presence of the Mitfmi-ew mutation (not shown). In fact, the S73A mutation would not be expected to lead to a phenotypic improvement when introduced as a second-site mutation in Mitfmi-ew, whose protein does not bind DNA (Hemesath et al. 1994), although it might improve the function of milder Mitf alleles.
The molecular analysis of the duplication and eventual resolution readily explains most if not all biological phenomena associated with the targeting. When both the duplication and the cassette are present (S73A-neo), small amounts of aberrant RNAs that are expected to encode proteins with premature stop codons or internal exon duplications are made. This observation, combined with the general reduction of Mitf mRNA levels, would seem to provide sufficient reason for impairment of melanocyte development and lead to white coats. More intriguing is the observation that these white mice still have normal-sized, although hypopigmented, eyes. A combination of a white coat with normal eye size is not without precedent among the many Mitf alleles, however, and in the case of Mitfmi-bw is associated with differential effects on mRNA isoforms. This allele leads to white animals with black eyes and is characterized by the insertion of a LINE element in intron 3 that eliminates the accumulation of M-Mitf mRNA, the melanocyte isoform, while leaving the accumulation of other RNA isoforms relatively unimpaired (Yajima et al. 1999). The normal eye sizes in S73A-neo mice can likewise be explained by differential effects on mRNA isoforms. As is evident from the gene rearrangement in S73A-neo mice, starting from the downstream 1H* and 1D* promoters, H- and D-Mitf mRNAs that are entirely normal except for the lack of exon 2B can be generated, and it is precisely the H- and D-isoforms that are relevant for eye development, and not the isoforms initiated from upstream promoters, that are expected to be present only in abnormal form in S73A-neo mice (K. Bharti et al., unpublished results). Also, because postresolution homozygotes (S73A1-neo) have normal eyes, we can conclude that exon 2B, which is largely missing in RNAs of these mice, is not required for eye development. Nevertheless, there remains a seeming paradox in the above arguments as they imply that S73A-neo mice are white despite the fact that they can make functional M-Mitf starting from the downstream exon 1M*, but that their eyes are normal because they can make functional H- and D-Mitf starting from the corresponding downstream exons. This paradox may be resolved, however, if we consider that neither the absolute nor the relative amounts of functionally normal and abnormal mRNAs need to be identical in melanocytes and the retinal pigment epithelium and that eye development is generally less sensitive to Mitf mutations than is development of melanocytes (Bharti et al. 2006).
Upon removal of the neomycin resistance cassette, white spotting was reduced and the coat became mostly gray. Although these mice (S73A-Δneo) likely make similar aberrant mRNAs and proteins as S73A-neo mice, the overall levels of their mRNAs were higher. In fact, they must be high enough to allow for the development of melanocytes, although not for rendering the hairs completely black. Interestingly, another Mitf allele, Mitfmi-x39, also leads to a gray coat when homozygous. This allele is characterized by a genomic deletion encompassing exon 4 and flanking regions, resulting in proteins that lack one of the activation domains (Hallsson et al. 2000). This phenotype has to be contrasted with that associated with Mitfmi-vga-9 whose M-Mitf coding region is unaltered but whose M-Mitf promoter is crippled. Mitfmi-vga-9 can lead to white spotting, in particular when combined with hypomorphic alleles such as Mitfmi-sp, but not to a gray coat (Steingrimsson et al. 2003). Hence, the appearance of the gray-coat phenotype is not simply due to changes in Mitf expression levels but seemingly requires the expression of abnormal proteins.
In sum, the varieties in coat-color phenotypes are due at least in part to changes in a transcription factor gene, Mitf, reflecting a subtle interplay between expression levels and alterations in protein-coding regions. All of this is overlaid in our mice, of course, by frequent recombination events that regenerate a codon-targeted, but otherwise entirely normal, gene. It will be a fascinating task not only to dissect the mechanisms by which such recombinations occur, but also to elucidate how the codon change alters splicing patterns and ultimately the expression of the set of target genes that control melanocyte proliferation, migration, and differentiation.
Acknowledgments
We thank Kapil Bharti for an Mitf plasmid and for help with sequence analysis, the National Institute of Neurological Disorders and Stroke (NINDS) sequencing facility for excellent services, and the Animal Health and Care Section of NINDS for outstanding animal care. This research was supported in part by the Intramural Research Program of the National Institutes of Health, NINDS, NHGRI, and by the Icelandic Research Fund, Reykjavik, Iceland.
References
- Arnheiter, H., L. Hou, M.-T. T. Nguyen, K. Bismuth, T. Csermely et al., 2006. Mitf—A Matter of Life and Death for the Developing Melanocyte. Humana Press, Totowa, NJ.
