Abstract
Grain yield is a major goal for the improvement of durum wheat, particularly in drought-prone areas. In this study, the genetic basis of grain yield (GY), heading date (HD), and plant height (PH) was investigated in a durum wheat population of 249 recombinant inbred lines evaluated in 16 environments (10 rainfed and 6 irrigated) characterized by a broad range of water availability and GY (from 5.6 to 58.8 q ha−1). Among the 16 quantitative trait loci (QTL) that affected GY, two major QTL on chromosomes 2BL and 3BS showed significant effects in 8 and 7 environments, with R2 values of 21.5 and 13.8% (mean data of all 16 environments), respectively. In both cases, extensive overlap was observed between the LOD profiles of GY and PH, but not with those for HD. QTL specific for PH were identified on chromosomes 1BS, 3AL, and 7AS. Additionally, three major QTL for HD on chromosomes 2AS, 2BL, and 7BS showed limited or no effects on GY. For both PH and GY, notable epistasis between the chromosome 2BL and 3BS QTL was detected across several environments.
DURUM wheat (Triticum durum Desf.) is an important crop for the human diet (e.g., pasta, couscous, bread, etc.), particularly in the Mediterranean basin where ∼75% of the world's durum grain is produced. Durum wheat is primarily grown under rainfed conditions where the frequent occurrence of drought combined with heat stress is the major factor limiting grain yield (Araus et al. 2002, 2003a,b; Condon et al. 2004). In the Mediterranean basin, durum wheat is cultivated across a number of macroenvironments that differ widely in the quantity of rainfall as well as in their thermo-pluviometrical patterns during the crop cycle (Leemans and Cramer 1991; Loss and Siddique 1994; Dunkeloh and Jacobeit 2003).
As compared to hexaploid wheat, durum wheat underwent a more limited selection until 1960, when more intense breeding programs based on innovative germplasm introgressions and multienvironment testing for wide adaptation were applied also to durum wheat. Accordingly, the genetic gains obtained after 1970 in grain yield (GY) of durum wheat are comparable to those obtained for hexaploid wheat. These gains have mainly been attributed to a balanced improvement in fertility because of higher allocation of assimilates to the growing tillers and ears concomitant with a general increase in total biomass production, with the harvest index remaining practically unchanged (Slafer and Andrade 1993; Slafer et al. 1996; Pfeiffer et al. 2000; De Vita et al. 2007; Slafer and Araus 2007). As suggested by Pfeiffer et al. (2000), GY components have reached a near-optimal balance in modern elite durums. While the improvement of GY under optimal growing conditions has prevailingly been attributed to increased spike fertility, under Mediterranean-like conditions the importance of traits at the basis of growth plasticity, such as early vigor and a finely tuned heading date that allows the plant to escape from terminal drought, has been universally recognized (Richards 2000; Spielmeyer et al. 2007).
The molecular tools necessary to identify the quantitative trait loci (QTL) governing GY and genotype × environment (G × E) interaction are now available for most crops, including wheat (Tuberosa et al. 2002a; Borevitz and Chory 2004; Qi et al. 2004; Dilbirligi et al. 2006; Maccaferri et al. 2006; Song et al. 2007). In the past decade, microsatellite markers (simple sequence repeats, SSRs) have been extensively exploited for the construction of wheat maps aligning hundreds of SSRs (Röder et al. 1998; Nachit et al. 2001; Blanco et al. 2004), the Consensus Ta-SSR-2004 map (Somers et al. 2004), and the Wheat-Composite 2004 map (http://wheat.pw.usda.gov/ggpages/map_summary.html).
