Abstract
Mitochondrial DNA (mtDNA) is one of the most popular population genetic markers. Its relevance as an indicator of population size and history has recently been questioned by several large-scale studies in animals reporting evidence for recurrent adaptive evolution, at least in invertebrates. Here we focus on mammals, a more restricted taxonomic group for which the issue of mtDNA near neutrality is crucial. By analyzing the distribution of mtDNA diversity across species and relating it to allozyme diversity, life-history traits, and taxonomy, we show that (i) mtDNA in mammals does not reject the nearly neutral model; (ii) mtDNA diversity, however, is unrelated to any of the 14 life-history and ecological variables that we analyzed, including body mass, geographic range, and The World Conservation Union (IUCN) categorization; (iii) mtDNA diversity is highly variable between mammalian orders and families; (iv) this taxonomic effect is most likely explained by variations of mutation rate between lineages. These results are indicative of a strong stochasticity of effective population size in mammalian species. They suggest that, even in the absence of selection, mtDNA genetic diversity is essentially unpredictable, knowing species biology, and probably uncorrelated to species abundance.
MITOCHONDRIAL DNA (mtDNA) is by far the most widely used population genetic marker in animals (Avise et al. 1987). The reasons typically invoked to justify the choice of mtDNA are its high level of variability, its clonal (maternal) inheritance, and its supposed nearly neutral mode of evolution. In a population at mutation/drift equilibrium, the expected level of genetic diversity of a neutral locus is proportional to the effective population size (and to the locus mutation rate Wright 1931). mtDNA diversity is therefore typically assumed to reflect demographic effects, i.e., variations in population size between species or populations, which makes it a popular tool for conservation purposes (e.g., Harrison 1989; Roman and Palumbi 2003).
Several reports, however, have recently questioned the relevance of mtDNA as a marker of population size and history (Ballard and Whitlock 2004; Hurst and Jiggins 2005; Bazin et al. 2006). Among these, the Bazin et al. (2006) study proposed that patterns of mtDNA diversity in animals are largely influenced by adaptive evolution. First, Bazin et al. (2006) showed that the average level of within-species mtDNA diversity is not correlated to factors presumably influencing the effective population size: invertebrate species, for example, are not more polymorphic than vertebrates, marine mollusks are not more polymorphic than continental ones, and small, planktonic crustaceans are not more polymorphic than large, benthic ones. Nuclear loci, in contrast, behave essentially as expected under the nearly neutral model. Second, Bazin et al. (2006) reported a higher fixation rate of amino-acid substitutions in species with a large population size, while the opposite would be expected under the nearly neutral theory. These results were interpreted as reflecting the recurrent fixation of adaptive changes in mtDNA, consistent with Gillespie's (2000, 2001) genetic draft theory. It was proposed that selective sweeps associated to the fixation of favorable mutations (Maynard Smith and Haigh 1974) lead to frequent drops of mtDNA variability, removing any detectable influence of effective population size on the mitochondrial diversity (Bazin et al. 2006).
By questioning the near-neutrality assumption, this and other studies cast doubts on the reliability of mtDNA as a marker of species demography: under the genetic draft model, the mitochondrial diversity of a given species is primarily determined by the time since the last selective sweep, irrespective of population size. If confirmed, this result may have important practical consequences and would call for a reappraisal of many population genetic studies based on mtDNA. Further assessment of this strong claim therefore appears worthwhile.
Several limitations of the Bazin et al. (2006) study stress the need for additional investigations. First, this study was done at a relatively high taxonomic level and therefore involved comparing the genetic diversity of highly divergent species (e.g., mammals and mollusks). Life-history traits are obviously not comparable between organisms that differ, so the link between the genetic diversity and the biology of species, if any, is difficult to make. The basic properties of the mitochondrial genome, furthermore, might vary between distantly related taxa. The Bazin et al. (2006) approach, for example, did not properly account for potential variations in mitochondrial and nuclear mutation rates across animal phyla (Martin and Palumbi 1993; Lynch et al. 2006), although several of their results are robust to this nuisance parameter.
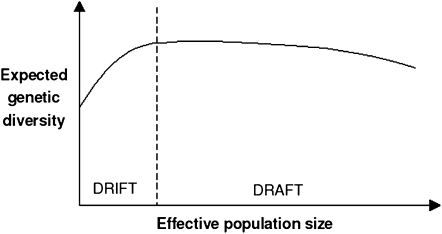
Second, and most importantly, it is unclear from Bazin et al. (2006) whether adaptive mitochondrial evolution is effective in all animal phyla. Gillespie's model predicts a merely flat relationship between genetic diversity and effective population size (Figure 1): small populations undergo genetic drift, and large populations undergo genetic draft. The population adaptive mutation rate is obviously higher in large populations, which compensates for the decrease in genetic drift. The results of Bazin et al. (2006) strongly suggest that the mtDNA diversity of invertebrate species is reduced by the recurrent fixation of advantageous mutations: invertebrates probably belong to the “draft zone” (Figure 1). But the location of vertebrate species in Gillespie's scheme is unclear. Although they do not exclude mitochondrial adaptation in vertebrates as well, the results of Bazin et al. (2006) do not appear incompatible with an evolutionary process dominated by genetic drift in this group. This is an important issue because a large fraction of the mtDNA-based population and conservation genetics literature focuses on vertebrates. The question has been preliminarily addressed by Mulligan et al. (2006). Reanalyzing the Bazin et al. data, these authors reported a positive correlation between the average mtDNA nucleotide diversity and allozyme heterozygosity of eight mammalian orders, suggesting that mammalian mtDNA could belong to the “drift zone” (see Figure 1).
Figure 1.—
Gillespie's genetic draft model. In small populations at mutation-drift equilibrium, the genetic diversity is proportional to the effective population size (drift zone). In large-enough populations, adaptive mutations impact the relationship by resulting in recurrent drops of diversity (draft zone). Compensation between the two forces leads to a merely flat relationship for a wide range of population sizes (redrawn after Gillespie 2001).