- Bennett, D. C., and M. L. Lamoreux, 2003. The color loci of mice: a genetic century. Pigment Cell Res. 16: 333–344. [DOI] [PubMed] [Google Scholar]
- Bharti, K., M. T. Nguyen, S. Skuntz, S. Bertuzzi and H. Arnheiter, 2006. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res. 19: 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilliant, M. H., Y. Gondo and E. M. Eicher, 1991. Direct molecular identification of the mouse pink-eyed unstable mutation by genome scanning. Science 252: 566–569. [DOI] [PubMed] [Google Scholar]
- Conrad, B., and S. E. Antonarakis, 2007. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 8: 17–35. [DOI] [PubMed] [Google Scholar]
- Demuth, J. P., T. De Bie, J. E. Stajich, N. Cristianini and M. W. Hahn, 2006. The evolution of mammalian gene families. PLoS ONE 1: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sepulveda, P., J. L. Guenet and J. J. Panthier, 1995. Phenotypic reversions at the W/Kit locus mediated by mitotic recombination in mice. Mol. Cell. Biol. 15: 5898–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra, A. S., E. Mignot and J. M. Puck, 1996. Gene localization and syntenic mapping by FISH in the dog. Cytogenet. Cell Genet. 74: 113–117. [DOI] [PubMed] [Google Scholar]
- Garraway, L. A., H. R. Widlund, M. A. Rubin, G. Getz, A. J. Berger et al., 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122. [DOI] [PubMed] [Google Scholar]
- Gondo, Y., J. M. Gardner, Y. Nakatsu, D. Durham-Pierre, S. A. Deveau et al., 1993. High-frequency genetic reversion mediated by a DNA duplication: the mouse pink-eyed unstable mutation. Proc. Natl. Acad. Sci. USA 90: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux et al., 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath, T. J., E. Steingrimsson, G. McGill, M. J. Hansen, J. Vaught et al., 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8: 2770–2780. [DOI] [PubMed] [Google Scholar]
- Hershey, C. L., and D. E. Fisher, 2005. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene 347: 73–82. [DOI] [PubMed] [Google Scholar]
- Hertwig, P., 1942. Neue Mutationen und Kopplungsgruppen bei der Hausmaus. Z. Indukt. Abstamm. Vererbungsl. 80: 220–246. [Google Scholar]
- Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrímsson, N. G. Copeland et al., 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74: 395–404. [DOI] [PubMed] [Google Scholar]
- Hughes, M. J., J. B. Lingrel, J. M. Krakowsky and K. P. Anderson, 1993. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 268: 20687–20690. [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2003. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3: 35–44. [PubMed] [Google Scholar]
- Minor, G., 1968. Mouse News Lett. 38: 25.
- Nakayama, A., M. T. Nguyen, C. C. Chen, K. Opdecamp, C. A. Hodgkinson et al., 1998. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 70: 155–166. [DOI] [PubMed] [Google Scholar]
- Opdecamp, K., A. Nakayama, M. T. Nguyen, C. A. Hodgkinson, W. J. Pavan et al., 1997. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development 124: 2377–2386. [DOI] [PubMed] [Google Scholar]
- Seperack, P. K., M. C. Strobel, D. J. Corrow, N. A. Jenkins and N. G. Copeland, 1988. Somatic and germ-line reverse mutation rates of the retrovirus-induced dilute coat-color mutation of DBA mice. Proc. Natl. Acad. Sci. USA 85: 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson, E., H. Arnheiter, J. H. Hallsson, M. L. Lamoreux, N. G. Copeland et al., 2003. Interallelic complementation at the mouse Mitf locus. Genetics 163: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson, E., N. G. Copeland and N. A. Jenkins, 2004. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38: 365–411. [DOI] [PubMed] [Google Scholar]
- Tachibana, M., Y. Hara, D. Vyas, C. A. Hodgkinson, J. Fex et al., 1992. Cochlear disorder associated with melanocyte anomaly in mice with a transgenic insertional mutation. Mol. Cell. Neurosci. 3: 433–445. [DOI] [PubMed] [Google Scholar]
- Tachibana, M., L. A. Perez-Jurado, A. Nakayama, C. A. Hodgkinson, X. Li et al., 1994. Cloning of MITF, the human homolog of the mouse microphthalmia gene and assignment to chromosome 3p14.1-p12.3. Hum. Mol. Genet. 3: 553–557. [DOI] [PubMed] [Google Scholar]
- Tallquist, M. D., and P. Soriano, 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26: 113–115. [DOI] [PubMed] [Google Scholar]
- Taylor, J. S., and J. Raes, 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson and R. C. Mulligan, 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65: 1153–1163. [DOI] [PubMed] [Google Scholar]
- Wu, M., T. J. Hemesath, C. M. Takemoto, M. A. Horstmann, A. G. Wells et al., 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14: 301–312. [PMC free article] [PubMed] [Google Scholar]
- Yajima, I., S. Sato, T. Kimura, K. Yasumoto, S. Shibahara et al., 1999. An L1 element intronic insertion in the black-eyed white (Mitf[mi-bw]) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum. Mol. Genet. 8: 1431–1441. [DOI] [PubMed] [Google Scholar]