Abstract
LINE-1 (L1) retrotransposons are the most abundant type of mammalian retroelement. They have profound effects on genome plasticity and have been proposed to fulfill essential host functions, yet it remains unclear where they lie on the spectrum from parasitism to mutualism. Their ubiquity makes it difficult to determine the extent of their effects on genome evolution and gene expression because of the relative dearth of animal models lacking L1 activity. We have isolated L1 sequences from 11 megabat species by a method that enriches for recently inserted L1s and have done a bioinformatic examination of L1 sequences from a 12th species whose genome was recently shotgun sequenced. An L1 extinction event appears to have occurred at least 24 million years ago (MYA) in an ancestor of the megabats. The ancestor was unusual in having maintained two highly divergent long-term L1 lineages with different levels of activity, which appear, on an evolutionary scale, to have simultaneously lost that activity. These megabat species can serve as new animal models to ask what effect loss of L1 activity has on mammalian genome evolution and gene expression.
RETROELEMENTS constitute a major fraction of mammalian genomes, with LINE-1 (L1) retrotransposons being the most common autonomous elements. Mammalian genomes appear to contain >100,000 copies of these elements, comprising 15–20% of the mass of the nuclear DNA (International Human Genome Sequencing Consortium 2001; Mouse Genome Sequencing Consortium 2002). Their existence in mammals seems to have preceded the mammalian radiation, and they are found in all species that have been examined (Furano 2000; Han and Boeke 2005). Yet oddly, there are relatively few potentially active L1s in the genomes that have been studied. The human and mouse genomes are estimated to contain ∼100 (Sassaman et al. 1997; Brouha et al. 2003) and 3000 (DeBerardinis et al. 1998) potentially active copies, respectively, and the vast majority of new insertions appear to emanate from very few of those potentially active copies. For this reason, and for other reasons not completely understood, recently transposed elements group into one or very few lineages of closely related copies (Deininger et al. 1992; Casavant et al. 1996; Furano 2000).
Major questions remain as to how L1s affect the genomes and organisms in which they reside. The null hypothesis is that they function simply as parasites. Young, full-length elements in humans have been shown to be subject to negative selection (Boissinot et al. 2006), yet it has been hypothesized that L1s may have evolved to furnish essential functions for their host. Proposed functions include a role in DNA double-strand break repair (Hutchison et al. 1989; Teng et al. 1996; Morrish et al. 2002) and in propagation of the X chromosome inactivation signal (Lyon 1998). It has also been suggested that L1 reverse transcriptase may be necessary for preimplantation development in mice (Beraldi et al. 2006) and that L1 retrotransposition may affect neuronal somatic diversification (Muotri et al. 2005).
Independent of any immediate essential function that L1s may have, it has become obvious that they affect both genome evolution and gene activity in multiple ways (Han and Boeke 2005; Hedges and Batzer 2005). They provide the molecular machinery for movement of SINEs and pseudogenes and mediate ectopic recombination (Furano 2000). They are estimated to have moved as much as 1% of the genome by 3′ transduction (Goodier et al. 2000; Pickeral et al. 2000). They have also been shown to have the potential to function as a “rheostat” for gene expression (Han et al. 2004). LINE-1 retrotransposition has additionally been shown to be associated with genomic instability (Gilbert et al. 2002; Symer et al. 2002).
The ubiquitous nature of these elements has made it difficult to determine the extent of their effects on genomes and led to the assumption that all mammalian species would be found to contain active L1s. However, our previous identification of a group of rodents in which L1s have become extinct showed that extinction might be rare but was possible (Casavant et al. 2000; Grahn et al. 2005). Although this finding has given an important model system for asking questions about the effects of these elements, from a larger perspective it represents only one of nature's experiments on the evolutionary effects in mammals of life without L1 activity.
As part of an extensive screen for active L1 elements in eutherian mammals, we investigated L1s in the bats (Chiroptera). The bats are the second most speciose order of mammals, surpassed only by the rodents, and among the mammals have unusually small genomes (Bachmann 1972; Redi et al. 2005). Since this difference in genome size is correlated with reduced copy number of interspersed and tandemly duplicated repetitive sequences in at least one bat species (Van den Bussche et al. 1995), we were interested in looking at the dynamics of L1 elements within this group.
In this study we have identified an independent L1 extinction in the megabats (family Pteropodidae), determined the phylogenetic distribution of this event, and shown that, on an evolutionary scale, two long-term L1 lineages within these animals appear to have become extinct simultaneously. These findings show that L1 extinctions may not be as unusual as thought. They also provide a new system to address the question of how L1s affect genome evolution and to explore the possibility of mammalian genomes using multiple routes for suppression of L1 activity and adaptation to the loss of that activity. This L1 extinction event can be viewed as another experiment of nature that can increase our understanding of this significant player in the organization of the mammalian genome.
MATERIALS AND METHODS
Specimens and DNA isolation:
Specimens used in this study and their accession numbers are listed in Table 1. Tissues were obtained from The Museum, Texas Tech University. Genomic DNA was extracted as previously described (Longmire et al. 1988).
TABLE 1.
Specimens examined, number of L1 sequences analyzed, and conserved amino acids
| No. of L1 clones
|
Changes at conserved AA sites | |||
|---|---|---|---|---|
| Specimens | Sequenced | In final data set | ||
| Order: Chiroptera | ||||
| Yinpterochiroptera | TK40884 | |||
| Rhinopoma hardwickei | ||||
| Megabats (Pteropodidae) | ||||
| Megaloglossus woermanni | TK21565 | 27 | 24 | 5.7 (2)b |
| Rousettus amplexicaudatus | TK20031 | 24 | 7 | 4.9 (3) |
| Cynopterus sphinx | TK21250 | 38 | 24 | 6.1 (2) |
| Megaerops niphanae | TK21085 | 37 | 14 | 5.1 (2) |
| Dobsonia moluccensis | TK20261 | 24 | 21 | 6.3 (3) |
| Macroglossus sp. | TK20306 | 31 | 22 | 5.6 (1) |
| Pteropus hypomelanus | TK20059 | 39 | 25 | 5.3 (1) |
| Pteropus macrotis | TK20310 | 36a | 28 | 5.1 (1) |
| Melonycteris melanops | TK20071 | 23 | 20 | 5.5 (2) |
| Hypsignathus monstrosus | TK21542 | 28 | 17 | 6.5 (2) |
| Nyctimene albiventer | TK20056 | 23 | 16 | 5.9 (2) |
| Yangochiroptera | ||||
| Tonatia saurophila bakeri | TK104519 | |||
| Artibeus jamaicensis | TK27682 | |||
| Order: Insectivora | ||||
| Notiosorex crawfordi | TK84584 | |||
An additional six sequences were obtained for a more 5′ region of ORF2 in P. macrotis, and all elements appeared to represent ancient insertions.
Average (and minimum) number of conserved amino acids altered per element after return to the correct reading frame.
Southern blot analysis:
Genomic DNAs were digested with RsaI, DdeI, or EcoRI then 1.5 μg of each was run on agarose gels and Southern blotted onto nylon membranes by standard procedures (Ausubel et al. 1989). A DNA probe from a recently inserted dog L1 was random prime labeled with 32P. The probe covers the same 575-bp region in L1 open reading frame 2 (ORF2) as that used for sequence analysis in this study. Prehybridization, hybridization, and washing were done under low stringency conditions as previously described (Casavant et al. 1996), but with the following buffers. Prehybridization buffer contained 0.8 mg/ml denatured salmon sperm DNA, 6× SSC, 10× Denhardt's solution, and 0.3% SDS. Hybridization buffer was the same as prehybridization buffer but contained no salmon sperm DNA. Wash buffer was 5× SSC, 0.1% SDS.
PCR, clone isolation, and DNA sequencing:
A degenerate PCR and colony color screening technique designed to enrich for L1 fragments retaining a single open reading frame was used to isolate a portion of ORF2 straddling the reverse transcriptase domain from a number of L1 elements from each species (Cantrell et al. 2000). The majority of elements were isolated using the previously described 8FDeg and 9RDeg primers, which amplify a 614-bp region in elements with no insertions or deletions, covering bases 4969–5583 of Mus L1Md-A2, GenBank accession no. M13002, yielding 575 bp between the primers for sequence analysis (see Figure 1 in Cantrell et al. 2000). These primers contain restriction enzyme cloning sites added to their 5′ ends, which permit ligation of PCR products after digestion with either EcoRI and BamHI, XhoI and BamHI, or XhoI and PstI. A small number of elements were also isolated by the above technique using a different set of primers 6FDeg (5′-GGG GTA CCT GTC GAC ATG AAY ATH GAY GCN AA-3′) and 3RDeg (5′-CGG GAT CCA ACT GCA GTM NAC DAT CAT RTC RTC-3′), which amplify ORF2 bases 4481–4995 of M13002 and introduce KpnI and BamHI cloning sites. DNA inserts from at least 12 blue and 12 white colonies were sequenced for each species. Double-strand sequencing was done with an Applied Biosystems 3730 DNA analyzer (Foster City, CA).
Figure 1.—
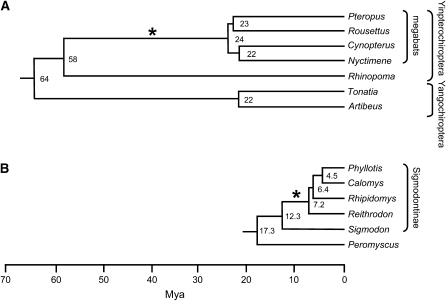
–Phylogenies of selected bat species (A) and rodent species (B). The phylogenies are adapted from Teeling et al. (2005) and from Steppan et al. (2004), respectively. The asterisk on each tree shows the branch on which L1 activity ceased. Branch lengths indicate the molecular time scale, and the numbers at nodes are the estimated times since divergence (millions of years ago, MYA). The megabats (family Pteropodidae) and the microbat genus Rhinopoma are within the suborder Yinpterochiroptera, while the genera Tonatia and Artibeus are microbat genera within the suborder Yangochiroptera. The four megabat species shown represent a broad phylogenetic range of that group. The rodent divergence times are the estimates Steppan et al. obtained using nonparametric rate smoothing. The sigmodontine rodents are indicated within the bracket.
The sensitivity of the degenerate PCR and colony screening technique for detection of young L1 elements was determined by spiking known amounts of genomic DNA from the megabat Rousettus amplexicaudatus with varied quantities of an active mouse L1 element before amplification and cloning. The mouse L1 element was in the plasmid pDB97 (provided by Sandra L. Martin, University of Colorado, Denver), which was digested with the restriction enzyme EcoRI to produce linear DNA before mixing and amplification with R. amplexicaudatus DNA. Aliquots of R. amplexicaudatus DNA were spiked with plasmid DNA equivalent to 1, 3, 10, 100, or 1000 young L1 copies per haploid genome. Amplification, cloning, and sequence analysis were done as with pure genomic DNA samples except that only clones from blue colonies were analyzed.
Alignments and sequence analyses:
Initial alignments and sequence analyses were performed using ClustalW and the LASERGENE analysis package (DNASTAR, Madison, WI). Final alignments were adjusted manually. Phylogenetic analyses were performed using PAUP* 4.0b10 (Swofford 2002) and Bayesian inference with MRBAYES version 3.0 (Huelsenbeck and Ronquist 2001) with gaps excluded.
Independent analyses were initially performed on each species to select the subset of elements to be used in the final data set. A total of 330 megabat elements were sequenced, none of which was found to contain complete open reading frames. One hundred twelve elements were eliminated through use of the following criteria. Elements that contained deletions >80 bp were removed. Criteria described previously (Grahn et al. 2005) were used to eliminate all but 1 element from any set of elements that appeared to be related by processes other than normal L1 retrotransposition (e.g., unequal crossing over, gene conversion, or alleles). A total of 24 elements fit these criteria by having at least one identical, or closely related partner. In all of these cases, each set of partners shared otherwise unique inactivating mutations showing that they had not been duplicated by normal retrotransposition. Only 1 element was retained from each of these groups.
The oldest elements were eliminated by examining the divergence from each element to its nearest neighbors within that species, the lineage-1 consensus within that species, and the overall lineage-2 consensus sequence. Construction of the lineage-specific consensus sequences is described below. Any element which showed an unadjusted divergence from other elements within that species of >17% plus divergence from the consensus sequence for its lineage of >12% was eliminated from further analysis. The majority of elements eliminated from each species were removed either because of great divergence from all the other elements in that species or secondarily because they contained large deletions.
The data set was searched for recombinants by the procedure Sawyer (1989) and by comparing phylogenetic trees generated from data sets after partitioning sequence segments into three separate data blocks of ∼190 bp each, as described previously (Grahn et al. 2005). Putative recombinants were eliminated from further analyses.
The number of mutations affecting conserved amino acids was determined by first returning each sequence to its original reading frame by removing insertions and replacing gapped positions with N's and then comparing the amino acid sequence to a consensus of conserved amino acids within this region as described in results.
The phylogeny of the final data set was estimated from the sequences returned to their original reading frame using Bayesian inference and the GTR+Γ nucleotide substitution model, which was selected using DT-ModSel (Minin et al. 2003). Four independent searches with MRBAYES were each run for 10 million generations. Parameter values were plotted vs. generation number to determine when stationarity was reached and the burn-in for each search was then discarded. The posterior probability of each clade was estimated as the percentage of trees that included the clade after the burn-in.
Construction of ancestral sequences:
Since there is a lack of phylogenetic signal from L1 sequences within each lineage after L1 extinction, the most recent active L1 ancestor was constructed by first obtaining the consensus sequence. Each consensus sequence was then corrected at CpG sites, which are recognizable because they show very high rates of specific adjacent mutations. Lineage 1 contains 12 CpG sites, while lineage 2 contains 4.
Bioinformatic analysis:
Genome sequences for Pteropus vampyrus and Myotis lucifugus were downloaded from NCBI's trace file database (ftp://ftp.ncbi.nih.gov/pub/TraceDB/). Depending on the species, there were 16 or 17 files that each contained ∼500,000 sequences. The L1 protein sequence for the megabat lineage-1 ancestral sequence including the PCR primer regions was used in a tBLASTn search of the trace files of the P. vampyrus sequences, translated in all six reading frames. The protein sequence of a recently inserted Myotis species L1 from the same 614-bp region of ORF2 was used to query the translated M. lucifugus sequences (Altschul et al. 1997). Default settings for the tBLASTn searches were used except that up to 10,000 alignments were saved for each file; this maximum number of hits was never reached in any file of 500,000 sequences. The output for each search was parsed using the BioPerl module SearchIO (http://www.bioperl.org), and the resultant table was loaded into a MySQL database (http://www.mysql.org). The database was then queried to determine sequentially how many files contained >573 bp (the length of the translated ORF used in phylogenetic analyses) of L1 sequence within this region, if it had a single high-scoring segment pair (indicating no frameshift mutations), and if it had no stop codons within the translated sequence. The protein sequences that met these criteria were then aligned using ClustalW, and the resultant alignments were edited in MacClade (Maddison and Maddison 2003) to determine the amino acid at each of the 38 conserved sites. To confirm that the megabat lineage-1 ancestral sequence would detect divergent L1 sequences, it was used in a BLASTn search of a database of 1764 full-length human LINEs, and the translated sequence was used in a tBLASTn search of the six reading frame translations of these same full-length L1s. All 1764 human sequences were detected in both queries.
RESULTS
Isolation and initial analysis of megabat L1 sequences:
As part of a continuing study of L1 elements from different mammalian species, we initially analyzed a 575-bp region of the ORF2 in L1s from the megabat species P. macrotis. The isolation procedure used is a degenerate PCR followed by a colony screening technique that preferentially yields more recently transposed (younger) elements from any mammalian species (Cantrell et al. 2000). L1 isolation by the above procedure normally yields a sizable percentage of elements with a single ORF throughout the region amplified (ORF+). Strong selection for younger elements comes from the preferential amplification of L1 sequences which, due to less mutation since insertion, still retain PCR primer binding sites encoding highly conserved amino acids. A secondary degree of enrichment for young elements is also normally achieved by a color screen that gives blue colonies when inserted L1 sequences, which still retain a single ORF, produce L1/betagalactosidase fusion proteins. However, only elements with stops in all reading frames (ORF−) were found from P. macrotis, suggesting that the sequences were derived from ancient insertion events (old L1s). A background of blue colonies containing ORF− L1 sequences is observed even in species that lack ORF+ L1s; these appear to be due to rare, cryptic, internal translation initiation sequences (Cantrell et al. 2000).
To evaluate the sensitivity of our screening technique for detecting young L1s that might be at low copy number in the genome, we spiked genomic DNA from a megabat with quantities of a cloned mouse L1 element equivalent to 1, 3, 10, 100, or 1000 young L1 copies per haploid genome. Amplification, cloning, screening, and sequencing was carried out for each aliquot of spiked DNA, and 16 clones from blue colonies were sequenced from each sample. No mouse L1s clones were found among the 16 sequenced from the megabat sample spiked with mouse L1 equivalent to 1 copy per haploid genome, but for the sample spiked with 3 copies per haploid genome 25% of the clones were found to contain the mouse L1. The number of mouse L1 clones among the 16 sequenced rose to 38, 94, and 100% for spiking equivalent to 10, 100, or 1000 young L1 copies per haploid genome, respectively. Thus this technique is very effective at enriching for young elements against a background of older degenerate copies.
Even though we have used the 8FDeg and 9RDeg primers to isolate young L1s from >130 species of mammals including all 18 orders of eutherians, 5 orders of marsupials, and 1 of monotremes, the formal possibility exists that the primers may have missed a divergent subfamily of L1s due to changes in active L1s at conserved amino acids encoded in the primer binding sites. In consideration of this possibility, we also sequenced six of the relatively rare blue colonies arising from PCR done with the alternative primers, 6FDeg and 3RDeg, covering a region more 5′ in ORF2. All of these L1 sequences also appeared to be derived from ancient insertion events, because none of them was ORF+ and there was great sequence divergence between all pairs.
By the same methods, we have isolated young L1 elements from a Rhinopoma species, which is within the suborder Yinpterochiroptera, and from members of the suborder Yangochiroptera, suggesting that the potential L1 extinction occurred within the Yinpterochiroptera, after the divergence of the megabats from their common ancestor with Rhinopoma (see Figure 1A). We chose 10 additional megabat species distributed throughout the Pteropodidae for more detailed analysis (Table 1). The purpose of this study was to see if a slowdown or extinction of L1 activity had occurred, and if so, to determine the phylogenetic distribution of this potential event.
At least 24 L1-containing clones were sequenced from each species. Preliminary analysis of each species, and then of the full data set, was carried out to remove the oldest sequences, which confound accurate alignment, and elements that appeared to have arisen by events other than retrotransposition (i.e., orthologous loci or tandem duplications, see materials and methods). Even among the remaining 218 elements, only L1 sequences that appeared to be old by several criteria were found. All of the sequences were ORF−, and most of them contained insertions and/or deletions.
Since mutation after transposition should eventually give rise to divergence among amino acids essential for L1 activity, one might ask whether any of these elements were transposed recently enough that they would still retain conserved amino acids after they had been returned to their original reading frame. We have previously identified 38 amino acids within the region studied here that are completely conserved in young L1 elements isolated from every order of placental mammals, five orders of marsupials, and one of monotremes (Grahn et al. 2005). The conserved amino acids begin at amino acid 714 of Mus L1Md-A2, GenBank accession no. M13002. The sequence is shown here, where the numbers in parentheses indicate the number of amino acids between conserved residues: Y(22)GYK(1)N(2)KS(20)F(8)YLG(9)L(3)N(14) W(4)C(1)W(1)G(2)NI(1)KM(2)LP(7)A(1)P(17)F(1)W(18)*GG(3)P(4)YY(1)A(3)K(2)WYW(3)R. The asterisk indicates a region that has undergone a one-codon deletion in the active L1s of Notiosorex crawfordi. To examine changes at these 38 sites, each sequence in the final megabat data set was returned to its original reading frame by removing insertions and replacing gapped positions in alignments with N's. Analysis of the conserved amino acid positions is summarized in Table 1. There was an average of 5.7 amino acid alterations per element at these positions, and there was no element in the entire data set retaining all 38 conserved amino acids, further suggesting that none of these elements is capable of transposition, and that substantial time has passed since any were transposed.
Southern hybridizations:
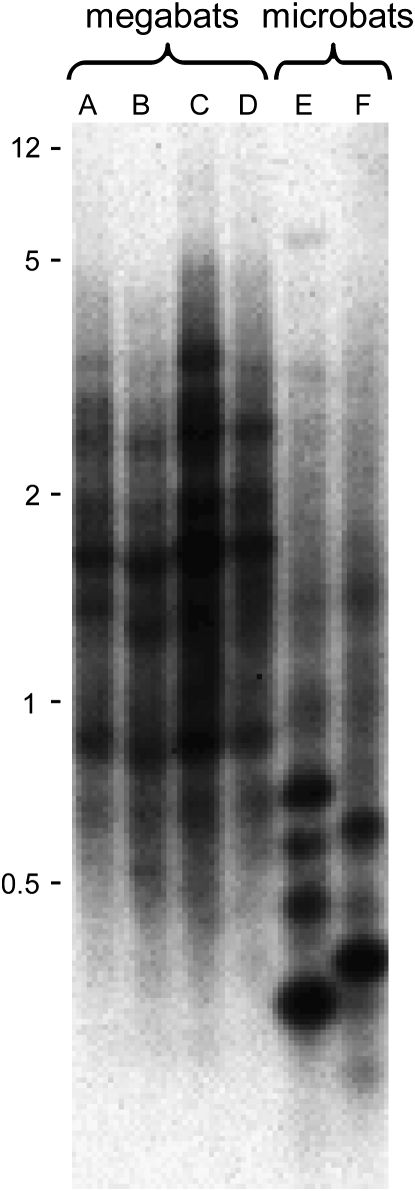
Analysis of genomic DNA by Southern hybridizations can give evidence of recent L1 activity independent of any sequence-dependent limitations of PCR. We have previously shown that mammalian species containing recently inserted L1 elements (L1-active species) show distinct taxon-specific bands upon Southern hybridization with an L1 probe, unless those species are closely related. On the other hand, species in which L1s have become extinct (L1-inactive species) fail to show taxon-specific bands (Casavant et al. 2000; Grahn et al. 2005). Four of the species that represent a wide phylogenetic range among the megabats were studied by Southern hybridizations. These four species, which are estimated to have diverged 22–24 MYA (Teeling et al. 2005), were compared with two microbat species in the Yangochiroptera that are estimated to have diverged 22 MYA (Figure 1A). Southern hybridizations were initially done after single enzyme digestion of genomic DNAs with three different restriction enzymes: DdeI, RsaI, and EcoRI, followed by gel electrophoresis. A recently inserted dog L1 probe was used to ensure that each of the species was an equal evolutionary distance from the probe. No species-specific bands were seen among the megabat species for any type of digest, and the RsaI digests produced the most informative Southerns (Figure 2). The two microbat species, which we have shown to contain recently inserted L1 elements, each show a number of intense species-specific bands. Each band represents an L1 restriction site-defined subfamily that has been amplified since divergence of the two species. In contrast, hybridization profiles for the four megabat species (Cynopterus sphinx, Nyctimene albiventer, P. hypomelanus, and R. amplexicaudatus) are very similar, suggesting a lack of L1 activity since species divergence. Even though each lane of the Southern blot received the same amount of DNA, slight differences in hybridization intensity among these four species may be due to genome-size differences. The genome sizes of the bat species used in these Southerns are unknown, but bats show considerable genome-size variation (Gregory 2006).
Figure 2.—
–Southern hybridization of genomic DNA digested with RsaI and probed with a dog L1. The four left lanes contain DNA from megabat species: (A) Cynopterus sphinx, (B) Nyctimene albiventer, (C) Pteropus hypomelanus, and (D) Rousettus amplexicaudatus. The two right lanes contain DNA from two microbat species: (E) Artibeus jamaicensis and (F) Tonatia saurophila bakeri. Numbers at the left show positions of size markers (kb).
Phylogenetic analysis:
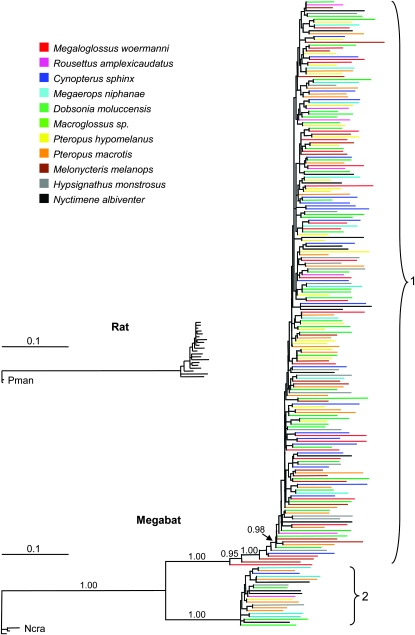
The 218 megabat L1 sequences were compared by combining alignments from all species after removal of insertions. Young L1s from the shrew, Notiosorex crawfordi, were used as the outgroup to produce the megabat Bayesian tree shown in Figure 3. The megabat elements show a drastically different topology compared to what is typical for species with active L1s (Scott et al. 2006). Specifically, megabat L1s exhibit long terminal branches, indicative of extensive mutation since insertion. Also, all megabat L1s form two large polytomies rather than species-specific clusters as is typical for active L1s (Grahn et al. 2005). For comparison, Figure 3 additionally shows an analysis of 20 rat L1 sequences drawn to the same scale. The major difference between terminal branch lengths is immediately evident. The rat sequences were isolated using the same procedure as was used for the megabat sequences. As is typical with this method, many of the rat L1s are ORF+ and have short terminal branches, suggesting recent insertion into the genome. The striking difference between the two trees is seen even though all sequenced L1s were included in the rat tree while only the most recently inserted L1s from each species were included in the megabat tree.
Figure 3.—
–Maximum a posteriori probability phylogeny of 218 megabat L1 sequences from 11 megabat species. Three recently transposed L1 sequences from Notiosorex crawfordi (Ncra) were used as the outgroup. Posterior probabilities are given above relevant branches to show the two supported long-term lineages (1 and 2) and significantly older elements. Names of individual elements have been removed, and terminal branches for megabat species have been color coded. The tree is rooted with the N. crawfordi elements. The small tree is of 20 Rattus norvegicus L1 sequences with two recently transposed Peromyscus maniculatus (Pman) elements used as the outgroup. The small tree was constructed using rat sequences from 10 blue colonies and 10 white colonies obtained by PCR as for the megabat sequences but with no subsequent removal of older elements. The large tree contains only the more recently transposed subset of megabat sequences from each species.
A notable feature of the megabat tree is the existence of two distinct megabat clades which show that there were two independent, long-term lineages before the L1 extinction. An additional striking feature of the tree is the fact that nearly all of the L1 sequences, except for a few older sequences within the major lineage (lineage 1) are united with neighbors by extremely short internal branches, essentially giving rise to two large polytomies. It is also significant that there are no supported, species-specific clades, but rather the species are mixed across the tree. This suggests that L1 extinction occurred before the divergence of these species, and according to the dating of the radiation of the megabats by Teeling et al. (2005) (Figure 1), would place extinction in a common ancestor at least 24 MYA.
Further support for loss of L1 activity before divergence of the species comes from comparison of ancestral sequences constructed for each species. Ancestral sequences were constructed without the six lineage-1 elements shown by the tree in Figure 3 to have inserted significantly prior to the extinction. The consensus sequence was constructed for each species, and CpG sites, which can be recognized because they mutate at an unusually high rate due to cytosine methylation, were returned to their ancestral states. In spite of the relatively ancient divergence of these species, the adjusted pairwise sequence distances between the species-specific ancestral sequences were extremely low, ranging from 0 to 0.0053 per site with an average of 0.0037 per site, which suggests a single extinction event for lineage 1. Accurate species-specific ancestral sequences for the lineage-2 elements could not be constructed because there were so few elements from each species.
Bioinformatic scanning of the P. vampyrus genome:
The recent whole-genome shotgun (WGS) sequencing of the megabat P. vampyrus has allowed us to use a bioinformatics approach to search for any evidence of L1 activity within that genome. The P. vampyrus WGS database was analyzed in parallel with the recently produced WGS database for the microbat M. lucifugus, chosen because we have shown a Myotis species to contain recently inserted L1s (E. Howell and H. Wichman, unpublished data) and both databases are unassembled trace file archives giving ∼2× coverage. The tBLASTn searches of the translated trace files, using either the megabat lineage-1 consensus sequence described in materials and methods or a recently inserted M. lucifugus L1 sequence covering the same region, detected ∼138,000 L1s in the megabat genome and 104,000 L1s in the microbat genome (Table 2). The L1 sequences from these trace files were then queried to see if they satisfy the three criteria we use to determine if PCR-derived L1 sequences originated from potentially active elements. Table 2 shows that slightly more L1 sequences greater than the 573-bp length necessary to contain all of the amino acids in the region of analysis were detected in the M. lucifugus database than in the P. vampyrus database. However, there were >13 times as many of these long sequences that had single ORFs in the M. lucifugus database than in the P. vampyrus database (919 vs. 69 elements). When the long ORF+ sequences were inspected for retention of the 38 amino acids conserved in this region, none of the P. vampyrus sequences was found to retain all of those conserved amino acids, while over half of the M. lucifugus sequences (586) retained all of the conserved amino acids. By these criteria, the M. lucifugus genome appears to contain hundreds of sequences derived from potentially active L1 elements, while the P. vampyrus genome appears to contain only inactive elements.
TABLE 2.
Summary of in silico analysis of Pteropus vampyrus and Myotis lucifugus L1s
| P. vampyrus WGSa | M. lucifugus WGSa | |
|---|---|---|
| Total trace files | 7,385,838 | 8,276,891 |
| L1s identified by query sequences covering the degenerate PCR regionb | 137,860 | 104,493 |
| L1s >573 bpcd | 2,323 | 2,533 |
| L1s with a single ORFd | 69 | 919 |
| L1s with all 38 conserved amino acidsd | 0 | 586 |
WGS, whole-genome shotgun sequence database.
The degenerate PCR region covers the 614 bp of the L1 ORF2 amplification product that includes primer/L1 homology sequences.
The length of the translated ORF used in phylogenetic analyses of amplified elements.
Sequence numbers in each of these rows were obtained by analysis of the L1s in the row immediately above.
Further analysis was carried out on the 69 P. vampyrus sequences shown to contain a single long ORF to search for any evidence of recent activity. As would be expected for a WGS database, a number of identical sequences and sequences that differed at only a few sites were found. These may represent multiple reads (with or without error) of the same sequence in a database with 2× coverage, alleles at the same locus, or sequences recently duplicated by retrotransposition or other mechanisms. Removal of duplicate sequences led to a data set containing 54 unique P. vampyrus elements, and phylogenetic analysis yielded a tree (supplemental Figure S1 at http://www.genetics.org/supplemental/) with a topology similar to the megabat tree shown in Figure 3. The tree contains primarily long terminal branches and two polytomies, one for each of the lineages. As with the elements isolated by PCR from the other species of megabats, there were several sets of two to three almost identical elements, but the elements in each set shared changes at amino acid positions that are conserved in L1s across Mammalia, suggesting that they had not been duplicated by normal L1 retrotransposition (see materials and methods and supplemental Figure S1 for more detail). Supplemental Figure S1 also shows a tree obtained after phylogenetic analysis of 54 unique M. lucifugus L1s, randomly chosen from the 919 sequences containing single long ORFs. The tree is strikingly different, showing the large number of closely related L1s indicative of a species retaining substantial L1 activity.
If L1 activity in P. vampyrus had maintained active retrotransposition with a very small number of the active elements that generated the polytomies seen in Figure 3, then a lineage-specific ancestral sequence generated using young elements from P. vampyrus should be substantially different from any ancestral sequence constructed above using PCR-derived elements. Lineage-1 and lineage-2 ancestral sequences were constructed from the 54 unique P. vampyrus long, ORF+ elements. Both the lineage-1 and lineage-2 species-specific P. vampyrus ancestral sequences differed from the megabat lineage-specific ancestral sequences at only 1 base in the 575-bp region, a difference less than that seen between the majority of the megabat species-specific ancestral sequences. The near identity of these ancestral sequences is strong evidence for a single L1 extinction in a common ancestor of all the megabat species.
Divergence within and between lineages:
The elements analyzed in Figure 3 were examined further to address the question of whether the two lineages died out at different times or at the same time. Since inactive elements should be mutating at the neutral mutation rate, and both lineages have come from the range of species, the average sequence distance between each element and its lineage-specific ancestor should be the same for lineage 1 as it is for lineage 2 if activity ceased in both lineages at the same time. Lineage-specific ancestral sequences were constructed after removal of the six significantly older lineage-1 elements. The average adjusted pairwise distance from each lineage-1 element to the lineage-1 ancestral sequence was 0.08895 per site, while the distance from each lineage-2 element to the lineage-2 ancestral sequence was 0.08900 per site. This near identity in pairwise sequence distances for each lineage suggests that there was simultaneous extinction in both lineages.
Comparison of the lineage-specific ancestral sequences gives a measure of how much the two lineages diverged before loss of L1 activity. Their adjusted pairwise sequence distance of 0.296 per site shows an unparalleled divergence of two L1 lineages from each other while still active.
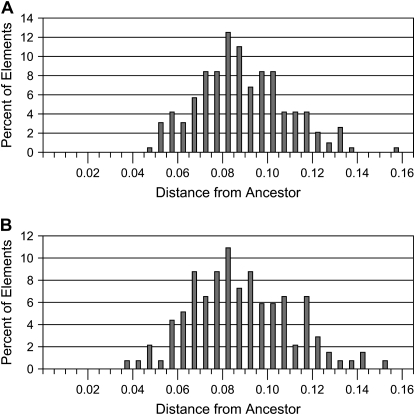
This megabat L1 extinction can be compared to the L1 extinction in the sigmodontine rodents because the same region was isolated in the same manner from the L1-inactive rodents (Grahn et al. 2005). Surprisingly, the average adjusted sequence distance of 0.089 per site for the above 212 megabat sequences to their ancestors is almost identical to the average sequence distance of 0.088 per site for the sigmodontine elements to their ancestor. Figure 4 shows the distributions of the sequence distances of the elements to their ancestors for each extinction. The distributions not only appear to be very similar, but they also show similar variances: 0.00040 for the megabat distances shown in Figure 4A and 0.00050 for the sigmodontine distances shown in Figure 4B. [We note that the variance previously reported for the sigmodontine extinction (Grahn et al. 2005) was incorrect due to a formula error in the spreadsheet.]
Figure 4.—
–Distance distribution of megabat lineage-1 elements (A) and sigmodontine elements (B) to ancestor. The adjusted pairwise sequence distance from each megabat lineage-1 (L1) element to the megabat lineage-1 ancestor and from each sigmodontine L1 element to the sigmodontine ancestor was determined. For each data set, the percentage of the total elements that were within each indicated distance range is shown.
Since loss of L1 activity appears to have occurred after the divergence of the megabats from the rest of the Yinpterochiroptera but before divergence of the extant megabat species, and all of the L1 divergence from the most recent ancestral sequences should reflect neutral mutation, estimates of bat divergence times (Teeling et al. 2005) shown in Figure 1 can be used to determine a rough estimate of the neutral substitution rate in this group. For their estimate of megabat divergence time, Teeling et al. used four megabat species that cover a broad phylogenetic range and probably the deepest split within the group. They estimated divergence of the megabats from a common ancestor at 24 MYA (95% credibility interval, 20–29 MYA) and the divergence of the megabats from the rest of the Yinpterochiroptera at 58 MYA (95% credibility interval, 53–63 MYA). If the average adjusted sequence distance of L1 elements to their ancestors of 0.089 per site is divided by the above times, the neutral substitution rate within the megabats is found to be 0.15–0.37%/MY (95% credibility interval for Teeling's estimates, 0.14–0.44%/MY).
DISCUSSION
Extinction of L1 activity:
It is difficult to prove that there has been complete loss of L1 activity in any mammalian species rather than a precipitous decline in activity, but three independent lines of inquiry contain multiple indicators that support L1 extinction in all megabat species studied here. First, L1 sequences obtained by a procedure that provides a highly sensitive selection for more recently inserted elements by both degenerate PCR and a colony color screen (Cantrell et al. 2000) contain six indicators that they were transposed long ago: (1) No L1 sequences retaining intact ORFs were found even though the region analyzed covered only 9% of a full-length element; (2) all sequences had suffered alterations in conserved amino acids; (3) nearly all sequences contained multiple insertions and deletions; (4) the most recent ancestral L1 sequences constructed for each species were nearly identical in spite of the relatively ancient divergence of some of the species; (5) phylogenetic analysis of the L1s from all of these species showed no evidence for species-specific clades but produced single large polytomies for each L1 lineage with L1s from each species dispersed throughout the tree; and (6) every element showed a long terminal branch indicative of the accumulation of many mutations and thus a long period of time since insertion.
In addition, the L1 screening procedure used here has allowed isolation of young elements from every order of eutherians, five orders of marsupials, and one of monotremes (K. Bush, M. Cantrell, I. Erickson, A. Keys, A. Martinez, L. Scott and H. Wichman, unpublished data) and is thus likely to reveal any active L1s present. Nevertheless, we used this same screening technique with degenerate primers designed to an alternative L1-ORF2 region to isolate a number of P. macrotis sequences in case the primer target sites for our standard primer pair were more diverged in megabats than in other mammals. These sequences also appeared to be derived from ancient insertions. We also evaluated the sensitivity of the technique by spiking megabat DNA with a cloned mouse L1 element. We found that the younger element was detected at reasonably high frequency even at copy numbers as low as three per haploid genome.
Second, Southern hybridizations showed species-specific bands for microbat species, indicative of L1 activity since divergence, but showed no species-specific bands for megabat species which are estimated to have diverged in the same time frame of ∼22 MYA (Teeling et al. 2005).
The third independent line of inquiry is based on whole-genome shotgun sequencing of the megabat species P. vampyrus. Comparison of the P. vampyrus genome sequences to a similar microbat genome database, that of M. lucifugus, showed much evidence for recent L1 activity in M. lucifugus, but only evidence for ancient L1 activity in P. vampyrus.
These multiple, independent lines of evidence strongly suggest that L1 activity has ceased in the megabats. Although the deep nodes of megabat phylogenies are not well resolved at this time (Hollar and Springer 1997; Alvarez et al. 1999; Giannini and Simmons 2005; Teeling et al. 2005), it is clear that the species used in this study represent a broad phylogenetic sampling of the group. Our results thus suggest a single extinction event in a common ancestor of all the megabats.
Comparison of the megabat L1 extinction with L1 activity in other species:
This is the second demonstrated case of L1 extinction; the first occurred in a large group of South American rodents, the sigmodontines (Casavant et al. 2000; Grahn et al. 2005). Different mammalian species show variations in rate of L1 deposition (Furano 2000), but there have been very few other reports of either loss of L1 activity or drastic reductions in activity among mammalian species. Putative L1 extinction events or quiescence of activity have been reported for deer mice and voles (Kass et al. 1992; Vanlerberghe et al. 1993), primates (Boissinot et al. 2004), and members of the Afrotherian and Xenarthran mammals (Waters et al. 2004). We have examined these putative events, and in each case our ORF screening technique has yielded closely related L1s that were ORF+ over the region of analysis (Grahn 2004). Thus, while some of these other cases may represent L1 quiescence, L1 extinction has not been convincingly demonstrated except in sigmodontines and now the megabats. In addition, we have determined the phylogenetic limits of each of these events by examining many genera within the two groups.
It is intriguing that both the megabat L1 sequences analyzed here and the same region in the elements isolated in the same manner from the L1-inactive rodents (Grahn et al. 2005) show such similar average sequence distances from their ancestors of 0.089 and 0.088 per site, respectively. While it might become difficult to detect very recent L1 extinctions because elements have not yet built up enough changes to lose ORFs, older extinctions should be readily detectable. Is the similar divergence in these two extinctions a coincidence, or might there be some limitation on divergence of L1 elements from an active ancestor before loss of a putative host function? If the latter is the case, then loss of L1 activity might be a positive event in the short term but a deleterious event for the host when played out over evolutionary time.
The distributions of sequence distances in each extinction event are also surprisingly similar. There is no skew in the distributions toward elements less divergent than the mean, making it unlikely that this similarity is an artifact of the technique used for isolation of the sequences. Thus, the low variance and similarity in the distributions of sequence distances implies loss of activity at the same evolutionary rate in the two independent events and raises the possibility that L1s in both situations suffered sudden extinctions.
L1 lineages and lineage divergence:
The vast majority of L1 elements are deposited as inactive pseudogenes which originate from the small number of active copies present in the genome at any one time (Furano 2000; Brouha et al. 2003). Multiple studies suggest that the active master elements give rise to families of related elements, and as older active elements are replaced by more recently deposited ones, successive subfamilies give rise to long-term L1 lineages (Casavant et al. 1998; DeBerardinis et al. 1998; Boissinot et al. 2004; Khan et al. 2006). It is quizzical that most mammalian species contain single long-term lineages in spite of the presence of multiple active elements. Khan et al. (2006) have shown that human ancestors contained multiple L1 lineages at times in the past, yet even in that case, multiple lineages eventually became extinct, giving rise to the present situation in humans of a single active lineage. Might there be constraints on the divergence attained between multiple active elements or multiple coexisting lineages before selection leads to lineage loss or return to a single lineage?
Single L1 lineages are reminiscent of the phylogenies of influenza (Buonagurio et al. 1986), raising the possibility that L1 evolution is dominated by some sort of arms race. One possibility is that the increasing divergence between active elements of separate, coexisting lineages results in higher transposition rates and increased selective pressure on the host to control L1 retrotransposition. From another viewpoint, competition between L1 elements in separate lineages may normally lead to long-term survival of only one lineage. Constraints on L1 lineage divergence could also arise if L1s indeed contain recognition sequences for propagation of X chromosome inactivation (Lyon 1998). Such scenarios lead to a prediction for limited divergence between lineages and invite examination of present and past divergences.
The published maximum ORF2 divergence between extant, coexisting L1 lineages was seen in Peromyscus maniculatus (Casavant et al. 1996). Young elements from each of the two lineages within that species had an adjusted sequence distance of 0.129 per site. Analysis of Khan's human L1 consensus sequences (Khan et al. 2006) suggests that the region studied here may have diverged in ancient coexisting lineages by an adjusted sequence distance of as much as 0.22 per site. Since the megabat lineage-specific ancestral sequences were reconstructed using the most recently inserted elements from each lineage, they give a snapshot of divergence between the two lineages just before L1 extinction. The striking sequence distance seen of 0.296 per site is over twice that found in P. maniculatus and significantly above the distances seen in ancient human lineages. This result shows that relatively large divergence in active L1 parasites within a host can be tolerated, but it remains unclear whether that divergence may have contributed to the demise of the L1 lineages in the megabats.
An interesting similarity between the two lineages in P. maniculatus and the two in the megabats is that in both cases, the lineage which appears to be more prolific is the lineage which is evolving more rapidly, as seen by the longer internal branches in the clades containing more elements. This may reflect the inherently high mutation rates elicited by reverse transcription as the more active lineage replaces its master elements more often, or it may reflect a more error-prone reverse transcriptase in the more active lineage.
Neutral substitution rate in megabats:
Nucleotide substitution rates have been estimated for a number of mammalian species, but we are unaware of any such estimates for the Chiroptera. Our identification of a large number of L1 elements deposited near the time of L1 extinction in the megabats gives a collection of neutrally evolving pseudogenes. By linking the divergence of these elements from their last common ancestor to the extinction time of L1s, we have estimated a range for the neutral substitution rate in megabats of 0.15–0.37%/MY (95% credibility interval, 0.14–0.44%/MY). This range is necessarily broad because we do not know where along the branch leading to the most recent common ancestor of the megabats the L1 extinction occurred. The range recognizes that L1 extinction could have occurred as recently as the divergence time of megabat species from each other or as far back as the divergence of the megabat lineage from the rest of the Yinpterochiroptera (Figure 1). This rate can be compared to the neutral substitution rates in human, mouse, and rat (Rat Genome Sequencing Project Consortium 2004), if the divergence of human from these rodent lineages is assumed to be 87 MYA and the divergence of mouse from rat is assumed to be 9–23 MYA (Adkins et al. 2001, 2003; Springer et al. 2003; Steppan et al. 2004). These divergence times give a relatively slow human rate of 0.15%/MY, and faster mouse and rat rates of 0.36–0.92%/MY, and 0.40–1.01%/MY, respectively. An independent estimate of megabat mutation rate would allow a better estimate of the timing of L1 extinction. However, given the virtually identical amount of divergence of elements after L1 extinction, and using the timing of L1 extinction in sigmodontine rodents and megabats (Figure 1), we can estimate that the neutral mutation rate in sigmodontine rodents is at least twofold higher than in the megabats.
Possible factors contributing to L1 extinction:
What factors might have led to the L1 extinction seen here? Was it a stochastic process, or was there some change in selective forces that increased the probability of an extinction? The genomes of bats are known to be smaller than the other major groups of mammals (Bachmann 1972; Redi et al. 2005). For example, the average haploid genome size (C-value) of the 83 Chiropteran species present in the Animal Genome Size Database, Release 2.0 (Gregory 2006) is 2.55 pg, while the C-values of the 517 other mammalian species within that database average 3.57 pg. When the two suborders of the Chiroptera within that database are examined, the 7 species of Yinpterochiroptera, which include the megabats, are found to have an average C-value of 2.16 pg, while the 76 species of Yangochiroptera have an average C-value of 2.59 pg. It is known that retroelements are major contributors to increases in genome size in mammals (Kidwell 2002), so it is reasonable that reduction in L1 activity might have been a factor contributing to reductions in genome size in the bats. Changes in L1 activity would also affect deposition of SINEs and pseudogenes, which are dependent upon the L1 molecular machinery. If the reduced genome size in the Chiroptera, and still greater reduction in C-value among the Yinpterochiroptera, reflect novel selective constraints within those groups, then loss of L1 activity may have been a route to higher fitness in an ancestor of the megabats.
We find it quite striking that on an evolutionary scale, both megabat L1 lineages died out at the same time. One might initially consider the possibility that a drastic reduction in population size in the lineage leading to the megabats led to loss of all active elements from the genome by genetic drift. However, even though the number of active elements is low relative to the total L1 copy number, there are still 100 to thousands of active elements per genome, at least in human and mouse (Sassaman et al. 1997; Brouha et al. 2003). Thus, the number of active elements in each genome constitutes a much higher effective population size for the L1s than for the host species and it seems unlikely for all active L1 elements to be lost solely due to a population bottleneck while the species itself survived.
An alternative reason for loss of L1 activity might have been mutation in the most active elements themselves, leading to reduced retrotransposition of those elements which produce the majority of progeny in each lineage. Such a shift in the delicate balance between sufficient levels of L1 activity to ensure production of new master elements before old masters become inactive due to mutation vs. host control against increased proliferation, could lead to loss of L1 activity. However, this explanation seems unlikely because it would require two separate sets of lineage-specific mutations to have arisen at roughly the same time to extinguish both lineages simultaneously.
A more parsimonious scenario for the simultaneous loss of activity in both lineages is a mutation in the host control machinery, resulting in greater repression of all L1 retrotransposition. It therefore appears likely that the loss of L1 activity seen in the megabats was due to changes in a host control system.
Are there common features in the genomes of these L1-inactive groups and their sister taxa retaining L1 activity that might shed light on the interplay of L1s with their hosts? We have identified a new family of endogenous retroviruses called mysTR elements in the L1-inactive sigmodontine rodents and their phylogenetic neighbors (Cantrell et al. 2005). Recently deposited mysTR elements are present at ∼1000–10,000 copies per genome in these species—unprecedented levels for an endogenous retrovirus family with such closely related members—but it is not clear whether there is a correlation between higher mysTR copy numbers and loss of L1 activity. It will be interesting to see if loss of L1 activity in the megabats is correlated with unusual deposition of any other retroelements.
Although transposable elements have classically been viewed as selfish parasites, there is an increasing perspective that it may be more accurate to consider the entire coevolutionary spectrum that different elements may occupy: from parasite to mutualist providing essential host functions (Miller et al. 1999; Kidwell and Lisch 2000; Gregory 2005). Our identification of model systems such as the megabats, and dissection of the components of their genomes, should allow us to clarify the placement of L1s within this spectrum, and as a result increase our understanding of how they affect their mammalian hosts. Such model systems may also help distinguish between potential roles played by the activity of L1 elements vs. the role of their sequences as part of the architecture of mammalian genomes.
Acknowledgments
We thank Robert J. Baker for aid in obtaining tissues used in this study. We are indebted to Darin R. Rokyta and Issac K. Erickson for helpful discussions and aid in phylogenetic analyses. This study was supported by a grant from the National Institutes of Health (GM38737 to H.A.W.). Analytical resources were provided by the Idea Network of Biomedical Research Excellence (INBRE, RR016454) and the Center of Biomedical Research Excellence (COBRE, RR016448) grants from the National Institutes of Health.
References
- Adkins, R. M., E. L. Gelke, D. Rowe and R. L. Honeycutt, 2001. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol. Biol. Evol. 18: 777–791. [DOI] [PubMed] [Google Scholar]
- Adkins, R. M., A. H. Walton and R. L. Honeycutt, 2003. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol. Phylogenet. Evol. 26: 409–420. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, Y., J. Juste, E. Tabares, A. Garrido-Pertierra, C. Ibanez et al., 1999. Molecular phylogeny and morphological homoplasy in fruitbats. Mol. Biol. Evol. 16: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1989. Current Protocols in Molecular Biology. Wiley-Interscience, New York.
- Bachmann, K., 1972. Genome size in mammals. Chromosoma 37: 85–93. [DOI] [PubMed] [Google Scholar]
- Beraldi, R., C. Pittoggi, I. Sciamanna, E. Mattei and C. Spadafora, 2006. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 73: 279–287. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., C. Roos and A. V. Furano, 2004. Different rates of LINE-1 (L1) retrotransposon amplification and evolution in New World monkeys. J. Mol. Evol. 58: 122–130. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., J. Davis, A. Entezam, D. Petrov and A. V. Furano, 2006. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl. Acad. Sci. USA 103: 9590–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley et al., 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonagurio, D. A., S. Nakada, J. D. Parvin, M. Krystal, P. Palese et al., 1986. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science 232: 980–982. [DOI] [PubMed] [Google Scholar]
- Cantrell, M. A., R. A. Grahn, L. Scott and H. A. Wichman, 2000. Isolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques 29: 1310–1316. [DOI] [PubMed] [Google Scholar]
- Cantrell, M. A., M. M. Ederer, I. K. Erickson, V. J. Swier, R. J. Baker et al., 2005. MysTR: an endogenous retrovirus family in mammals undergoing recent amplifications to unprecedented copy numbers. J. Virol. 79: 14698–14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant, N. C., A. N. Sherman and H. A. Wichman, 1996. Two persistent LINE-1 lineages in Peromyscus have unequal rates of evolution. Genetics 142: 1289–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant, N. C., R. N. Lee, A. N. Sherman and H. A. Wichman, 1998. Molecular evolution of two lineages of L1 (LINE-1) retrotransposons in the California mouse, Peromyscus californicus. Genetics 150: 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant, N. C., L. Scott, M. A. Cantrell, L. E. Wiggins, R. J. Baker et al., 2000. The end of the LINE?: lack of recent L1 activity in a group of South American rodents. Genetics 154: 1809–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis, R. J., J. L. Goodier, E. M. Ostertag and H. H. Kazazian, Jr., 1998. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat. Genet. 20: 288–290. [DOI] [PubMed] [Google Scholar]
- Deininger, P. L., M. A. Batzer, C. A. Hutchison, III and M. H. Edgell, 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8: 307–311. [DOI] [PubMed] [Google Scholar]
- Furano, A. V., 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acid Res. Mol. Biol. 64: 255–294. [DOI] [PubMed] [Google Scholar]
- Giannini, N. P., and N. B. Simmons, 2005. Conflict and congruence in a combined DNA-morphology analysis of megachiropteran bat relationships (Mammalia: Chiroptera: Pteropodidae). Cladistics 21: 411–437. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., S. Lutz-Prigge and J. V. Moran, 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325. [DOI] [PubMed] [Google Scholar]
- Goodier, J. L., E. M. Ostertag and H. H. Kazazian, Jr., 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9: 653–657. [DOI] [PubMed] [Google Scholar]
- Grahn, R. A., 2004. Evolutionary dynamics of LINE-1 transposable elements in rodents. Ph.D. Thesis, University of Idaho, Moscow, ID.
- Grahn, R. A., T. A. Rinehart, M. A. Cantrell and H. A. Wichman, 2005. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 110: 407–415. [DOI] [PubMed] [Google Scholar]
- Gregory, T. R., 2005. The Evolution of the Genome. Elsevier Academic Press, Burlington, MA.
- Gregory, T. R., 2006. Animal Genome Size Database. http://www.genomesize.com.
- Han, J. S., and J. D. Boeke, 2005. LINE-1 retrotransposons: Modulators of quantity and quality of mammalian gene expression? BioEssays 27: 775–784. [DOI] [PubMed] [Google Scholar]
- Han, J. S., S. T. Szak and J. D. Boeke, 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268–274. [DOI] [PubMed] [Google Scholar]
- Hedges, D. J., and M. A. Batzer, 2005. From the margins of the genome: mobile elements shape primate evolution. BioEssays 27: 785–794. [DOI] [PubMed] [Google Scholar]
- Hollar, L. J., and M. S. Springer, 1997. Old World fruitbat phylogeny: evidence for convergent evolution and an endemic African clade. Proc. Natl. Acad. Sci. USA 94: 5716–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hutchison, III, C. A., S. C. Hardies, D. D. Loeb, W. R. Shehee and M. H. Edgell, 1989. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome, pp. 593–617 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, DC.
- International Human Genome Sequencing Consortium, 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Kass, D. H., F. G. Berger and W. D. Dawson, 1992. The evolution of coexisting highly divergent LINE-1 subfamilies within the rodent genus Peromyscus. J. Mol. Evol. 35: 472–485. [DOI] [PubMed] [Google Scholar]
- Khan, H., A. Smit and S. Boissinot, 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., and D. R. Lisch, 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15: 95–99. [DOI] [PubMed] [Google Scholar]
- Longmire, J. L., A. K. Lewis, N. C. Brown, J. M. Buckingham, L. M. Clark et al., 1988. Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2: 14–24. [DOI] [PubMed] [Google Scholar]
- Lyon, M. F., 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80: 133–137. [DOI] [PubMed] [Google Scholar]
- Maddison, D. R., and W. P. Maddison, 2003. MacClade 4.0, Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA.
- Miller, W. J., J. F. McDonald, D. Nouaud and D. Anxolabehere, 1999. Molecular domestication–more than a sporadic episode in evolution. Genetica 107: 197–207. [PubMed] [Google Scholar]
- Minin, V., Z. Abdo, P. Joyce and J. Sullivan, 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52: 674–683. [DOI] [PubMed] [Google Scholar]
- Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato et al., 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31: 159–165. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Muotri, A. R., V. T. Chu, M. C. Marchetto, W. Deng, J. V. Moran et al., 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. [DOI] [PubMed] [Google Scholar]
- Pickeral, O. K., W. Makalowski, M. S. Boguski and J. D. Boeke, 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium, 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521. [DOI] [PubMed] [Google Scholar]
- Redi, C. A., H. Zacharias, S. Merani, M. Oliveira-Miranda, M. Aguilera et al., 2005. Genome sizes in afrotheria, xenarthra, euarchontoglires, and laurasiatheria. J. Hered. 96: 485–493. [DOI] [PubMed] [Google Scholar]
- Sassaman, D. M., B. A. Dombroski, J. V. Moran, M. L. Kimberland, T. P. Naas et al., 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16: 37–43. [DOI] [PubMed] [Google Scholar]
- Sawyer, S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Scott, L., A. Kuroiwa, M. Matsuda and H. A. Wichman, 2006. X accumulation of LINE-1 retrotransposons in Tokudaia osimensis, a spiny rat with the karyotype XO. Cytogenet. Genome Res. 112: 261–269. [DOI] [PubMed] [Google Scholar]
- Springer, M. S., W. J. Murphy, E. Eizirik and S. J. O'Brien, 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100: 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan, S., R. Adkins and J. Anderson, 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53: 533–553. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost et al., 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327–338. [DOI] [PubMed] [Google Scholar]
- Teeling, E. C., M. S. Springer, O. Madsen, P. Bates, S. J. O'Brien et al., 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307: 580–584. [DOI] [PubMed] [Google Scholar]
- Teng, S. C., B. Kim and A. Gabriel, 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383: 641–644. [DOI] [PubMed] [Google Scholar]
- Van den Bussche, R. A., J. L. Longmire and R. J. Baker, 1995. How bats achieve a small C-value: frequency of repetitive DNA in Macrotus. Mamm. Genome 6: 521–525. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, F., F. Bonhomme, C. A. Hutchison, III and M. H. Edgell, 1993. A major difference between the divergence patterns within the lines-1 families in mice and voles. Mol. Biol. Evol. 10: 719–731. [DOI] [PubMed] [Google Scholar]
- Waters, P. D., G. Dobigny, A. T. Pardini and T. J. Robinson, 2004. LINE-1 distribution in Afrotheria and Xenarthra: implications for understanding the evolution of LINE-1 in eutherian genomes. Chromosoma 113: 137–144. [DOI] [PubMed] [Google Scholar]