To improve our understanding of the determinism of within-species mtDNA diversity, we focus here on a more recent model taxon, namely mammals (as in Mulligan et al. 2006). Mammals were chosen because (i) they share a common body plan and physiology; (ii) life-history traits in this group are well documented, fairly variable, and comparable across species; (iii) their mitochondrial genomes share a common structure, gene order, and replication mode; (iv) their phylogeny is reasonably well known; and (v) they belong to vertebrates, for which the issue of the near-neutrality of mtDNA is still pending and crucial.
Our primary goal was to reproduce the Bazin et al. (2006) approach to check whether evidence for mitochondrial adaptive evolution is detected at this lower taxonomic level. Does mtDNA diversity correlate to nuclear diversity at this scale? Is there an excess of fixation of nonsynonymous changes? Then we planned to investigate whether the mtDNA diversity of mammalian species can be predicted from life-history and ecological variables, as would be expected if the effective population size was a strong determinant of the mtDNA diversity. Are small mammals more polymorphic, on average, than large ones? Are carnivorous predator species less diverse than herbivorous prey? The relationship between life-history traits and genetic diversity in mammals was explored decades ago using allozymes (Ryman et al. 1980; Wooten and Smith 1985; Sage and Wolff 1986) and yielded equivocal results. A reanalysis using sequence data would appear timely.
MATERIALS AND METHODS
Mitochondrial data:
An mtDNA data set was built from Polymorphix (Bazin et al. 2005), a within-species sequence variation database. Polymorphix contains within-species homologous sequence alignments (families) built from EMBL/GenBank thanks to suitable similarity and bibliographic criteria. To obtain a homogeneous data set, a single mitochondrial gene was used, namely the cytochrome b. This marker represents 76% of mammalian coding sequence mitochondrial families in Polymorphix. We extracted from Polymorphix every mammalian family for which any >100-bases-long cytochrome b fragment was available in six individuals or more. Sequence alignments were inspected by eye and corrected when required. Dubious sequences were manually removed. This includes potential nuclear pseudogenes and sequences with many gaps or undetermined nucleotides. We obtained a data set of 277 species.
Having observed through a preliminary analysis that the level of nucleotide diversity varies between regions of the cytochrome b gene, we decided to analyze a common fragment in all species. For this purpose, sequences were aligned using ClustalW (Thompson et al. 1994) and widely available fragments in the database were selected. Three data sets exploring various ratios of sequence length/number of species were built:
Data set 1: 89 species, 1140 nucleotides (complete cytochrome b).
Data set 2: 138 species, 401 nucleotides (positions 1–401).
Data set 3: 176 species, 180 nucleotides (positions 146–325).
These data sets include members of all major orders of placental mammals. Data set 2, for example, spans 32 families and 9 orders, among which four contain >10 species: Rodentia (66), Chiroptera (24), Eulipothypla (13), and Cetartiodactyla (11). A good representation of the morphological and ecological diversity of mammals is achieved (data available at http://kimura.univ-montp2.fr/data). The taxonomic sampling, for example, spans five orders of magnitude for body mass. Species names were taken from Polymorphix, and higher taxonomic information is from Wilson and Reeder (2005).
Allozyme data:
Estimates of allozyme heterozygosity in 184 mammalian species were obtained from the Nevo et al. (1984) compilation. Thirty species are represented in both the mtDNA and allozyme data set. Allozyme heterozygosities were averaged over >10 (typically 20–30) loci, and the most popular allozyme loci were used in many species. Allozyme heterozygosities are therefore fairly comparable between species.
Sampling quality control:
Polymorphix families are extracted from GenBank and are therefore potentially heterogeneous. In particular, no control of the sampling strategy is made. To address this problem, we checked 60 species randomly chosen from our mtDNA data set. Among the 46 species for which we could access the corresponding publication, 37 (80%) were sampled over their entire geographic range. This proportion was unrelated to taxonomy or body mass. This suggests that these data essentially measure diversity at the species level, as intended.
Polymorphism sequence data analyses:
Two measures of molecular genetic variability were used, namely the nucleotide diversity π (Tajima 1983) and Watterson's statistics θw (Watterson 1975). In the case of the haploid, maternally transmitted mtDNA, both statistics are unbiased estimates of the Nefμ product under the assumption of neutrality and mutation/drift equilibrium, where Nef is the effective population size of females and μ is the locus mutation rate. π and θw were calculated from the total length of the analyzed fragments and expressed in per-site level of diversity (after being divided by sequence length).
The neutrality index (NI; Rand and Kann 1996) was calculated for data set 1 when outgroups were available. This index aims at comparing the ratio of nonsynonymous (i.e., amino acid changing) to synonymous (silent) changes within species (Pn/Ps) and between species (dn/ds): The NI is 1 when evolution is neutral, >1 under purifying selection, and <1 in case of adaptation. The significance was assessed using the McDonald–Kreitman test (McDonald and Kreitman 1991). Estimates of π, θw, and NI were obtained using the Bio++ library (Dutheil et al. 2006).
Ecological and life-history variables:
For each species in the data set, five life-history traits, eight ecological variables, and The World Conservation Union (IUCN)-threatened species categories (IUCN, 2001) were compiled from multiple published sources (see Table 1 for explanation of variables and http://kimura.univmontp2.fr/data for references). The order, family, and species taxonomical levels were assigned to conform to recent taxonomical assessment (Wilson and Reeder 2005), and a fourth level, namely a supra-order, was assigned following recent molecular phylogenies (Murphy et al. 2001; Springer et al. 2004). The five life-history variables were (i) body mass, (ii) sexual maturity (age of female sexual maturity in months), (iii) reproduction frequency (time between two litters in months), (iv) maximal longevity (estimating from record longevity in years), and (v) fecundity (number of young per litter). Body mass is defined as the mean of the male and female adult body weights. All the maximal longevity values were retrieved from the AnAge database (De Magalhaes et al. 2005). We do not have any information about population structure (i.e., details on how many populations have been sampled) for all species in our data set. Therefore we used the Berlin et al. (2007) data set. Berlin et al. (2007) computed FST on the basis of cytochrome b polymorphism data for 30 genera, among which 17 genera match with our data set.
TABLE 1.