Several QTL experiments have been carried out in cereals to unravel the genetic basis of GY and the morphophysiological traits known to determine yield under nonstressed and stressed conditions. Reviews summarizing the findings of QTL studies have been published for barley (Cattivelli et al. 2002), wheat (Gupta et al. 1999, 2006), rice (Zeng et al. 2006), and maize (Tuberosa et al. 2002b; Schaeffer et al. 2006). For each crop, tens of QTL have been found in different genetic backgrounds. This notwithstanding, the knowledge gained so far for grain yield determinants is still incomplete. In fact, QTL for yield and yield-related traits most frequently account for between ∼2 and 10% of the total phenotypic variation; major QTL with R2 values ≥15% have seldom been described, especially when evaluating segregating materials obtained from elite accessions (Quarrie et al. 2005; Dilbirligi et al. 2006). The identification of QTL with major and environmentally stable additive effects is even more desirable when targeting drought-prone environments where the spatial and temporal phenotypic variation (including G × E effects) is usually larger than that observed in favorable environments (Lanceras et al. 2004), a condition that lowers the heritability of target traits. Under such a contrasting scenario, the identification of major QTL characterized by a limited G × E is highly desirable to enhance productivity and facilitate their cloning (Bortiri et al. 2006; Tuberosa and Salvi 2006). Accordingly, the stable expression of a QTL across a broad range of agrometeorological conditions is a critical factor when breeding for wide adaptation and yield stability. Examples of such QTL have been reported in bread wheat (Quarrie et al. 2007), rice (Wan et al. 2005, 2006), pearl millet (Yadav et al. 2002), and maize (Landi et al. 2007).
The genetics of GY and other agronomically important traits (e.g., heading date, plant height, etc.) are frequently complicated by the occurrence of epistatic interactions among the multiple QTL/genes controlling the target trait (Li et al. 1997, 2001, 2003; Kusterer et al. 2007). The relevance of the epistatic interactions can reach levels comparable to those observed for the additive QTL effects. Suggestions for planning and conducting QTL analysis able to more accurately detect epistatic interactions have been presented (Malmberg and Mauricio 2005; Zeng 2005; Stich et al. 2007). An essential prerequisite to ensure robust inferences on the epistatic interactions among QTL is the availability of mapping populations with a sufficiently large number of progenies (Schön et al. 2004).
In this study, a durum wheat population of 249 recombinant inbred lines (RILs) was evaluated in 16 Mediterranean environments to identify QTL for grain yield, heading date, and plant height. The RILs were tested across a wide range of growing conditions, with different thermo-pluviometrical patterns and especially water availability from heading to harvest. We report on the identification of two major QTL for grain yield and on the importance of their epistatic interaction.
MATERIALS AND METHODS
Plant materials:
A population of 249 RILs was produced by Società Produttori Sementi (Bologna, Italy), applying the single-seed descent method to progenies (from F2 to F6 generation) of the cross between the cultivars (cvs.) Kofa and Svevo. Seeds were bulked in the F7 generation. Kofa, a Southwestern United States cv. released by Western Plant Breeders (Arizona) was obtained from a population based on multiple parents (dicoccum alpha pop-85 S-1) mainly related to the American and CIMMYT germplasm, with the inclusion of emmer accessions. Svevo, an Italian cv. released by Società Produttori Sementi, has been obtained from the cross between a CIMMYT line (pedigree rok/fg//stil/3/dur1/4/sapi/teal//hui), related to the widely utilized Yavaros79 genetic background (Jori/Anhinga//Flamingo), and the cv. Zenit originating from a cross between Italian and American accessions (Valriccardo/Vic). Both Kofa and Svevo are well adapted to the Mediterranean climate and can be classified as early-flowering genotypes in such conditions.
Field trials and phenotypic traits:
Sixteen field trials were carried out over 2 years in different Mediterranean locations (8 trials in 2004 and 8 trials in 2005) in the following countries: Italy, Spain, Morocco, Tunisia, Syria, and Lebanon. Abbreviations have been used to identify the field experiments: the first three letters indicate the geographical location of the trial [Italy, Itl (Cerignola); Lebanon, Lbn (Tel Amara); Morocco, Mrc (Sidi El Aidi); Spain, Spn (Spn1 for Granada and Spn2 for Lleida); Syria, Syr (Tel Hadya); and Tunisia, Tns (Koudiat and Kef for the irrigated and rainfed trials in 2004, respectively]; the fourth letter (separated by a hyphen from the first three letters) indicates the water regime of the trial (irrigated, i; and rainfed, r), while the number indicates the year of the trial (2004, 04; and 2005, 05). Details on the locations and experimental conditions of each field trial are reported as supplemental data (supplemental data sets 1 and 2 at http://www.genetics.org/supplemental/) (Figure 1 and Table 1). The analysis of these supplemental data indicates that a broad range of environmental conditions was explored. Large differences among trials were evident for water input (rainfall plus supplementary irrigations), particularly from heading to harvest, with water input ranging from 8 to 281 mm. During the critical stage of grain filling, the total water input ranged from 0 to 159 mm. Soil moisture averaged over the grain-filling stage also varied largely among environments (from 5.7 to 23.0%). Consequently, a wide variation in crop-cycle length was observed across environments (for instance, the emergence-to-heading period ranged from 93 to 143 days).
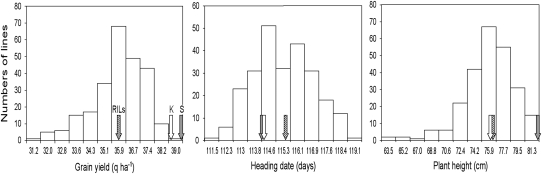
Figure 1.—
Frequency distributions of grain yield (GY), heading date (HD), and plant height (PH) of the Kofa × Svevo RILs based on the mean values across 16 Mediterranean environments. Means for RILs, Kofa (K), and Svevo (S) are indicated with arrows.
TABLE 1.
Mean phenotypic values of parents and RILs and heritability values across environments
| Parental cvs.a
|
RILsb
|
||||
|---|---|---|---|---|---|
| Trait | Svevo | Kofa | Mean | Mean | h2 |
| Grain yield (q ha−1) | 39.6 | 39.0 | 39.3 | 35.9 | 0.67 |
| Heading datec (days) | 114.3 | 114.2 | 114.3 | 115.3 | 0.95 |
| Plant height (cm) | 79 | 76 | 78 | 76 | 0.91 |
Means of the parental cultivars (Svevo and Kofa) and means and heritability values (h2) of the 249 RILs for grain yield, heading date, and plant height are shown. The summary statistics were calculated using the data from the 16 environments.
Statistics calculated from the average of 20 replicated plots per experiment, as in the augmented design experimental scheme adopted in this study.
Statistics calculated from the best linear unbiased predictor data (field experiments with unreplicated plots).
Days from emergence.
The following phenological variables and environmental factors have been considered: (i) phenology, with two variables, length of the periods from emergence to heading date and from heading to harvest; (ii) water available to the crop, including rainfall and irrigations, from emergence to heading, from heading to harvest, and, most critical under Mediterranean environments, during the grain-filling stage (from 2 weeks after heading to physiological maturity); (iii) thermal range at heading (maximum and mean temperatures averaged over the 10 days around heading) and grain filling, as defined above; (iv) thermal time to heading, from heading to harvest, and across grain filling, as calculated in growing degree days (GDD) by cumulating mean daily temperatures and considering a base temperature of 0° (Gallagher 1979); and (v) average soil moisture (0- to 30-cm deep) during the grain-filling stage.
The RILs were tested in unreplicated field trials, adopting a modified augmented design as a field experimental scheme, including three checks (cvs. Kofa, Svevo, and Vitron) distributed in each row of the field scheme. Vitron is a high-yielding cv. developed from CIMMYT germplasm (cross Jori/Anhinga//Flamingo), released in 1985, and characterized by high-yield stability in the Mediterranean basin (Pfeiffer et al. 2000). Seeds of the RILs and checks used in the field trials were increased in a single location (Lucera, Italy).
Before planting, seed viability was determined on a sample of 100 seeds/RIL. On the basis of these results, sowing was carried out with 400 viable seeds m−2. To prevent attacks from seed-transmitted fungal diseases, seed was treated with Vitavax FLO NF (Carboxin plus Thiram). Plot size was 4 m2 (eight 2.5-m-long rows, spaced 0.20 m apart). Trials were fertilized following the standard agricultural practices for each location and were treated with fungicides to avoid the development of fungal pathogens; weeds were chemically and mechanically controlled. To ensure an adequate protection against the various diseases and weeds affecting wheat, chemical treatments were carried out as necessary with the fungicide and herbicide active compounds recommended by the standard agriculture practices in each location and country.
Field data:
The following traits were considered: GY (in quintals per hectare, adjusted at 14% moisture), heading date (HD) (in days), and plant height (PH) (in centimeters). Heading date was recorded as the number of days from the emergence to the time when the ears of ