Abstract
Phylogenetic and population genetic analyses of DNA sequence data from 10 nuclear loci were used to test species divergence hypotheses within Passerina buntings, with special focus on a strongly supported, but controversial, sister relationship between Passerina amoena and P. caerulea inferred from a previous mitochondrial study. Here, a maximum-likelihood analysis of a concatenated 10-locus data set, as well as minimize-deep-coalescences and maximum-likelihood analyses of the locus-specific gene trees, recovered the traditional sister relationship between P. amoena and P. cyanea. In addition, a more recent divergence time estimate between P. amoena and P. cyanea than between P. amoena and P. caerulea provided evidence for the traditional sister relationship. These results provide a compelling example of how lineage sorting stochasticity can lead to incongruence between gene trees and species trees, and illustrate how phylogenetic and population genetic analyses can be integrated to investigate evolutionary relationships between recently diverged taxa.
INFERRING the species tree of recently diverged taxa can be a challenging exercise (Maddison 1997; Felsenstein 2004; Maddison and Knowles 2006). The difficulty arises in part because the stochastic sorting of ancestral polymorphisms after speciation can result in gene trees that are discordant with species trees (Pamilo and Nei 1988; Takahata 1989; Maddison 1997; Rosenberg 2002, 2003; Maddison and Knowles 2006). For recently diverged taxa that have not yet attained complete reproductive isolation, gene flow introduces another source of discordance between the gene trees and the species tree (Takahata and Slatkin 1990; Takahata 1995; Wakeley 1996; Wakeley and Hey 1998). The impact of these processes on species tree reconstruction has long been recognized (Gillespie and Langley 1979; Nei and Li 1979), but it is only recently that a model-based phylogenetic framework has integrated one of them (ancestral sorting) into the species tree inference. Because postspeciation, interspecific gene flow has yet to be incorporated into a phylogenetic framework, nonphylogenetic methods must be used to infer species relationships indirectly. Here, phylogenetic and divergence population genetic analyses of a multilocus nuclear data set are used to examine a controversial sister species relationship in Passerina buntings.
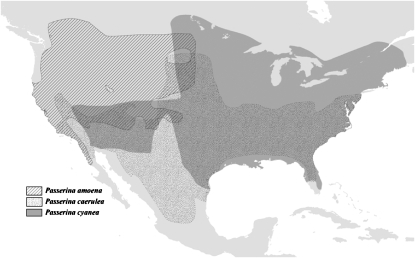
Over most of its taxonomic history, the genus Passerina (Aves: Cardinalidae) has been composed of six species of small (13–20 g), sexually dichromatic songbirds. The collective breeding ranges of these species encompass most of Mexico, the United States, and southern Canada (Figure 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/). Within the genus, Passerina cyanea and P. amoena have typically been considered sister species because they are morphologically similar and form a well-known hybrid zone where their breeding distributions overlap in the western Great Plains and eastern foothills of the Rocky Mountains in North America (Sibley and Short 1959; Emlen et al. 1975; Kroodsma 1975; Baker and Baker 1990; Baker and Boylan 1999). While the monophyly of Passerina has never been questioned (Hellack and Schnell 1977; Tamplin et al. 1993), there have been attempts to subsume Guiraca caerulea and some species of the genus Cyanocompsa into the genus (Phillips et al. 1964; Blake 1969; Mayr and Short 1970; Paynter 1970).
Figure 1.—
Breeding distributions of Passerina amoena, P. cyanea, and P. caerulea. Digital maps (Ridgely et al. 2003) for each species were downloaded from NatureServe (2006). Breeding distributions of the other four members of the genus Passerina are available in supplemental Figure 1 at http://www.genetics.org/supplemental/.
Klicka et al. (2001) addressed the evolutionary relationships of the traditional six-member genus and closely related species, including G. caerulea and three Cyanocompsa species, using 1143 bp of sequence data from the mitochondrial cytochrome b gene. An unexpected result of the study was that P. amoena and P. cyanea, the two morphologically similar species that form a hybrid zone in the Great Plains, were not sister species. Instead G. caerulea (hereafter, P. caerulea) was sister to P. amoena, a relationship that had never before been hypothesized. P. cyanea was placed as the most basal member of the now seven-member genus. As expected, the three Cyanocompsa species fell outside of Passerina.
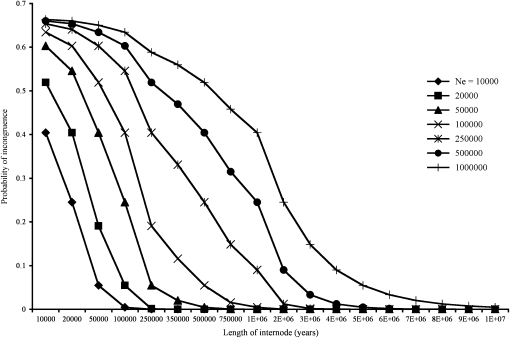
The branching structure of the mitochondrial tree and the demography of Passerina buntings suggest that the unexpected P. amoena/P. caerulea sister relationship could reflect discordance between the mitochondrial gene tree and the species tree. The internodes in the mitochondrial phylogeny are short, possibly indicating rapid divergences, and the population sizes for some of the species, including P. cyanea, P. amoena, and P. caerulea, are large—on the order of tens of millions of individuals (Payne 1992; Greene et al. 1996). This combination of short internodes and large population sizes is among the most difficult scenarios for inferring phylogenies from single locus data sets given the elevated risk of discordance between a species tree and its underlying gene trees (Figure 2). Using the calculation of Hein et al. (2005) for the probability of species tree/gene tree discordance and realistic parameter estimates for Passerina buntings, the probability of discordance is estimated to be as high as 55% (see Comparing mtDNA and nuclear DNA signals in discussion).
Figure 2.—
The probability a gene tree will be incongruent with a rooted three-member species tree as a function of the effective population size and the length of the internode in years. Effective population sizes and internode lengths were chosen to bracket realistic values for Passerina buntings.
Here, phylogenetic and divergence population genetics methods are used to test speciational hypotheses in Passerina buntings, with special focus on the controversial sister relationship uncovered by the mitochondrial study. Using the Passerina data set, the utility of combining phylogenetic and population genetics methods to uncover the evolutionary history of closely related species where hybridization may complicate evolutionary history is examined. Specifically, support for the following two alternative hypotheses was assessed:
Hypothesis 1: P. cyanea and P. amoena are more closely related to each other than either is to P. caerulea. This is the traditional sister relationship.
Hypothesis 2: P. amoena and P. caerulea are more closely related to each other than either is to P. cyanea. This is the sister relationship found in the mitochondrial tree.
MATERIALS AND METHODS
Sampling, amplification, and sequencing:
From museum collections we obtained vouchered frozen tissue samples from two individuals of each focal species, P. cyanea, P. amoena, and P. caerulea (see appendix for sampling localities). We also included one individual from two additional species: P. rositae and Cyanocompsa cyanoides. C. cyanoides is a member of a genus closely related to Passerina (Klicka et al. 2001) and was included here as an outgroup. We extracted total DNA from ∼25 mg of pectoral muscle using a DNeasy tissue kit (QIAGEN, Valencia, CA), following the manufacturer's recommended protocol. Each individual was amplified at 10 nuclear loci (Table 1) using the following PCR conditions in a 25 μl amplification reaction: ∼50 ng template DNA (2.5 μl of QIAGEN DNA extracts), 1 μl of 10 mm dNTPs (2.5 mm each dATP, dTTP, dCTP, and dGTP), 1 μl of each primer (10 mm, Table 1), 2.5 μl 10× buffer with MgCl2 (15 mm), 0.1 μl Taq (5 units/μl AmpliTaq DNA polymerase, Applied Biosystems, Foster City, CA), and 16.9 μl sterile dH2O. The thermocycling profile was as follows: an initial 95° denaturation for 2 min followed by 35 cycles consisting of a 30-sec, 95°-denaturation step, a 30-sec, locus-specific temperature primer annealing step (Table 1), and a 2-min, 72°-extension step, and a final extension of 5 min at 72°. We checked for amplification by electrophoresing 2.5 μl of each PCR amplicon on a 1% agarose gel and then used 20% polyethylene glycol (PEG) to purify the remaining 22.5 μl of the PCR product from successful amplifications.
TABLE 1.
PCR conditions, sequence length, primer sequences, and EMBL/GenBank accession numbers for 10 nuclear loci used in all analyses
| Locus | Annealing temp | Length (bp)a | PCR/sequencing primersb | Internal sequencing primersc | Accession nos. |
|---|---|---|---|---|---|
| AETC | 60° | 299 | For: TGGACTTCAAATCCCCCGATGATCCCAGC | NA | EU191758–765 |
| Rev: CCAGGCACCCCAGTCTACCTGGTCAAA | |||||
| βact3 | 62° | 423 (255) | For: GGCAATGAGAGGTTCAGGT | NA | EU191766–773 |
| Rev: TGGTACCACCAGACAGCAC | |||||
| GADPH | 62° | 303 | For: ACCTTTCATGCGGGTGCTGGCATTGC | NA | EU191774–781 |
| Rev: CATCAAGTCCACAACACGGTTGCTGTA | |||||
| MC1R | 62.5° | 774 | For: CTGGCTCCGGAAGGCRTAGAT | For: SGCRTAGAAGATGGTGATGTAGC | EU191782–789 |
| Rev: AYGCCAGYGAGGGCAACCA | Rev: GTBGACCGCTACATCACCAT | ||||
| MPLPR | 63.5° | 293 (144) | For: TACATCTACTTTAACACCTGGACCACCTG | NA | EU191790–797 |
| Rev: TTGCAGATGGAGAGCAGGTTGGAGCC | |||||
| Myo2 | 58° | 684 | For: GCCACCAAGCACAAGATCCC | NA | EU191798–805 |
| Rev: GCAAGGACCTTGATAATGACTT | |||||
| ODC | 65° | 609 | For: GACTCCAAAGCAGTTTGTCGTCTCAGTGT | NA | EU191806–813 |
| Rev: TCTTCAGAGCCAGGGAAGCCACCACCAAT | |||||
| Rho1 | 60° | 855 | For: TGCTACATCGAGGGCTTCTT | For: GTGCCTAAAAATGTCCTCT | EU191814–821 |
| Rev: GCAGTGACCAGAGAGCGATT | Rev: GCTGGCATGGGGAGGAGAG | ||||
| TGFβ2 | 62° | 544 (283) | For: GAAGCGTGCTCTAGATGCTG | NA | EU191822–829 |
| Rev: AGGCAGCAATTATCCTGCAC | |||||
| Tropo6 | 60° | 448 | For: AATGGCTGCAGAGGATAA | NA | EU191830–837 |
| Rev: TCCTCTTCAAGCTCAGCACA |
Sequence lengths inside parentheses indicate length of longest independently segregating block.
PCR and external sequence primer references: AETC, Primmer et al. (2002); βact3, B. Marks (personal communication); GADPH, Primmer et al. (2002); MC1R, Cheviron et al. (2006); MPLPR, Primmer et al. (2002); Myo2, Heslewood et al. (1998) (For) and Slade et al. (1993) (Rev); ODC, Primmer et al. (2002); Rho1, Primmer et al. (2002); TGFβ2, Primmer et al. (2002); Tropo6, Primmer et al. (2002).
Internal sequencing primer references: MC1R, Cheviron et al. (2006); Rho1, this study.
Both strands of the PEG-purified PCR amplicons were cycle-sequenced using 1.5 μl of 5× sequencing buffer (Applied Biosystems), 1 μl of 10 mm primer, 2–3 μl template (depending on the intensity of the PCR bands on the agarose gels), 0.5 μl Big Dye Terminator cycle-sequencing kit v 3.1 (Applied Biosystems), and 1–2 μl sterile dH2O (depending on amount of template added), for a total volume of 7 μl. The longer loci were also cycle-sequenced with internal sequencing primers using the same protocol (Table 1). We cleaned cycle-sequencing products on Sephadex (G-50 fine) columns and electrophoresed the cleaned products on a 3100 Genetic Analyzer (Applied Biosystems). In those cases where direct sequencing of purified PCR amplicons revealed more than one heterozygous site within a sequence, we phased the haplotypes by cloning the PCR products using the TA Topo cloning kit according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) and sequencing five clones. All sequences were edited and assembled using Sequencher v 4.1 and 4.6 (GeneCodes, Ann Arbor, MI).
Phylogenetic analyses:
Sequence alignments of all 10 nuclear loci were unambiguous and checked visually. In all phylogenetic analyses, gaps were alternately classified as a fifth state or as missing data. Phylogenetic analyses were conducted on a concatenated data set since partition homogeneity tests revealed no significant conflicts between the data from each of the individual loci. Tests for recombination (see Population genetic analyses below) indicated that three loci (βact3, MPLPR, and TGFβ2) had undergone recombination. Therefore, phylogenetic analyses were conducted on both the entire concatenated data set (5232 bp) and on a reduced concatenated data set containing only the largest independently segregating block of sequence for βact3, MPLPR, and TGFβ2 (4668 bp). Using PAUP* v 4.0b10 (Swofford 1998) we estimated a neighbor-joining tree from the p-distance matrix and calculated likelihood scores on a series of nested substitution models. These were then evaluated using ModelTest v 3.8 (Posada and Crandall 1998). Due to its advantages over hierarchical likelihood-ratio tests (Posada and Buckley 2004), we used the Bayesian Information Criterion (BIC) to determine the best-fit model of sequence evolution for the concatenated data set as well as for each locus independently (Table 2).
TABLE 2.
Summary of model parameters used in all phylogenetic analyses as determined by the Bayesian information criterion (BIC) as implemented in ModelTest 3.8 (Posada and Crandall 1998)
| Data set | Model | Base frequenciesa | ti/tv ratio | Rate matrixb | Pinv |
|---|---|---|---|---|---|
| AETC | K80 | Equal | 5.8 × 1036 | — | — |
| βact3 | K80 | Equal | 6.1 × 1036 | — | — |
| GADPH | JC | Equal | — | — | — |
| MC1R | F81 | 0.1533, 0.3795, 0.2348, 0.2304 | — | — | — |
| MPLPR | F81 | 0.2097, 0.2600, 0.3476, 0.1827 | — | — | — |
| Myo2 | JC | Equal | — | — | — |
| ODC | F81 | 0.2790, 0.1677, 0.2127, 0.3406 | — | — | — |
| Rho1 | JC | Equal | — | — | — |
| TGFβ2 | JC | Equal | — | — | — |
| Tropo6 | F81 | 0.1959, 0.2227, 0.1714, 0.4057 | — | — | — |
| 8-terminal-taxac | K81uf+I | 0.2231, 0.2429, 0.2462, 0.2876 | — | 1.0, 4.424, 1.8284, 1.8284, 4,424, 1.0 | 0.9295 |
| 16-terminal-taxac | K81uf+I | 0.2364, 0.2287, 0.2616, 0.2633 | — | 1.0, 7.150, 2.243, 2.243, 7.150, 1.0 | 0.9307 |
Order of base frequencies is A, C, G, T.
Order of rate matrix is A to C, A to G, A to T, C to G, C to T, and G to T.
These data sets contain the concatenated sequences from all 10 loci.
To investigate species-level relationships between these taxa we conducted a variety of phylogenetic analyses on the concatenated data set using the model of sequence evolution identified by ModelTest v 3.8. First, we analyzed an 8-terminal-taxa data set composed of sequence data from either one (P. rositae and C. cyanoides) or two individual(s) per species (P. cyanea, P. amoena, and P. caerulea) where double peaks in chromatograms were inferred to be heterozygous sites and coded as polymorphisms using the standard IUPAC degeneracy codes. Second, we analyzed a 16-terminal-taxa data set that consisted of two phased haplotypes, determined by sequencing cloned PCR products from each individual for a total of four sequences per species of P. cyanea, P. amoena, and P. caerulea and two sequences per species for P. rositae and C. cyanoides. These data sets of concatenated sequences will be referred to hereafter as the 8-terminal-tip data set and the 16-terminal-tip data set.
Using parameters estimated on a neighbor-joining tree (Saitou and Nei 1987), we used PAUP* v 4.0b10 (Swofford 1998) to perform heuristic searches [tree bisection–reconnection (TBR) branch swapping; 10 random-addition replicates] under the maximum-likelihood (ML) optimality criterion for both the 8- and 16-terminal-tip concatenated data sets. We assessed nodal support using 100 ML heuristic bootstrap replicates with the search parameters described above. We also performed a variety of constrained ML searches (Table 3) on the 8- and 16-terminal-tip data sets using the sequence evolution model and search parameters, except without bootstrap replicates, as described above. Using a Shimodaira–Hasegawa test (SH) (Shimodaira and Hasegawa 1999) with full optimization and 1000 bootstrap replicates, we assessed whether the likelihoods of the unconstrained and constrained trees were significantly different.
TABLE 3.
Likelihood scores for various topological constraints
| Constrainta
|
|||
|---|---|---|---|
| Data set | (((Pam, Pcy), Pca), Pro, Ccy)b | (((Pam, Pca), Pcy), Pro, Ccy) | (((Pcy, Pca), Pam), Pro, Ccy) |
| 8-terminal-tip concatenatedc | −8148.9118 | −8151.7215 | −8155.0551 |
| 8-terminal-tip concatenatedd | −7239.1485 | −7240.0937 | −7243.9608 |
| 16-terminal-tip concatenatedc | −9140.8436 | −9142.4041 | −9147.5444 |
Likelihood scores (ln L) were estimated using a heuristic search (TBR branch swapping, 10 random addition replicates). Underlining indicates topology with greatest likelihood. The constraint trees are also the species toplogies used in the minimize-deep-coalescences (Maddison and Knowles 2006) and COAL analyses (Degnan and Salter 2005).
Species name abbreviations: Pam, P. amoena; Pcy, P. cyanea; Pca, P. caerulea; Pro, P. rositae; Ccy, Cyanocompsa cyanoides.
This was also the topology recovered in all the unconstrained searches, which also gave identical ln L scores.
These data sets included all sequence data from all loci.
This data set included only the largest independently segregating block of sequence for those loci showing evidence of recombination.
We also assessed nodal support for the 8- and 16-terminal-tip topologies using posterior probabilities generated using MrBayes v 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). We used the same general model (GTR + I) as in the ML searches and uninformative priors and ran two replicate analyses of four Markov chains for 1,000,000 generations. Additionally, we ran a mixed-model analysis using MrBayes in which the 8-terminal-tip concatenated data set was partitioned into the 10 individual loci, each partition with the appropriate model of sequence evolution (Table 2). In both the one-model and mixed-model searches, we sampled trees every 100 generations and calculated final posterior probabilities after excluding the first 250,000 generations as burn-in. Plots of likelihood scores during the run were examined visually to insure stationarity had been reached. In all searches, the average standard deviation of split frequencies was <0.005, further evidence stationarity had been reached.
Minimize-deep-coalescences analyses:
Because well-supported species trees inferred from concatenated data sets can be discordant with the true species tree, especially if the species divergence was recent (Degnan and Rosenberg 2006; Edwards et al. 2007; Kubatko and Degnan 2007), we inferred a species tree using a method that considers the individual gene histories. Specifically, we assessed the fit of the gene trees onto three possible rooted species trees for P. cyanea, P. amoena, and P. caerulea, using the minimize-deep-coalescences method as implemented in the program Mesquite (Maddison 1997; Maddison and Knowles 2006). This method attempts to reconcile gene trees contained within species trees by minimizing the number of deep coalescences, those cases where the coalescence of two gene copies predates the speciation event. Using a parsimony optimality criterion, the “best” species tree is the one that requires the fewest deep-coalescent events in contained gene genealogies. The number of deep-coalescent events, or costs, can be summed across multiple markers to accommodate multilocus data sets. Using Mesquite (Maddison and Maddison 2004), we compared the fit of the genealogies from each locus into the possible species trees using methods slightly modified from the recently described method of Maddison and Knowles (2006).
We used MrBayes v 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) to sample the tree space of the gene trees for each locus independently, using completely phased haplotypes and the same model parameters with uninformative priors as in the ML tree searches and the same MrBayes run parameters as described above. After discarding the first 250,000 generations as burn-in, we used PAUP* to calculate a majority-rule consensus gene tree from each locus-specific search. We then overlaid each unrooted locus-specific gene tree onto the three possible species trees (Table 3) and tallied the number of deep- coalescent events required to reconcile the gene trees with the species trees. The total cost of each species tree was determined by summing the locus-specific tallies (Table 4), with the species tree having the lowest cost being inferred as the correct species tree.
TABLE 4.
Deep coalescent events required to reconcile unrooted locus-specific gene trees, inferred using Bayesian tree searches, with three possible species topologies
| Species topologiesa
|
|||
|---|---|---|---|
| Locus | (((Pam, Pcy), Pca), Pro, Ccy) | (((Pam, Pca), Pcy), Pro, Ccy) | (((Pcy, Pca), Pam), Pro, Ccy) |
| AETC | 5 | 7 | 7 |
| βact3 | 6 | 6 | 6 |
| GADPH | 3 | 3 | 3 |
| MC1R | 1 | 0 | 1 |
| MPLPR | 12 | 11 | 10 |
| Myo2 | 3 | 3 | 3 |
| ODC | 7 | 7 | 6 |
| Rho1 | 0 | 1 | 1 |
| TGFβ2 | 5 | 7 | 7 |
| Tropo | 0 | 1 | 1 |
| Total Cost | 42 | 46 | 45 |
Underlining indicates species topology with fewest deep coalescent events for each locus as well as for the lowest total summed over all locus-specific gene trees.
Species name abbreviations: Pam, P. amoena; Pcy, P. cyanea; Pca, P. caerulea; Pro, P. rositae; Ccy, Cyanocompsa cyanoides.
Calculating probabilities of gene trees given possible species trees:
Recent software developments give researchers increased ability to evaluate the variation in lineage sorting among loci and how it influences phylogenetic inferences. The program COAL (Degnan and Salter 2005), which extends earlier work (Takahata and Nei 1985; Rosenberg 2002), can calculate the probability of a gene tree given a species tree for any number of taxa. This represents a significant step toward developing a method capable of estimating the likelihood of sequence data given a species tree, which can be estimated as follows:
 |
(Maddison 1997). The first probability statement is analogous to the log-likelihood scores generated by phylogenetic inference software packages such as PAUP* (Swofford 1998). The program COAL can calculate the second probability statement. Unfortunately, complete evaluation of the above equation is hampered because the summation step requires summing over all possible gene trees, which includes both topology and branch lengths (Maddison 1997). Currently, no method is capable of performing that summation because of the near infinite number of possible gene trees. It will likely require a method that employs some type of importance sampling of the gene tree space to effectively evaluate that step of the overall equation (J. Degnan, personal communication).
In light of the current methodological limitations, we focused on calculating the conditional probability of a gene tree given a species tree. Locus-specific gene trees were inferred in three ways. First, we randomly chose a single individual to represent P. amoena, P. cyanea, and P. caerulea, which along with one individual from each outgroup taxon (P. rositae and C. cyanoides), resulted in a data set with five terminal tips. For each locus, we estimated a gene tree under the maximum-likelihood search criterion as described above and then used COAL to calculate the probability of each ML gene tree given three possible species trees (Table 3) using COAL. ML inferences for Tropo6 produced >100 equally likely gene trees and so we eliminated Tropo6 from the COAL analysis.
We also inferred locus-specific gene trees from the 8-terminal-tip data set. Since COAL requires fully resolved bifurcating gene trees and since locus-specific ML searches of this 8-terminal-tip data set produced numerous equally likely topologies with unresolved polytomies, we inferred locus-specific neighbor-joining gene trees, using uncorrected p-distances. In the Tropo6 data set, some of the sequences were identical across individuals. Accordingly, neighbor-joining analyses failed to produce a fully resolved tree for Tropo6 and we excluded it from COAL calculations.
In an attempt to incorporate a larger sample of the possible gene topologies and branch lengths, we sampled a collection of gene trees from the locus-specific Bayesian tree searches described above (see Minimize-deep-coalescences analyses). Our approach was to calculate the probability of each of the three possible species trees (Table 3) given each of the last 1000 gene trees from the Bayesian searches, using COAL, and average the probabilities of those gene trees to generate a more robust estimate of the likelihoods of the three species trees. Unfortunately, in each locus-specific Bayesian tree search some gene trees contained unresolved polytomies, which COAL cannot handle; therefore, we abandoned this method of inferring gene trees.
We replicated the COAL calculations, for both the five- and eight-terminal tip data sets, using three different internal:terminal branch-length ratios (1:1, 1:2, and 1:10). All internal:terminal branch-length ratios, with the exception of 1:1, correspond to topologies with internode lengths that are short, relative to the terminal branches, which appears to be the general pattern observed in Passerina buntings. To assess the difference in probabilities of the gene trees if the species tree united P. cyanea and P. amoena as sisters (hypothesis 1) with one in which P. caerulea and P. amoena were sisters (hypothesis 2), we conducted a test of proportions (Sokal and Rohlf 1995) whereby we tallied the number of gene trees that were more probable under hypothesis 1 and compared that with a tally of the number of gene trees that were more probable under hypothesis 2.
Population genetic analyses:
Both phylogenetic methods described above assume that discordance between gene trees and the species tree is due to genetic drift (i.e., lineage sorting stochasticity). Because of the well-documented hybrid zone between P. amoena and P. cyanea, it would be ideal if a gene flow parameter could be incorporated into the phylogenetic analysis. This is not currently possible. We thus examined the utility of divergence population genetics for inferring species relationships indirectly. The Isolation with Migration (IM) software package (Hey and Nielsen 2004; Hey 2005) can estimate pairwise divergence times between P. cyanea, P. amoena, and P. caerulea, while jointly estimating pairwise migration rates. A sister relationship is inferred to be between the species with the most recent divergence estimate.
IM combines coalescent theory with Bayesian methodologies to simultaneously estimate multiple population genetic parameters for two diverging populations. Briefly, using a Markov chain Monte Carlo (MCMC) approach, IM generates posterior probability distributions of six parameters (θpopulation1, θpopulation2, θancestral, m1, m2, t). These parameters can then be used to calculate population demographic estimates, i.e., effective population sizes of the two populations (Ne1 and Ne2), the ancestral population size (i.e., the population size prior to initial divergence = NeA), migration rates between the two populations (m1 and m2, which allows for asymmetrical migration rates), and the time since divergence of the two populations (t). A more recent version of IM (Hey 2005) also allows for the inclusion of a population splitting parameter, s, which allows for exponential population growth or decline in each descendant population.
IM assumes selective neutrality of each marker and no intralocus recombination. We tested for neutrality by calculating Tajima's D (Tajima 1983) for each locus using Arlequin v 3.0 (Excoffier et al. 2005). None of the 10 loci showed a significant departure from neutrality (each P > 0.25). To investigate intralocus recombination, we used TOPALi v 2 (McGuire and Wright 1998, 2000), which uses a sliding-window approach to search for putative recombination breakpoints along an alignment of DNA sequences. Using the difference in sum-of-squares (DSS) method, with a 100-bp window and a 10-bp increment, we looked for signals of recombination, manifested by a significant DSS peak along the sequence alignment. The statistical significance of DSS peaks was assessed using 100 parametric bootstraps. Significant DSS peaks were detected in 3 loci, βact3, MPLPR, and TGFβ2 (Table 1). For each of those 3 loci, we only included the longest independently segregating block of sequence from the phased haplotype data in the IM runs along with the total available phased haplotype sequences from each of the other seven markers.
Using IM, we estimated three pairwise divergence times, one between P. cyanea and P. amoena, one between P. caerulea and P. amoena, and one between P. cyanea and P. caerulea. Initial IM runs, conducted using the infinite sites (IS) model (Kimura 1969) and employing unrealistically large maximum priors (t = 20, q1 = q2 = qa = 500, m1 = m2 = 50, or m1 = m2 = 0 for parameter estimation without gene flow) were used to establish appropriate parameter priors for all subsequent analyses (Won and Hey 2005). Once appropriate priors were identified, we ran IM three times with identical starting conditions, with the exception of the random number seed, to assess convergence. All runs began with a burn-in period of 100,000 steps and were allowed to continue until the minimum effective sample size (ESS) was >100 (Hey 2005). Because runs with the same starting conditions produced qualitatively similar results, we only present data from the longest run (>3 × 109 steps). For each pairwise estimate of divergence time, we performed a variety of IM analyses with varying numbers of parameters. The simplest model included four parameters (θ1, θ2, θA, and t); other models included five (θ1, θ2, θA, t, and m—equal migration between populations or θ1, θ2, θA, t, and s—allowing for population growth) or six parameters (θ1, θ2, θA, t, m1, and m2—allowing for different, directional migration between populations). All model parameter estimates in IM are scaled to the mutation rate, μ. To translate scaled divergence time estimates to real time, we included mutation rates for each locus calculated as the sequence length multiplied by 3.6 × 10−9 substitutions/site/year (Axelsson et al. 2004).
RESULTS
Phylogenetic analyses:
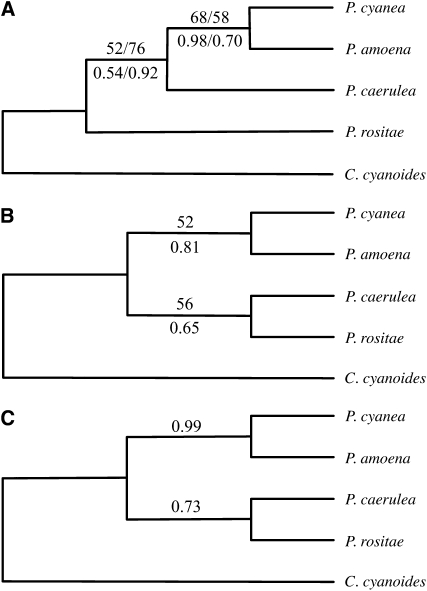
Maximum-likelihood analysis of the entire concatenated 8-terminal-tip data set, based on 5232 bp of sequence data from each individual, produced one strongly supported tree (Figure 3A). In it, the traditional sister relationship between P. amoena and P. cyanea (hypothesis 1) was recovered. This unconstrained tree was more likely than either of the constrained topologies, regardless of the data set, although the differences in likelihood scores were not significant (SH test, P > 0.05, Table 3). Support values from ML bootstrap analyses and Bayesian posterior probabilities, calculated under a single model of sequence evolution for the entire data set, indicated the topology recovered in the unconstrained searches was relatively well supported (Figure 3). The Bayesian analysis of the 8-terminal-tip data set using a mixed model, in which a sequence evolution model was specified for each locus, uncovered the same topology as the Bayesian analysis of the 16-terminal-tip data set (Figure 3, B and C). All of the above phylogenetic analyses were rooted using C. cyanoides as an outgroup. Replicate analyses with the reduced concatenated data set (4668 bp) produced the same topologies as the entire concatenated data set (Figure 3A).
Figure 3.—
Species trees inferred from the entire concatenated data set (5332 bp), except where noted. In all phylogenies, multiple sequences from each species were collapsed into a single “representative.” Only bootstrap support values >50 (at the top of the branches) and Bayesian posterior probabilities >0.50 (at the bottom of the branches) are illustrated. (A) Phylogeny inferred from the 8-terminal-tip data set. For both bootstrap support values and Bayesian posterior probabilities the first number refers to the concatenated data set and the second to a reduced concatenated data set containing only the largest independently segregating block of sequence from those loci for which recombination was detected. (B) ML and Bayesian species tree inferred from the 16-terminal-tip data set with bootstrap support values at the top of the branches and posterior probabilities at the bottom. (C) Posterior probabilities of Bayesian phylogeny inferred from the 8-terminal-tip data set with a mixed model of sequence evolution.
Phylogenies inferred from the 16-terminal-tip data set were also similar to the ML and Bayesian phylogenies inferred from the 8-terminal-tip data set (Figure 3). Both, ML and Bayesian, 16-terminal-tip phylogenies contained a well-supported sister clade between P. cyanea and P. amoena. The placement of P. caerulea with respect to P. rositae and the P. cyanea/P. amoena clade varied depending on the data set, 8- or 16-terminal tip, used to infer the phylogeny. Both ML and Bayesian inferences uncovered a weakly supported clade containing P. cyanea, P. amoena, and P. caerulea, when tree searches were conducted on the 8-terminal-tip data set. When based on the 16-terminal-tip data set, both analyses recovered a sister relationship between P. rositae and P. caerulea, and that clade was sister to the P. cyanea/P. amoena clade (Figure 3C).
The topologies inferred from the tree searches of the concatenated data sets did not differ when gaps were coded as missing data or as a fifth state (topologies not shown).
Minimize-deep-coalescences analyses:
Overlaying the majority-rule consensus gene trees from the Bayesian tree searches on the three possible species trees resulted in three different deep coalescent costs for each gene—one cost for each species tree (Table 4). For four locus-specific gene trees (AETC, Rho1, TGFβ2, and Tropo6) the fewest number of deep coalescent events were observed when the species topology united P. amoena and P. cyanea as sister species. The fewest deep coalescent events for one locus (MC1R) occurred when P. amoena and P. caerulea were sister species, and the P. cyanea/P. caerulea sister species tree contained the fewest deep coalescences when overlaid with the ODC and MPLPR gene trees.
Summing the number of deep coalescent events needed to fit the gene trees into the species tree across loci produced a total cost of 42 for the P. amoena/P. cyanea sister species tree (Table 4). Fitting the gene trees into the other two species trees required more deep coalescences: 46 for the P. amoena/P. caerulea tree and 45 for the P. cyanea/P. caerulea tree.
Calculating probabilities of gene trees given possible species trees:
After randomly choosing one individual to represent P. amoena, P. cyanea, and P. caerulea, and eliminating Tropo6 from further analyses, 14 ML gene topologies were inferred from phylogenetic tree searches conducted on each locus separately (Table 5). Multiple gene trees with equal likelihoods were inferred from three loci (AETC—three gene trees, βact3—two, and MPLPR—three). Replicate COAL calculations that differed only in the ratio of internal to terminal branch lengths (1:1, 1:2, and 1:10) all gave qualitatively similar results. When terminal branch lengths were longer than internal branch lengths (1:2 and 1:10), 3 of the 14 gene topologies were equally probable whether the given species tree had P. cyanea and P. amoena as sister species or a sister species relationship between P. amoena and P. cyanea (Table 5). Those 3 gene topologies (one of two βact3 gene trees and two of three MPLPR gene trees) were excluded from further analyses, because with respect to these relationships, these topologies are uninformative. Of the remaining 9 ML gene topologies (one of the three equally likely AETC gene trees was chosen at random), 8 were more probable given a species tree uniting P. cyanea and P. amoena as sisters, and 1 was more probable given a P. amoena–P. caerulea species tree (Table 5, test of proportions, nobs = 9, P = 0.05). When the internal:terminal branch length ratio was 1:1, 4 of the 14 gene topologies were equally probable given either species tree and were subsequently eliminated. Seven of the remaining 8 (1 AETC gene tree chosen at random) ML gene topologies were more probable with P. cyanea/P. amoena as sister species (Table 4, test of proportions, nobs = 8, P = 0.1).
TABLE 5.
Probabilities of ML gene trees given two alternative species topologies with differing internal:terminal branch length ratios (1:1, 1:2, and 1:10)
| Probability
|
|||||
|---|---|---|---|---|---|
| Locusa | ML gene treeb | Species topologyb | 1:1 | 1:2 | 1:10 |
| AETC | ((Pam, Pcy), ((Ccy, Pca), Pro)) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00082 | 0.00495 | 0.01511 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00018 | 0.00199 | 0.01157 | ||
| ((Pam, Pcy), ((Pro, Ccy), Pca)) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00082 | 0.00495 | 0.01511 | |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00018 | 0.00199 | 0.01157 | ||
| ((Pam, Pcy), (Ccy, (Pro, Pca))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.01663 | 0.03091 | 0.02587 | |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00366 | 0.01270 | 0.01997 | ||
| βact3 | ((((Pam, Ccy), Pcy), Pro), Pca) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00000 | 0.00487 | 0.01103 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00000 | 0.00191 | 0.00748 | ||
| (((Pam, Ccy), (Pcy, Pro)), Pca) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00002 | 0.00051 | 0.00647 | |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00002 | 0.00051 | 0.00647 | ||
| GADPH | ((((Pam, Pcy), Ccy), Pca), Pro) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00082 | 0.00487 | 0.01103 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00018 | 0.00191 | 0.00748 | ||
| MC1R | (((Pam, Pca), Pcy), (Ccy, Pro)) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.01075 | 0.02075 | 0.02104 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.07336 | 0.07322 | 0.03321 | ||
| MPLPR | ((Pam, ((Pro, Pca), Pcy)), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00059 | 0.00259 | 0.00532 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00059 | 0.00259 | 0.00532 | ||
| (((Pam, (Pro, Pca)), Pcy), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00059 | 0.00259 | 0.00532 | |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00059 | 0.00259 | 0.00532 | ||
| (((Pam, Pcy), (Pro, Pca)), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.06330 | 0.05614 | 0.02232 | |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.01366 | 0.02188 | 0.01605 | ||
| Myo2 | (((Pam, Pcy), (Pro, Pca)), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.06330 | 0.05614 | 0.02232 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.01366 | 0.02188 | 0.01605 | ||
| ODC | ((((Pam, Pcy), Pca), Pro), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.40808 | 0.17006 | 0.02217 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.05686 | 0.04358 | 0.01230 | ||
| Rho1 | ((((Pam, Pcy), Pca), Pro), Ccy) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.40808 | 0.17006 | 0.02217 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.05686 | 0.04358 | 0.01230 | ||
| TGFβ2 | ((((Pam, Pcy), Ccy), Pca), Pro) | ((((Pcy, Pam), Pca), Pro), Ccy) | 0.00082 | 0.00487 | 0.01103 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 0.00018 | 0.00191 | 0.00748 | ||
Underlining indicates probabilities that are greater with a P. amoena/P. cyanea species topology than with a P. amoena/P. caerulea species topology. Italic numbers indicate the opposite pattern. All probabilities calculated using COAL (Degnan and Salter 2005).
Gene tree searches based on Tropo6 sequence data from one randomly chosen individual from each species resulted in >100 ML gene trees and so that locus was dropped from the COAL analysis.
Species names abbreviations: Pam, P. amoena; Pcy, P. cyanea; Pca, P. caerulea; Pro, P. rositae; Ccy, C. cyanoides.
Nine gene topologies, one for each locus with the exception of Tropo6, were inferred from the eight-terminal tip data set using neighbor-joining analyses (Table 6). In the COAL calculations with internal:terminal branch lengths of 1:1 and 1:2, there was no difference in the number of locus-specific gene topologies that were more probable with P. cyanea and P. amoena as sisters in the species tree and the number of gene topologies that were more probable with a P. amoena/P. caerulea sister relationship (Table 6, test of proportions, nobs = 9, P > 0.05). When the internal:terminal branch length ratio of the species trees was 1:10, all nine gene topologies were more probable when the species tree placed P. cyanea and P. amoena as sisters (Table 6, test of proportions, nobs = 9, P = 0.02).
TABLE 6.
Probabilities of neighbor-joining gene topologies, inferred from the eight terminal tip data set, given two alternative species trees with differing internal:terminal branch length ratios (1:1, 1:2, and 1:10)
| Probability
|
|||||
|---|---|---|---|---|---|
| Locusa | NJ treeb | Species topologyb | 1:1 | 1:2 | 1:10 |
| AETC | (Pcy1, (((Pam1, Pcy2), Pam2), ((Ccy, Pro), (Pca1, Pca2)))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 2.365E-09 | 4.78789E-07 | 1.8218E-05 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 1.62E-10 | 4.6316E-08 | 1.82953E-06 | ||
| βact3 | (Pcy1, (((Pam1, Pcy2), Ccy), ((Pro, (Pca1, Pca2)), Pam2))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 5.05E-10 | 2.77914E-07 | 2.1178E-05 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 3.99E-10 | 1.07017E-07 | 2.62828E-06 | ||
| GADPH | (Pcy1, (((Pam1, Pam2), (Pro, (Pca1, Pca2))), (Ccy, Pcy2))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 3.143E-09 | 1.8008E-06 | 0.00018838 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 2.7306E-08 | 2.72713E-06 | 3.75984E-05 | ||
| MC1R | (Pcy1, ((((Pam1, Pam2), (Pca1, Pca2)), (Ccy, Pro)), Pcy2)) | ((((Pcy, Pam), Pca), Pro), Ccy) | 2.124E-09 | 1.30936E-06 | 0.000149501 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 1.02979E-07 | 5.10169E-06 | 3.50167E-05 | ||
| MPLPR | (Pcy1, ((((Pam1, Pam2), Pcy2), Ccy), ((Pro, Pca1), Pca2))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 5.136E-09 | 9.52899E-07 | 2.77842E-05 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 3.98E-10 | 5.10169E-06 | 1.74861E-05 | ||
| Myo2 | (Pcy1, ((((Pam1, Pam2), Pcy2), Ccy), (Pro, (Pca1, Pca2)))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 1.53135E-06 | 6.8394E-05 | 0.000690821 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 1.28294E-06 | 2.86844E-05 | 9.5464E-05 | ||
| ODC | (Pcy1, ((((Pam1, Pcy2), (Ccy, Pro)), (Pca1, Pca2)), Pam2)) | ((((Pcy, Pam), Pca), Pro), Ccy) | <1.0E-12 | 1.393E-09 | 2.7787E-06 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | <1.0E-12 | 5.27E-10 | 3.8284E-07 | ||
| Rho1 | (Pcy1, ((((Pam1, Pam2), (Pca1, Pca2)), (Ccy, Pro)), Pcy1)) | ((((Pcy, Pam), Pca), Pro), Ccy) | 2.124E-09 | 1.30936E-06 | 0.000149501 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 1.02979E-07 | 5.10169E-06 | 3.50167E-05 | ||
| TGFβ2 | (Pcy1, ((Pam1, (Ccy, (Pcy2, Pam2))), (Pro, (Pca1, Pca2)))) | ((((Pcy, Pam), Pca), Pro), Ccy) | 1.4E-11 | 3.7386E-08 | 1.2647E-05 |
| ((((Pam, Pca), Pcy), Pro), Ccy) | 1.1E-11 | 6.71486E-07 | 1.4255E-08 | ||
Underlining indicates probabilities that are greater with a P. amoena/P. cyanea species topology than with a P. amoena/P. caerulea species topology. Italic numbers indicate the opposite pattern. All probabilities calculated using COAL (Degnan and Salter 2005).
NJ gene tree search based on Tropo6 sequence data produced an unresolved topology and was dropped from COAL analyses.
Species names abbreviations: Pam, P. amoena; Pcy, P. cyanea; Pca, P. caerulea; Pro, P. rositae; Ccy, C. cyanoides. Numbers following species abbreviations in NJ topologies refer to the different individuals sampled. All species were constrained to be monophyletic in the species topologies.
Population genetic analyses:
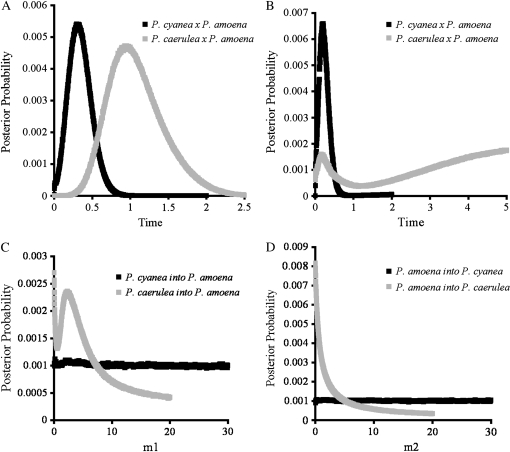
After excluding all gaps in the sequence data and retaining only the largest independently segregating block of sequence for those loci that showed evidence of recombination, 4721 bp of sequence data were included in the IM analyses. When divergence times were estimated in the absence of migration, analyses uncovered a more recent splitting between P. cyanea and P. amoena than between either of those two species and P. caerulea (Figure 4A, Table 7). Estimates for θ1, θ2 and θA were inconclusive; the distributions of the posterior probabilities were nearly flat in all comparisons (data not shown). Allowing for growth or decline in the descendant populations did not qualitatively alter the divergence time estimates for any of the pairwise comparisons; the most recent divergence was between P. amoena and P. cyanea, with divergence times later for both the P. caerulea/P. amoena and P. caerulea/P. cyanea comparisons (Table 7).
Figure 4.—
The posterior probability distributions of model parameters (t, m1, and m2) estimated using IM for pairwise comparisons P. cyanea × P. amoena and P. caerulea × P. amoena. (A) Pairwise divergence time estimates with no postdivergence gene flow. (B) Pairwise divergence time estimates allowing for postdivergence gene flow. (C and D) Migration-rate estimates. All parameter estimates are scaled to the neutral mutation rate (μ).
TABLE 7.
Maximum-likelihood estimates (MLE) and 90% highest posterior density (HPD) intervals of divergence time estimates for pairwise comparisons between P. amoena, P. cyanea, and P. caerulea
| Constant populationb
|
Variable populationc
|
With migration (initial peak)d
|
With migration (high point)d
|
|||||
|---|---|---|---|---|---|---|---|---|
| Comparison | t | t (yr)e | t | t (yr)e | t | t (yr)e | t | t (yr)e |
| P. cyanea × P. amoena | ||||||||
| MLEa | 0.2105 | 138,000 | 0.207 | 136,000 | 0.211 | 138,000 | 0.211 | 138,000 |
| 90% HPDa | 0.0375–0.4345 | 25,000–286,000 | 0.037–0.443 | 24,000–291,000 | 0.033–0.445 | 21,700–292,000 | 0.033–0.445 | 21,700–292,000 |
| P. caerulea × P. amoena | ||||||||
| MLEa | 0.7185 | 486,000 | 0.4825 | 327,000 | 0.1825 | 103,000 | 4.9975 | 3,380,000 |
| 90% HPDa | 0.3555–1.3125 | 241,000–888,000 | 0.2475–1.5325 | 167,000–1,037,000 | NAf | NAf | NAf | NAf |
| P. caerulea × P. cyanea | ||||||||
| MLEa | 0.537 | 363,000 | 0.4935 | 334,000 | 0.0735 | 49,700 | 4.9975 | 3,380,000 |
| 90% HPDa | 0.177–1.091 | 120,000–738,000 | 0.1505–1.3265 | 102,000–898,000 | NAf | NAf | NAf | NAf |
MLEs are the locations of the peaks of the posterior probabilities in Figure 4; HPDs are the values along the x-axes in Figure 4 that contain 90% of the area of the histograms.
Divergence time estimates for IM runs under the constant population size model.
Divergence time estimates for IM runs under the variable population-size model, which includes the population splitting parameter s.
Divergence time estimates for IM runs under constant population-size model while allowing for migration. In all comparisons (except P. cyanea × P. amoena) there was an initial peak and a nonconverging, nonzero tail (high point). See Figure 4.
Mutation scaled estimates of divergence time (t) were translated into years using a mutation rate of 3.6 × 10−9 substitutions/site/year (Axelsson et al. 2004).
These runs failed to converge so 90% HPDs are not presented.
When divergence time was estimated jointly with migration, the high points of the posterior probability distributions suggested a different pattern. The divergence time pattern was the same regardless of whether the direction of gene flow was constrained to be symmetrical (five-parameter model) or asymmetrical (six-parameter model); thus, only the divergence times estimated under the six-parameter model are discussed. In the P. cyanea/P. amoena comparison, the divergence time (0.211), estimated jointly with migration, was nearly identical to the divergence time estimates using a model without migration (0.2105; Table 7). For both the P. amoena/P. caerulea and P. cyanea/P. caerulea comparisons, the divergence time estimate with the highest posterior probability was 4.9975, much greater than the estimates in the absence of migration (0.7185 and 0.537, respectively; Table 7). However, each of the posterior probability distributions of divergence time estimates for the P. amoena/P. caerulea and P. cyanea/P. caerulea comparisons, which never converged to zero, also had peaks much closer to the peak of the P. amoena/P. cyanea distribution (Figure 4B, Table 7).
Estimates of migration rates between the species varied. The posterior probability distributions of m1 and m2 between P. cyanea and P. amoena were flat, but nonzero (Figure 4, C and D). In contrast, migration into P. caerulea from both P. cyanea and P. amoena was effectively zero (m2 in both comparisons, Figure 4D). However, the parameter distributions for migration into P. cyanea and P. amoena from P. caerulea both show evidence for nonzero gene flow (Figure 4C). The posterior probability distributions for m1 were also similar for both comparisons involving P. caerulea, one peak at zero and a secondary, nonzero peak indicating gene flow into either P. cyanea or P. amoena from P. caerulea (Figure 4C).
While the divergence time parameter estimates did change with the inclusion of the gene flow parameter (Figure 4 and Table 7), the data were not significantly more likely under the more complex model in any of the pairwise comparisons. In all three comparisons twice the difference between the likelihoods of the data under the complex model (allowing for directional migration) and the simple model (no migration) were less than the critical value (χ = 5.991, d.f. = 2, P = 0.05)
DISCUSSION
Both phylogenetic and population genetic analyses of the 10-locus data set supported the traditional sister relationship between P. amoena and P. cyanea. Interestingly, the P. caerulea/P. amoena sister relationship proposed by Klicka et al. (2001) was supported by analyses of the MC1R data (Tables 4–6). Mutations in MC1R are known to correlate with variations in melanin-based plumage in a variety of taxa, including birds (Mundy 2005; Hoekstra 2006). Within Passerina, P. amoena and P. caerulea are the only species with putatively melanin-based plumages (Hill and McGraw 2006) and selection acting on MC1R might explain the differences we found between inferences based on MC1R and those based on other loci. There is a nonsynonymous change at amino acid residue 102 (Val → Leu) that unites P. caerulea and P. amoena (Val) to the exclusion of P. cyanea (Leu), which does not have any melanistic plumage. However, P. rositae and C. cyanoides (neither of which have melanin-based plumage) share the Val allele with P. amoena and P. caerulea indicating that the allele (Leu) found in P. cyanea is likely derived and probably not related to the differences in plumage patterns between P. caerulea, P. amoena, and P. cyanea. Therefore, while we cannot completely rule out the possibility that selection on the melanin-based plumages of P. amoena and P. caerulea has resulted in the observed patterns, the available data suggest selection is an unlikely cause.
Below, we discuss the benefits of using the two types of analyses for resolving sister relationships in Passerina and then with respect to closely related taxa in general.
Comparing mtDNA and nuclear DNA signals:
The sister relationship between P. cyanea and P. amoena supported by the multilocus analyses conflicts with a well-supported P. caerulea/P. amoena sister relationship inferred from mitochondrial data (Klicka et al. 2001). A possible explanation for the discrepancies between the mtDNA and nuclear results is differential lineage sorting of the mtDNA with respect to the true species tree (Braun and Brumfield 1998). Assuming ((P. cyanea, P. amoena), P. caerulea) is the correct topology, one can estimate the probability of incongruence between a gene tree and the species tree (Felsenstein 2004; Hein et al. 2005). Incorporating a range of internode values based on the divergence time estimates from IM and a range of potential effective ancestral population sizes, the chance a gene tree differs from the true species tree in the Passerina system ranges from 0% (Ne = 10,000 individuals, internode length = 100,000 years) to 65% (ancestral Ne = 1,000,000, internode length = 100,000 years, Figure 2). Using an internode length of 350,000 years, which is the difference in the maximum-likelihood estimates of the divergence time for the P. cyanea × P. amoena (∼138,000 years ago) and the P. caerulea × P. amoena (∼486,000 years ago) comparisons (Table 7), and assuming no postspeciation gene flow, the probabilities of incongruence range from 0% (ancestral Ne = 10,000) to 55% (ancestral Ne = 1,000,000) (Figure 2). While an accurate estimate of the ancestral population size is unavailable, an effective size of 10,000 or 20,000 may be unrealistically small. Current population census estimates for P. cyanea are on the order of 20–80 million individuals (Payne 1992). Furthermore, while some coalescent-based ancestral population-size estimates tend to be relatively small (Hey 2005; Won and Hey 2005), an estimate in birds based on sequence data from multiple nuclear markers suggested an ancestral population size on the order of hundreds of thousands of individuals (Jennings and Edwards 2005). Large population sizes and short internode lengths combine to increase the chance of incomplete and incorrect lineage sorting of gene copies, confusing the phylogenetic relationships of the focal taxa. The analyses of a multilocus data set presented herein suggest that the stochastic nature of lineage sorting of the mitochondrial genome resulted in a gene tree that is discordant with the species tree.
In closely related taxa, gene flow, in addition to lineage sorting problems, may limit the ability to correctly infer the species tree from gene sequences. P. cyanea and P. amoena currently hybridize where their breeding ranges overlap, although the amount of introgression is currently unknown, and hybrid specimens between other members of the genus have been reported. The samples of P. amoena and P. cyanea included in this study were collected far from the Great Plains contact zone (appendix) to minimize the chance that recent introgression between the two species might be obscuring the true evolutionary relationships between P. cyanea, P. amoena, and P. caerulea. Ongoing studies of the genetic structure of the hybrid zone between P. cyanea and P. amoena suggest little mitochondrial or nuclear introgression is occurring (M. Carling, unpublished data). The data from the hybrid zone, along with the nonsignificant difference in the likelihood of the data to the more complex model in IM, indicate that gene flow between P. cyanea and P. amoena has probably not had a strong influence on the speciational history inferred in this study.
Implications for uncovering phylogenetic signals in closely related taxa:
Integrating top-down phylogenetic methods with bottom-up population genetic methods was successful in that the same sister relationship was supported by both types of analyses. The data set of 10 loci sampled from either one (P. rositae and C. cyanoides) or two individuals per species (P. cyanea, P. amoena, and P. caerulea) generated a relatively well-supported species tree despite the low level of variation at each locus. One possible solution to increase the resolution of individual gene trees is to increase the length of the sampled sequence. Theoretically, lengthening the sampled sequence should increase the chance of detecting variable sites, which may then offer additional support for a particular topology (Doyle 1992). In practice however, lengthening loci may not be a straightforward solution to a problem of poor resolution in phylogeny reconstruction. As the length of the sampled locus increases, so does the chance of sampling sites that have undergone recombination. We detected recombination in 3 of the 10 loci, a ratio similar to a recent multilocus study investigating the speciational history of Australian grass finches (Jennings and Edwards 2005). Both studies sampled loci between 200 and 900 bp, which demonstrates that potential effects of recombination should be considered in any population genetic or phylogenetic study employing sequence data from nuclear loci. Here, the results of the phylogenetic analyses were similar regardless of whether we analyzed the entire sequence length or just the longest independently segregating block of sequence from the 3 loci (βact3, MPLPR, and TGFβ2) that showed evidence of recombination (Table 3, Figure 3).
The sampling design used here closely mimicked the design that resulted in the greatest accuracy of species tree inference in Maddison and Knowles's 2006 investigation into the utility of using the minimize-deep-coalescences method for phylogenetic reconstruction. Using simulated data sets that differed both in the number of individuals sampled and the number of loci sampled, they assessed how often the minimize-deep-coalescences method recovered the true species tree, that is, the tree on which the sequences were simulated. A sampling design of nine loci and three individuals per species proved to be quite successful in recovering the correct species tree. The results of the modified minimize-deep-coalescences method used here provide empirical support for their best sampling design and serve as a useful reference point for researchers designing sampling protocols with the purpose of inferring phylogenies of closely related taxa.
Conclusions:
Using DNA sequence data from a suite of nuclear loci, phylogenetic, and population genetic analyses supported the traditionally accepted evolutionary relationships between three North American passerines. In contrast to the findings of an earlier study that used data from the mitochondrial gene cytochrome b to infer the phylogeny of the genus (Klicka et al. 2001), we found, using both phylogenetic and population genetic-based methods, P. cyanea and P. amoena to be more closely related to each other than either is to P. caerulea. These results underscore the importance of sampling multiple individuals and multiple loci when estimating multiple demographic parameters simultaneously under complicated evolutionary scenarios, e.g., when allowing for postdivergence gene flow. Unfortunately, IM and other coalescent-based methods available for inferring population genetic parameters are often limited in some ways (e.g., how many populations they can handle, the sequence evolution models employed, etc). The continued improvement of these methods will facilitate the combined analytical approach we advocate in studies with a greater number of focal taxa.
Acknowledgments
We thank D. Dittmann and S. Birks of the Louisiana State University Museum of Natural Science and University of Washington Burke Museum, respectively, for generously providing tissue samples for this research. We are also grateful to the following people for critical comments that significantly improved this manuscript: C. Burney, Z. Cheviron, A. Cuervo, R. Eytan, J. Maley, R. Nielsen, and two anonymous reviewers. J. Hey and J. Degnan provided assistance with IM and COAL, respectively. C. Burney helped produce Figure 1 and supplemental Figure 1. B. Marks allowed us to use primers he developed for βact3. This work was supported by National Science Foundation grants DEB-0543562 and DBI-0400797 to R.T.B.
APPENDIX: SAMPLING LOCALITIES FOR INDIVIDUALS INCLUDED IN THE STUDY TESTING SPECIES DIVERGENCE HYPOTHESES IN PASSERINA BUNTINGS
| Species | ID no. | Locality |
|---|---|---|
| Passerina amoena | B-4005 | Douglas County, Washington |
| B-24769 | San Bernadino County, California | |
| P. cyanea | B-20575 | Iberville Parish, Louisiana |
| B-20773 | Cass County, Michigan | |
| P. caerulea | B-20598 | Cameron Parish, Louisiana |
| B-21812 | Jeff Davis County, Texas | |
| P. rositae | 69937a | Monte Bonito, Chiapas, Mexico |
| Cyanocompsa cyanoides | B-12708 | Velasco, Santa Cruz Department, Bolivia |
Tissue obtained through loan from the University of Washington Burke Museum. All other tissue samples were obtained from the Genetic Resources Collection at the Louisiana State University Museum of Natural Science.
References
- Axelsson, E., N. G. C. Smith, H. Sundstrom, S. Berlin and H. Ellegren, 2004. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 21: 1538–1547. [DOI] [PubMed] [Google Scholar]
- Baker, M. C., and A. E. M. Baker, 1990. Reproductive-behavior of female buntings - isolating mechanisms in a hybridizing pair of species. Evolution 44: 332–338. [DOI] [PubMed] [Google Scholar]
- Baker, M. C., and J. T. Boylan, 1999. Singing behavior, mating associations and reproductive success in a population of hybridizing Lazuli and Indigo Buntings. Condor 101: 493–504. [Google Scholar]
- Blake, C. H., 1969. Notes of the Indigo Bunting. Bird Banding 40: 133–139. [Google Scholar]
- Braun, M. J., and R. T. Brumfield, 1998. Enigmatic phylogeny of skuas: an alternative hypothesis. Proc. R. Soc. Lond. Ser. B 265: 995–999. [Google Scholar]
- Cheviron, Z. A., S. J. Hackett and R. T. Brumfield, 2006. Sequence variation in the coding region of the melanocortin-1 receptor gene (MC1R) is not associated with plumage variation in the Blue-crowned Manakin (Lepidothrix coronata). Proc. R. Soc. Lond. Ser. B 273: 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan, J. H., and N. A. Rosenberg, 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2: 0762–0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan, J. H., and L. A. Salter, 2005. Gene tree distributions under the coalescent process. Evolution 59: 24–37. [PubMed] [Google Scholar]
- Doyle, J. J., 1992. Gene trees and species trees—molecular systematics as one-character taxonomy. Syst. Bot. 17: 144–163. [Google Scholar]
- Edwards, S. V., L. Liu and D. K. Pearl, 2007. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. USA 104: 5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen, S. T., J. D. Rising and W. L. Thompson, 1975. A behavioral and morphological study of sympatry in the Indigo and Lazuli buntings of the Great Plains. Wilson Bull. 87: 145–179. [Google Scholar]
- Excoffier, L., G. Laval and S. Schneider, 2005. Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol. Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 2004. Inferring Phylogenies. Sinauer Associates, Sunderland, MA.
- Gillespie, J. H., and C. H. Langley, 1979. Are evolutionary rates really variable? J. Mol. Evol. 13: 27–34. [DOI] [PubMed] [Google Scholar]
- Greene, E., V. R. Muehter and W. Davison, 1996. Lazuli Bunting (Passerina amoena), pp. 1–24 in The Birds of North America, No. 232, edited by A. Poole and F. Gill. The Academy of Natural Sciences, Washington, DC/The American Ornithologists' Union, Philadelphia.
- Hein, J., M. K. Schierup and C. Wiuf, 2005. Gene Genealogies, Variation and Evolution. Oxford University Press, New York.
- Hellack, J. J., and G. Schnell, 1977. Phenetic analysis of the subfamily Cardinalinae using external and skeletal characters. Wilson Bull. 89: 130–148. [Google Scholar]
- Heslewood, M. M., M. S. Elphinstone, S. C. Tidemann and P. R. Baverstock, 1998. Myoglobin intron variation in the Gouldian Finch Erythrura gouldiae assessed by temperature gradient gel electrophoreses. Electrophoresis 19: 142–151. [DOI] [PubMed] [Google Scholar]
- Hey, J., 2005. On the number of New World founders: a population genetic portrait of the peopling of the Americas. PLoS Biol. 3: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and R. Nielsen, 2004. Multilocus methods for estimating population sizes, migration rates, and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, G. E., and K. J. McGraw, 2006. Bird Coloration, Vol. I. Harvard University Press, Cambridge, MA.
- Hoekstra, H. E., 2006. Genetics, development and the evolution of adaptive pigmentation in vertebrates. Heredity 97: 222–234. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jennings, W. B., and S. V. Edwards, 2005. Speciational history of Australian Grass Finches (Poephila) inferred from thirty gene trees. Evolution 59: 2033–2047. [PubMed] [Google Scholar]
- Kimura, M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicka, J., A. J. Fry, R. M. Zink and C. W. Thompson, 2001. A cytochrome-b perspective on Passerina bunting relationships. Auk 118: 611–623. [Google Scholar]
- Kroodsma, R. L., 1975. Hybridization in buntings (Passerina) in North Dakota and eastern Montana. Auk 92: 66–80. [Google Scholar]
- Kubatko, L. S., and J. H. Degnan, 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56: 17–24. [DOI] [PubMed] [Google Scholar]
- Maddison, W. P., 1997. Gene trees in species trees. Syst. Biol. 46: 523–536. [Google Scholar]
- Maddison, W. P., and L. L. Knowles, 2006. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55: 21–30. [DOI] [PubMed] [Google Scholar]
- Maddison, W. P., and D. R. Maddison, 2004. Mesquite: a modular system for evolutionary analysis, Version 1.01. http://mesquiteproject.org.
- Mayr, E., and L. L. J. Short, 1970. Species taxa of North American birds, a contribution to comparative systematics. Pub. 9, Nuttall Ornithological Club, Cambridge, MA.
- McGuire, G., and F. Wright, 1998. TOPAL: recombination detection in DNA and protein sequences. Bioinformatics 14: 219–220. [DOI] [PubMed] [Google Scholar]
- McGuire, G., and F. Wright, 2000. TOPAL 2.0: improved detection of mosaic sequences within multiple alignments. Bioinformatics 16: 130–134. [DOI] [PubMed] [Google Scholar]
- Mundy, N. I., 2005. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. Lond. Ser. B 272: 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NatureServe, 2006. NatureServe Explorer: An Online Encyclopedia of Life, Version 6.1. http://www.natureserve.org/explorer.
- Nei, M., and W. H. Li, 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo, P., and M. Nei, 1988. Relationships between gene trees and species trees. Mol. Biol. Evol. 5: 568–583. [DOI] [PubMed] [Google Scholar]
- Payne, R. B., 1992. Indigo Bunting, pp. 1–24 in The Birds of North America, No. 4, edited by A. Poole, P. Stettenheim and F. Gill. The Academy of Natural Sciences, Washington, DC/The American Ornithologists' Union, Philadelphia.
- Paynter, R. A., 1970. Subfamily Cardinalidae, pp. 216–245 in Check-list of Birds of the World, edited by R. A. Paynter and R. W. Storer. Museum of Comparative Zoology/Heffernan Press, Worcester, MA.
- Phillips, A. R., J. T. Marshall and G. Monson, 1964. The Birds of Arizona. University of Arizona Press, Tucson, AZ.
- Posada, D., and T. R. Buckley, 2004. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53: 793–808. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Primmer, C. R., T. Borge and G. P. Saetre, 2002. Single-nucleotide polymorphism characterization in species with limited available sequence information: high nucleotide diversity revealed in the avian genome. Mol. Ecol. 11: 603–612. [DOI] [PubMed] [Google Scholar]
- Ridgely, R. S., T. F. Allnutt, T. Brooks, D. K. McNicol, D. W. Mehlman et al., 2003. Digital distribution maps of the birds of the Western hemisphere, version 1.0. NatureServe, Arlington, VA.
- Ronquist, F., and J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosenberg, N. A., 2002. The probability of topological concordance of gene trees and species trees. Theor. Popul. Biol. 61: 225–247. [DOI] [PubMed] [Google Scholar]
- Rosenberg, N. A., 2003. The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution 57: 1465–1477. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Shimodaira, H., and M. Hasegawa, 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16: 1114–1116. [Google Scholar]
- Sibley, C. G., and L. L. J. Short, 1959. Hybridization in the buntings (Passerina) of the Great Plains. Auk 76: 443–463. [Google Scholar]
- Slade, R. W., C. Moritz, A. Heideman and P. T. Hale, 1993. Rapid assessment of single-copy nuclear DNA variation in diverse species. Mol. Ecol. 2: 359–373. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry. W. H. Freeman, New York.
- Swofford, D. L., 1998. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1983. Evolutonary relationships of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., 1989. Gene genealogy in three related populations—consistency probability between gene and population trees. Genetics 122: 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., 1995. A genetic perspective on the origin and history of humans. Annu. Rev. Ecol. Syst. 26: 343–372. [Google Scholar]
- Takahata, N., and M. Nei, 1985. Gene genealogy and variance of interpopulational nucleotide differences. Genetics 110: 325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., and M. Slatkin, 1990. Genealogy of neutral genes in two partially isolated populations. Theor. Popul. Biol. 38: 331–350. [DOI] [PubMed] [Google Scholar]
- Tamplin, J. W., J. W. Demastes and J. V. Remsen, 1993. Biochemical and morphometric relationships among some members of the Cardinalinae. Wilson Bull. 105: 93–113. [Google Scholar]
- Wakeley, J., 1996. Pairwise differences under a general model of population subdivision. J. Genet. 75: 81–89. [Google Scholar]
- Wakeley, J., and J. Hey, 1998. Testing speciation models with DNA sequence data, pp. 157–167 in Molecular Approaches to Ecology and Evolution, edited by R. DeSalle and R. Schierwater. Birkhauser Verlag, Basel, Switzerland.
- Won, Y. J., and J. Hey, 2005. Divergence population genetics of chimpanzees. Mol. Biol. Evol. 22: 297–307. [DOI] [PubMed] [Google Scholar]