Discrete variables and categories
| Variables
|
||||
|---|---|---|---|---|
| Biogeography | Environment | Geographic range | Diet | |
| Categories | ||||
| Nearctic | Terrestrial | Limiteda | Vegetalivorousb | |
| Neotropical | Marine | Regionalc | Grazer | |
| Ethiopian | Extendedd | Folivorous | ||
| Palearctic | Worldwide | Nectarivorous | ||
| Oriental | Frugivorous | |||
| Australian | Carnivorous | |||
| Atlantic | Insectivorous | |||
| Pacific | Plankton eating | |||
| Indian | Omnivorous | |||
| Arctic | ||||
| Antarctic | ||||
| Cosmopolitane | ||||
| Variables
|
||||
| Habitat | Way of life | Mating system | Migrantf | Threatened statusg |
| Categories | ||||
| Aquatic | Subterraneanh | Monogamous | Migrant | Critically endangered |
| Forest | Fossoriali | Polygamous | Not migrant | Endangered |
| Desert | Terrestrial | Variable | Vulnerable | |
| Otherj | Arboreal | LR/CDk | ||
| Aquatic | LR/NTl | |||
| Flying | Least concern | |||
| Data deficient | ||||
| Not evaluated | ||||
Endemic or/and very narrow range size.
Eat different vegetal elements (leaf, graze, fruit, root, bulb).
Part of a biogeographic province.
Full biogeographic province.
Species are present in two or more biogeographic provinces.
Regular seasonal movement between a reproductive area and a distinct nonreproductive area.
IUCN–The World Conservation Union Red List threatened species cateorization system; see http://www.redlist.org for details.
Never eat on the ground.
Burrow well, but can also eat on the ground.
Other habitats (savannas, meadow, bush like, or nonspecialized).
Lower risk/conservation dependent.
Lower risk/near threatened.
To control our life-history variables, we compared them to existing databases. The body mass data of Smith et al. (2003) and the sexual maturity data of the AnAge database are highly correlated with ours (body mass: n = 128, r2 = 0.9905, P < 0.0001; maturity: n = 42, r2 = 0.9081, P < 0.0001). Moreover, the levels of correlation detected between our life-history variables—e.g., sexual maturity vs. body mass (n = 79, r2 = 0.4679, P < 0.0001), longevity vs. body mass (n = 62, r2 = 0.4771, P < 0.0001), reproduction frequency vs. body mass (n = 89, r2 = 0.3675, P < 0.0001)—are comparable with other studies (e.g., Millar and Zammuto 1983).
Statistical analyses:
Genetic diversity measures were arcsine transformed (Sokal and Rohlf 1981) and analyzed under the general linear model assumptions using R (Ihaka and Gentleman 1996). Quantitative life-history variables were log transformed. Because of the large number of factors, we could not perform a single full analysis. First, the effect of each ecological and life-history variable on genetic diversity was tested using independent regression or one-way ANOVA. Then we tested all combinations of two factors with interaction except for population structure because of the weak sample size (two-way ANOVA, multiple regression, ANCOVA).
Phylogenetic control:
A significant correlation between variables across species can arise just because of their shared phylogenetic history, even if the variables evolve independently (Felsenstein 1985). Phylogeny must therefore be regressed out of the analyses using the so-called comparative methods. Before doing this it must be shown that the variable of interest—in this study, genetic diversity—exhibits significant phylogenetic inertia (e.g., Ashton 2001). For this purpose, we tested for a taxonomic effect by using supra-order, order, and family as explanatory factors in one-way ANOVAs. Having detected a significant taxonomic effect, we performed a nested-hierarchical ANOVA to optimally decompose the variance across taxonomic levels. We also measured the phylogenetic autocorrelation of genetic diversity using Moran's I statistics (Gittleman and Kot 1990, implemented in APE package, Paradis et al. 2004).
When significant phylogenetic inertia was detected, we controlled for phylogeny in two ways. First, we used the residuals of the nested-hierarchical ANOVA for reanalyzing the effects of ecological and life-history traits. Second, we employed the phylogenetic eigenvector regression (PVR) method to correct for phylogenetic inertia (Diniz-Filho et al. 1998; Desdevises et al. 2003). This method can be applied to both continuous and discrete variables. The method starts by performing a principal coordinate analysis on a matrix of between-species patristic distances (implemented in ADE4-R package, Chessel et al. 2004). Following Desdevises et al. (2003), all the principal coordinates that were significantly correlated to the dependent variable were selected. Traits under analysis were then regressed on the retained eigenvectors. The estimated values express phylogenetic trends in the data, and residuals express independent evolution of each species. The proportion of genetic diversity variance explained exclusively by the ecological and life-history variables was quantified using multiple linear regressions, and its significance was assessed by using a regular parametric F-test (the permutation test used by Desdevises et al. 2003 gave similar results). Finally, the contrast method (Felsenstein 1985) was also used on continuous variables, yielding qualitatively equivalent results (not shown).
The phylogenetic tree was manually built according to the literature (Murphy et al. 2001; Huchon et al. 2002; Adkins et al. 2003; Hassanin and Douzery 2003; Springer et al. 2004; Steppan et al. 2004; Teeling et al. 2005; tree available as supplemental material at http://www.genetics.org/supplemental/). One sequence per species was randomly chosen to estimate branch lengths using the maximum-likelihood method (PHYML; Guindon and Gascuel 2003, model TN93 + Γ).
Substitution rate analysis:
The phylogenetic tree was also used to measure lineage-specific evolutionary rates. We performed a molecular clock analysis using the Multidivtime program (Thorne et al. 1998). To get sufficiently accurate estimates, we used only species for which the complete cytochrome b was available (data set 1, 89 species). The Bayesian approach in Multidivtime simultaneously estimates the rate and age for each node of a given tree, assuming a prior distribution of evolutionary rates along branches, and, optionally, constraints on the age of certain internal nodes. We used five fossil calibration points with upper and lower bounds: Cervidae/Bovidae (16.4–23.8 MY, Hassanin and Douzery 2003), Feliformia/Caniformia (23.8–42.8 MY, Benton and Donoghue 2007), Primates/rodents (61.5–95.5 MY, Benton and Donoghue 2007), Pan/Homo (6.5–10.0 MY, Benton and Donoghue 2007), Mus/Rattus (10.0–13.0 MY, Benton and Donoghue 2007). For each species, we stored the relative rate estimated in the corresponding terminal branch of the tree.
The mitochondrial DNA of mammals is characterized by a notoriously high amount of homoplasy. This effect decreases the efficiency of mitochondrial markers for phylogenetic analyses (see Springer et al. 2001; Galewski et al. 2006) and presumably also affects substitution rate estimates. To limit the effect of saturation, we chose to focus on transversions only, which are much less prone to generate homoplasy than transitions.
RESULTS
Testing the neutrality of mtDNA in mammals:
A significant, positive correlation was found between the allozyme heterozygosity and mtDNA nucleotide diversity of 29 mammalian species for which both measures are available (Figure 2, r2 = 0.187, P-value = 0.0191). This result contrasts with the absence of correlation between these two variables at the Metazoa level reported by Bazin et al. (2006). It indicates that adaptive evolution, if any, is not frequent enough to remove the influence of demographic effects, i.e., population size variations.
Figure 2.—
mtDNA genetic diversity and allozyme heterozygosity (29 species). Nucleotide diversity (π) and allozyme heterozygosity are arc-sine transformed.
Mulligan et al. (2006) also reported a positive correlation between the average mtDNA and allozyme diversity of eight mammalian orders. We did not, however, detect such a significant correlation at the order level with our data set (Kendall test, T = 3, P-value = 0.3333). The discrepancy between the two results is due to minor differences between the data sets. Mulligan et al. (2006) used all the mammalian data points from Bazin et al. (2006) (46 species), including genes other than cytochrome b, and various fragments of the cytochrome b, whereas our data set is standardized (see materials and methods). If only the cytochrome b gene is kept in the Mulligan et al. (2006) data set, the significance of the effect is strongly reduced (Kendall test, T = 0.73, P-value = 0.0555; P-value = 0.0027 with all data). The correlation at the species level, finally, is only marginally significant when the whole Mulligan et al. data set is used (n = 46, T = 0.198, P-value = 0.0552). Although they differ in minor aspects, the two studies essentially agree with the fact that allozyme heterozygosity and mtDNA diversity are (weakly) correlated in mammals.
The mtDNA neutrality index was calculated for 76 species and varied between 0.216 and 26.7. Two-thirds of the mtDNA data sets analyzed showed a NI value >1, which is indicative of a predominant effect of purifying selection. NI was not correlated to any of the life-history and ecological variables analyzed here or to allozyme heterozygosity or taxonomy. No clear pattern emerged that could distinguish the 25 species showing a NI value <1 from a random sample of the data set. This contrasts again with the Bazin et al. (2006) study in which a significantly lower average NI was reported in (presumably more abundant) invertebrates than (presumably less abundant) vertebrates, in agreement with the genetic draft model. Only two species, Pteronotus parnellii (Chiroptera, P-value = 0.007) and Microcebus rufus (Primates, P-value = 0.024) showed a significantly <1 NI, which is not more than expected just by chance, given the number of independent tests performed. Thirteen species, however, showed a significantly >1 NI. It should be noted that this result is robust to a potential underestimation of the synonymous divergence due to multiple hits; in the case of too distant outgroups, underestimating ds leads to an underestimate of NI.
Overall, these results do not support a prominent role of adaptive evolution in the mitochondrial genome of mammals. Although adaptation of course may occasionally occur, overall patterns of mtDNA diversity across taxa appear compatible with a nearly neutral mode of evolution, in which demographic effects are the major determinants of mtDNA variation. Mammalian species may well belong to Gillespie's “drift zone” (see Figure 1).
Neither life-history traits nor ecology correlate with mtDNA diversity:
Assuming that mtDNA diversity is governed mainly by population size variations, it is expected that the most abundant species would tend to be, on average, the genetically most diverse. Direct estimates of population size are available for a few mammalian species only and involve very large confidence intervals (Nei and Graur 1984). We can, however, make use of biological descriptors of species potentially correlated to population size. A total of 14 variables were used to explore as widely as possible the potential determinism of mtDNA diversity in mammals.
Analyses using nucleotide diversity (π), Watterson's statistic (θw), or synonymous nucleotide diversity (πs) and any of the three mtDNA data sets gave similar results. Except when indicated, we present only the results obtained from data set 2 (401 nucleotides, 138 species) using the nucleotide diversity (other results are available on request). The mean per-site nucleotide diversity is 0.025 ± 0.022 (mean synonymous nucleotide diversity: 0.097 ± 0.090). The most variable species is the Southern pocket gopher (Thomomys umbrinus, Geomyidae, Rodentia, π = 0.106) and the least variable one is the gray-brown mouse lemur (Microcebus griseorufus, Cheirogaleidae, Primates, π = 0.0004).
We first analyzed every explanatory variable separately (Table 2). Among continuous variables, body mass, sexual maturity, and longevity had a significant effect. In spite of the great variability of body mass in the data set (from 3.25 g for the common shrew, Sorex cinereus, to 666.5 kg for the yak, Bos grunniens), this variable explained only 6% of mtDNA polymorphism variability. This result is similar to Wooten and Smith's (1985) allozyme study (r2 = 0.04, P-value = 0.034). Two other continuous variables had a significant effect: sexual maturity (r2 = 0.1252, P-value = 0.0059) and longevity (r2 = 0.1260, P-value = 0.0091). These variables are strongly correlated to body mass (r2 = 0.4254, P-value < 0.0001 for sexual maturity and r2 = 0.4548, P-value < 0.0001 for longevity). The effects of body mass, sexual maturity, and longevity were removed when phylogenetic inertia was controlled for (see Table 2), indicating that the influence of life-history variables on mtDNA genetic diversity, if any, is very low.
TABLE 2.
Effects of life-history and ecological variables on mtDNA diversity
| No phylogenetic control
|
Taxonomic controla
|
PVRb
|
||||||
|---|---|---|---|---|---|---|---|---|
| N | Slope | r2 | P-value | r2 | P-value | r2 | P-value | |
| Body mass | 107 | −0.0049 | 0.0566 | 0.0136 | 0.0165 | 0.1877 | 0.0273 | 0.0802 |
| Sexual maturity | 59 | −0.0181 | 0.1252 | 0.0059 | 0.0185 | 0.3050 | 0.0018 | 0.6676 |
| Reproduction | 71 | −0.0318 | 0.0334 | 0.1271 | ||||
| Longevity | 53 | −0.0289 | 0.1260 | 0.0091 | 0.0650 | 0.0655 | 0.05 | 0.0841 |
| Fecundity | 87 | 0.0178 | 0.0080 | 0.4105 | ||||
| Biogeography | 138 | 0.1154 | 0.0504 | |||||
| Diet | 107 | 0.1947 | 0.0005 | 0.0533 | 0.4641 | 0.0894 | 0.1033 | |
| Geographic range | 117 | 0.0446 | 0.1590 | |||||
| Habitat | 108 | 0.0010 | 0.7927 | |||||
| Way of life | 112 | 0.1058 | 0.0346 | 0.0616 | 0.2335 | 0.0896 | 0.0703 | |
| Mating system | 71 | 0.0453 | 0.2064 | |||||
| Migrant | 94 | 0.0020 | 0.6629 | |||||
| Environment | 111 | 0.0065 | 0.4014 | |||||
| Threatened status | 138 | 0.0277 | 0.8119 | |||||
P-values less than the 0.05 threshold are in italics.
Residuals of nested ANOVA on taxonomy.
Phylogenetic eigenvector regression control.
Only two ecological variables had a significant effect, namely diet and “way of life.” Diet is the ecological variable best correlated to mtDNA polymorphism (r2 = 0.1947, P-value = 0.0005). The diet effect is essentially caused by the low genetic diversity of Carnivora. The “way of life” effect comes from the high genetic diversity of arboreal species, a result for which we have no obvious biological interpretation. As for life-history traits, ecological effects appear to be largely due to the phylogenetic inertia. Diet is strongly linked to taxonomy (e.g., most of the carnivores belong to the Carnivora order). Not surprisingly, phylogenetic controls removed the diet effect (Table 2, P-value = 0.1033) and the “way of life” effect (Table 2, P-value = 0.070).
We detected a positive correlation between genetic diversity and FST (P-value = 0.02, Kendall no parametric test). This correlation, however, is largely explained by a higher average FST in rodents. It was removed by a taxonomic control (P-value = 0.1263). Moreover, we did not find any correlation when we used the total Berlin et al. (2007) data set (including genera missing in our data set, N = 30).
Then we considered every two-factor combination of the explanatory variables. We present the result only for several models (Table 3; detailed results available on request). In two-way ANOVAs, no significant effect was found, except the body mass effect in one analysis, which did not resist the phylogenetic control (see Table 2).
TABLE 3.
Effects of life-history and ecological variables on mtDNA diversity: two-way ANOVAs
|
P-value
|
|||||||
|---|---|---|---|---|---|---|---|
| No phylogenetic control
|
PVRa: Model | ||||||
| Model | N | r2 | Model | Variable 1 | Variable 2 | Interaction | |
| Body mass + diet | 95 | 0.255 | 0.012 | 0.223 | 0.016 | 0.004 | 0.1243 |
| Body mass + way of life | 102 | 0.209 | 0.023 | 0.019 | 0.090 | 0.282 | 0.1059 |
| Body mass + range | 100 | 0.176 | 0.035 | 0.032 | 0.476 | 0.201 | 0.1316 |
| Body mass + threatened | 107 | 0.098 | 0.163 | 0.020 | 0.604 | 0.055 | |
| Threatened + range | 117 | 0.083 | 0.389 | 0.617 | 0.133 | 0.560 | |
P-values less than the 0.05 threshold are in italics.
Phylogenetic eigenvector regression control.
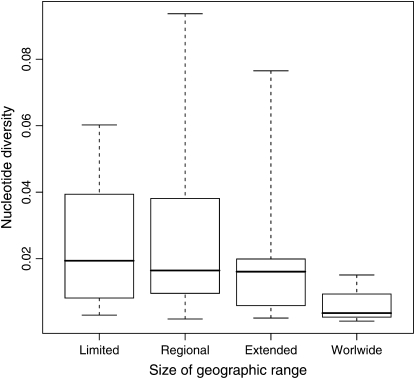
These results contradict many of our intuitive expectations. Figure 3 shows that the size of species geographic range does not influence mtDNA diversity: endemic species are not less polymorphic than cosmopolitan ones. Another surprising result is the absence of influence of IUCN categorization, since the system of categorization is partly linked to an evaluation of the population size. The effects of the few variables significantly influencing mtDNA diversity in one-way ANOVA are weak, disappear after the phylogenetic control (Table 2), and decrease when several variables are combined (Table 3). Overall, our results contradict the prediction that ecological and life-history traits influence the mitochondrial genetic diversity in mammals. Small, widely distributed, short-lived species are not more diverse than large, geographically restricted, threatened ones.
Figure 3.—
mtDNA diversity and geographic range (117 species) and nucleotide diversity (π). The median, quartiles, and extrema of the distribution of π are given for four classes of geographic range.
A strong taxonomic effect:
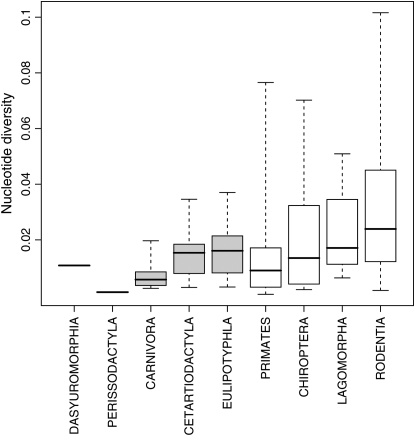
In a surprising way, taxonomy explained a large part of the variability in cytochrome b diversity across species, with highly significant effects (Table 4). One-way ANOVAs indicated that most of the variation (44%) is explained by the family level. The order level explains 18% of the variability. A nested ANOVA gave similar results (Table 4). Rodentia is the most polymorphic order (average π = 0.033). Carnivora is the least polymorphic order represented by more than one species in the data set (average π = 0.008, Figure 4). Supra-order average levels of diversity are also significantly different from each other, Euarchontoglire species being more diverse than Laurasiatheria. Similarly, we calculated the phylogenetic autocorrelation using Moran's I statistic (Gittleman and Kot 1990) and the phylogenetic inertia using PVR (Diniz-Filho et al. 1998; Desdevises et al. 2003). Genetic diversity had a highly significant autocorrelation (I = −0.01943, P-value < 0.0001) and the PVR method indicated a strong phylogenetic inertia on this variable (r2 = 0.319, P-value < 0.0001, after 10,000 random permutations). This explains why the few significant effects of ecological and life-history traits that we detected vanished when phylogenetic inertia was controlled for.
TABLE 4.
Taxonomic effect on mtDNA diversity
| Taxonomic level | One-way ANOVA
|
Nested ANOVA: Variance component | ||
|---|---|---|---|---|
| N | r2 | P-value | ||
| Supra-order | 138 | 0.1001 | 0.0009 | 0.050 |
| Order | 138 | 0.1780 | 0.0011 | 0.103 |
| Family | 138 | 0.4398 | <0.0001 | 0.273 |
Figure 4.—
mtDNA diversity and taxonomy (138 species) and nucleotide diversity (π). The median, quartiles, and extrema of the distribution of π are given for nine mammalian orders. Dasyuromorphia, Marsupialia. Shaded bars, Laurasiatheria; open bars, Euarchontoglires.
The taxonomic effect is explained by lineage-specific variations of mutation rate:
The above analyses show that neither life-history traits nor ecology influence the mitochondrial polymorphism, but reveal an unexpected link between taxonomy and genetic diversity. How to explain such a relationship? Variations of mtDNA mutation rate between mammalian lineages have been reported and could explain our results (Gissi et al. 2000; Spradling et al. 2001). We do not expect mutation rate to be a good predictor of species genetic diversity at mutation/drift equilibrium: between-species variations of mutation rates are probably weaker than variations of population sizes. In nonequilibrium conditions, however, mutation rates could matter. Mutation rates predominantly influence the speed of polymorphism recovery after a sudden loss of variability due to, for example, population bottlenecks (Nei and Graur 1984). If the stochasticity of mammalian effective population sizes was such that the mutation/drift equilibrium is rarely or never reached, then the mutation rate could be a determinant of species genetic diversity.
This hypothesis predicts that the average genetic diversity of taxonomic groups should be correlated with their current mutation rates. The mutation rate is usually unknown, but can be approached by estimating lineage-specific substitution rates; under neutrality, the two are proportional. We found that species' genetic diversity was positively correlated to their relative transversion substitution rate as estimated with Multidivtime (r2 = 0.110, P = 0.0016; Figure 5). This result is robust to the removal of non-neutral first and second codon positions (r2 = 0.044, P = 0.048) and to a taxonomic control—i.e., the average family rate is correlated to the average family diversity (N = 22, P = 0.04, Kendall test). Our mtDNA substitution rate estimator explains only 11% of the variance of mtDNA diversity across species, which can be explained by the high variability in genetic diversity resulting from demographic (or selective) stochasticity and/or by inaccurate estimation of evolutionary rates due to saturation and incomplete fossil information.
Figure 5.—
mtDNA diversity and transversion substitution rate (89 species). Nucleotide diversity (π) is arc-sine transformed; substitution rate is log transformed.
DISCUSSION
A detailed analysis of mtDNA diversity across a large sample of mammalian species revealed that (i) mtDNA in mammals does not reject the nearly neutral model, in contrast to a similar study conducted at the Metazoa level (Bazin et al. 2006); (ii) mtDNA diversity, however, is unrelated to any of the 14 life-history and ecological variables that we analyzed, including body mass, geographic range, and IUCN categorization; (iii) mtDNA diversity is highly variable between mammalian orders and families; and (iv) this variation is most likely explained by variations of mutation rate between lineages.
The claim of a prominent role for adaptive evolution in animal mtDNA is therefore not supported at the Mammalia level: mtDNA diversity is correlated to allozyme diversity, and neutrality indices mostly support purifying selection. Genetic drift, not genetic draft, is probably the main determinant of species' genetic diversity in this group. This can be considered good news for users of mtDNA as a demographic marker in mammals. Although the occurrence of occasional adaptive events cannot be excluded, mammalian mtDNA appears little affected by frequent selective sweeps. The question is difficult to assess in other vertebrate taxa because their ecology and life history have not been studied as deeply as in mammals. Fish, for example, in which larger population sizes are expected and for which the mtDNA diversity apparently does not correlate to the nuclear one (Bazin et al. 2006), could be worth investigating. Birds are also interesting because of their weaker average mtDNA diversity (as compared to mammals), perhaps due to the linkage between mtDNA and the W chromosome (Berlin et al. 2007).
Given this result, it came as a surprise that we could not detect an effect of potential population size indicators on mtDNA genetic diversity. Of course, it is possible that the life-history and ecological variables that we used are not appropriate markers of species effective population size. But this would mean that species abundance is not correlated to body size, geographic range, IUCN categorization, or to any combination of these and other variables, which seems quite unlikely. The relationship between body mass and abundance, for example, is well supported in many groups (White et al. 2007), including mammals (Damuth 1987). Bazin et al. (2006) detected a significant effect of indicators of population size on allozyme heterozygosity using much smaller data sets and arguably less sophisticated variables (marine vs. terrestrial mollusks, “large” vs. “small” crustaceans).
Similarly, the results obtained by Mulligan et al. (2006) are difficult to explain by a population size effect. We agree that the order showing the greatest mtDNA and allozyme variability (Rodentia) is one with large expected populations, whereas the least polymorphic order (Carnivora) is predicted to include species with smaller populations (higher-order predators). The ranking of orders with intermediate levels of diversity, however, is difficult to reconcile with intuition about populations sizes: Artiodactyla and Cetacean species are more polymorphic, on average, than Chiroptera and Eulipotyphla. It is interesting to note, finally, that Rodentia is the fastest-evolving mammalian order and Carnivora one of the slowest evolving (Multidivtime analysis; results not shown), which is in agreement with our mutation-effect hypothesis. Finally, Berlin et al. (2007) reported a significant correlation between body mass and mitochondrial diversity in birds and mammals. They did not, however, perform any phylogenetic control. When we did so using their data, the body mass effect was removed, as in this study.
The detection of a mutation rate effect but no population size effect suggests that this pattern is due to a strong stochasticity of population size in mammals. Generalizing Nei and Graur (1984), Iizuka et al. (2002) showed that when population size randomly fluctuates between two distinct values, the mutation rate becomes the major determinant of genetic diversity (for a small-enough minimal population size). A metapopulation model with local extinction and recolonization also predicts that diversity becomes independent from effective population size when the extinction rate is higher than the migration rate (Pannell and Charlesworth 1999). The literature provides examples of population size fluctuations caused by global climatic changes, for example, during the past 3 million years (Hewitt 2000), or by pandemic diseases (Harding et al. 2002). Our results suggest that stochastic variations in effective population size are probably a general process of mammalian species dynamics, strong enough to remove any effect of (current) species abundance on mtDNA diversity.
This conclusion is apparently in conflict with previous reports of ecological/life history correlates of nuclear genetic diversity. Nevo et al. (1984) conducted a meta-analysis of allozyme data and reported a significant effect of six variables (including range size, habitat, and longevity) on allozyme heterozygosity in mammals. These are only a few of the many ecological and life-history variables that they used, however. The percentages of explained variance were low and similar to those that we report in this study, and no phylogenetic control was performed. We reanalyzed the allozyme data controlling for taxonomy. Of the 21 variables considered by Nevo et al. (1984), 6 significantly influence allozyme diversity, among which 5 are still significant after taxonomic control: geographic range size, specialist vs. generalist, aridity, species size, and young dispersal; only 1 effect (longevity) was removed. The allozyme results appear more robust to taxonomical control than mitochondrial ones. We see two potential explanations for this pattern. First, allozyme heterozygosity is obtained by averaging over several loci, thus reducing the variance of the coalescent process. Second, mammalian mitochondrial DNA could occasionally experience selective sweeps, although we did not detect any NI effect. Although Nevo et al. (1984) put the emphasis on the detected correlations, our view is that this study is largely consistent with ours in suggesting a weak, if any, influence of ecology and life history on genetic diversity in mammals.
One conflict between mtDNA and nuclear data that we consider significant is the IUCN status effect. Spielman et al. (2004) reported that the nuclear heterozygosity of threatened taxa is, on average, lower than that of nonthreatened relatives. The effect is rather strong in mammals, in which 84% of endangered species are less diverse than their nonendangered relatives. This is true for both allozymes and microsatellite markers, and this contrasts with our mtDNA results. Even by applying the pairwise approach used by Spielman et al. (2004), we failed to recover any effect of the IUCN status: <50% of the endangered species in our mtDNA data set are less polymorphic than their nonendangered relatives. This could be due to a lack of power: our data set includes only 22 endangered species (63 in Spielman et al. 2004) and relies on a single locus, the mitochondrial one, while nuclear estimates of diversity are obtained by averaging over several loci, thus reducing the variance of the coalescent process. The mtDNA mutation rate effect that we detect, moreover, might add some noise and further decrease the power of the analysis.
So, even in the absence of strong adaptive effects, mtDNA diversity in mammals is largely unpredictable knowing species biology, and probably reflects in the first place the product of the mitochondrial mutation rate by the time elapsed since the last event of diversity reduction. This does not preclude the use of mtDNA to infer past demographic history, but still suggests that mtDNA diversity is a poor indicator of current population size and health.
Acknowledgments
We thank J. Dutheil and J. Claude for helpful discussions on phylogenetic control methods, S. Berlin for sharing published data, and two anonymous reviewers for helpful comments. This work was supported by the Centre National de la Recherche Scientifique, Université Montpellier II, and project MITOSYS of Agence Nationale pour la Recherche. This is publication ISE-M 2007-082.
References
- Adkins, R. M., A. H. Walton and R. I. Honeycutt, 2003. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol. Phylogenet. Evol. 26: 409–420. [DOI] [PubMed] [Google Scholar]
- Ashton, K.G., 2001. Body size variation among mainland populations of the western rattlesnake (Crotalus viridis). Evolution 55: 2523–2533. [DOI] [PubMed] [Google Scholar]
- Avise, J. C., J. Arnold, R. M. Ball, E. Bermingham, T. Lamb et al., 1987. Intraspecific phylogeography: the mitochondrial-DNA bridge between population-genetics and systematics. Annu. Rev. Ecol. Syst. 18: 489–522. [Google Scholar]
- Ballard, J. W. O., and M. C. Whitlock, 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13: 729–744. [DOI] [PubMed] [Google Scholar]
- Bazin, E., L. Duret, S. Penel and N. Galtier, 2005. Polymorphix: a sequence polymorphism database. Nucleic Acids Res. 33: D481–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin, E., S. Glémin and N. Galtier, 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. [DOI] [PubMed] [Google Scholar]
- Benton, M. J., and P. C. Donoghue, 2007. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24: 889–891. [DOI] [PubMed] [Google Scholar]
- Berlin, S., D. Tomaras and B. Charlesworth, 2007. Low mitochondrial variability in birds may indicate Hill-Robertson effects on the W chromosome. Heredity 99: 389–396. [DOI] [PubMed] [Google Scholar]
- Chessel, D., A. B. Dufour and J. Thioulouse, 2004. The ADE4 package—I: One-table methods. RNews 4(1): 5–10 (http://cranr-projectorg/doc/Rnews/). [Google Scholar]
- Damuth, J., 1987. Interspecific allometry of population-density in mammals and other animals: the independence of body-mass and population energy use. Biol. J. Linn. Soc. 31: 193–246. [Google Scholar]
- de Magalhaes, J. P., J. Costa and O. Toussaint, 2005. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 33: D537–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdevises, Y., P. Legendre, L. Azouzi and S. Morand, 2003. Quantifying phylogenetically structured environmental variation. Evolution 57: 2647–2652. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho, J. A., C. E. R. de Sant'ana and L. M. Bini, 1998. An eigenvector method for estimating phylogenetic inertia. Evolution 52: 1247–1262. [DOI] [PubMed] [Google Scholar]
- Dutheil, J., S. Gaillard, E. Bazin, S. Glémin, V. Ranwez et al., 2006. Bio++: a set of C++ libraries for sequence analysis, phylogenetics, molecular evolution and population genetics. BMC Bioinformatics 7: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Phylogenies and the comparative method. Am. Nat. 125: 1–15. [Google Scholar]
- Galewski, T., M. Tilak, S. Sanchez, P. Chevret, E. Paradis et al., 2006. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol. Biol. 6: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55: 2161–2169. [DOI] [PubMed] [Google Scholar]
- Gissi, C., A. Reyes, G. Pesole and C. Saccone, 2000. Lineage-specific evolutionary rate in mammalian mtDNA. Mol. Biol. Evol. 17: 1022–1031. [DOI] [PubMed] [Google Scholar]
- Gittleman, J. L., and M. Kot, 1990. Adaptation statistics and a null model for estimating phylogenetic effects. Syst. Zool. 39: 227–241. [Google Scholar]
- Guindon, S., and O. Gascuel, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Harding, K. C., T. Harkonen and H. Caswell, 2002. The European seal plague: epidemiology and population consequences. Ecol. Lett. 5: 727–732. [Google Scholar]
- Harrison, R. G., 1989. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 4: 6–11. [DOI] [PubMed] [Google Scholar]
- Hassanin, A., and E. J. P. Douzery, 2003. Molecular and morphological phylogenies of Ruminantia and the alternative position of the Moschidae. Syst. Biol. 52: 206–228. [DOI] [PubMed] [Google Scholar]
- Hewitt, G., 2000. The genetic legacy of the quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Huchon, D., O. Madsen, M. Sibbald, K. Ament, M. J. Stanhope et al., 2002. Rodent phylogeny and a timescale for the evolution of glires: evidence from an extensive taxon sampling using three nuclear genes. Mol. Biol. Evol. 19: 1053–1065. [DOI] [PubMed] [Google Scholar]
- Hurst, G. D. D., and F. M. Jiggins, 2005. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc. Biol. Sci. 272: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R., and R. Gentleman, 1996. R: a language for data analysis and graphics. J. Comp. Stat. 5: 299–314. [Google Scholar]
- Iizuka, M., H. Tachida and H. Matsuda, 2002. A neutral model with fluctuating population size and its effective size. Genetics 161: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., B. Koskella and S. Schaack, 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311: 1727–1730. [DOI] [PubMed] [Google Scholar]
- Martin, A. P., and S. R. Palumbi, Sr., 1993. Body size, metabolic-rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. USA 90: 4087–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Millar, J. S., and R. M. Zammuto, 1983. Life histories of mammals: an analysis of life tables. Ecology 64: 631–635. [Google Scholar]
- Mulligan, C. J., A. Kitchen and M. M. Miyamoto, 2006. Comment on “population size does not influence mitochondrial genetic diversity in animals.” Science 314: 1390. [DOI] [PubMed] [Google Scholar]
- Murphy, W. J., E. Eizirik, S. J. O'Brien, O. Madsen, M. Scally et al., 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348–2351. [DOI] [PubMed] [Google Scholar]
- Nei, M., and D. Graur, 1984. Extent of protein polymorphism and the neutral mutation theory. Evol. Biol. 17: 73–118. [Google Scholar]
- Nevo, E., A. Beiles and R. Ben-Shlomo, 1984. The evolutionary significance of genetic diversity: ecological, demographic and life-history correlates, pp. 13–295 in Evolutionary Dynamics of Genetic Diversity (Lecture Notes in Biomathematics, Vol. 53), edited by G. S. Mani. Springer-Verlag, Berlin.
- Pannell, J. R., and B. Charlesworth, 1999. Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution 53: 664–676. [DOI] [PubMed] [Google Scholar]
- Paradis, E., J. Claude and K. Strimmer, 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Rand, D. M., and L. M. Kann, 1996. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 13: 735–748. [DOI] [PubMed] [Google Scholar]
- Roman, J., and S. Palumbi, 2003. Whales before whaling in the north Atlantic. Science 301: 508–510. [DOI] [PubMed] [Google Scholar]
- Ryman, N., C. Reuterwall, K. Nygren and T. Nygren, 1980. Genetic-variation and differentiation in Scandinavian moose (Alces alces): Are large mammals monomorphic? Evolution 34: 1037–1049. [DOI] [PubMed] [Google Scholar]
- Sage, R. D., and J. O. Wolff, 1986. Pleistocene glaciations, fluctuating ranges, and low genetic-variability in a large mammal (Ovis dalli). Evolution 40: 1092–1095. [DOI] [PubMed] [Google Scholar]
- Smith, F. A., S. K. Lyons, S. K. M. Ernest, K. E. Jones, D. M. Kaufman et al., 2003. Body mass of late quaternary mammals. Ecology 84: 3403. [Google Scholar]
- Sokal, R., and F. Rohlf, 1981. Biometry. W. H. Freeman, San Francisco.
- Spielman, D., B. W. Brook and R. Frankham, 2004. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA 101: 15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, T., M. Hafner and J. Demastes, 2001. Differences in rate of cytochrome-b evolution among species of rodents. J. Mammal. 82: 65–80. [Google Scholar]
- Springer, M., R. Debry, C. Douady, H. Amrine, O. Madsen et al., 2001. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol. Biol. Evol. 18: 2132–2143. [DOI] [PubMed] [Google Scholar]
- Springer, M. J., M. J. Stanhope, O. Madsen and W. W. de Jong, 2004. Molecules consolidate the placental mammal tree. Trends Ecol. Evol. 19: 430–438. [DOI] [PubMed] [Google Scholar]
- Steppan, S. J., R. M. Adkins and J. Anderson, 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53: 533–553. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling, E. C., M. S. Springer, O. Madsen, P. Bates, S. J. O'Brien et al., 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307: 580–584. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibbson, 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, J. L., H. Kishino and I. S. Painter, 1998. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 15: 1647–1657. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- White, E. P., S. K. M. Ernest, A. J. Kerkhoff and B. J. Enquist, 2007. Relationships between body size and abundance in ecology. Trends Ecol. Evol. 22: 323–330. [DOI] [PubMed] [Google Scholar]
- Wilson, D., and D. Reeder, 2005. Mammal Species of the World. Smithsonian Institution Press, Washington, DC.
- Wooten, M. C., and M. H. Smith, 1985. Large mammals are genetically less variable. Evolution 3: 210–212. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]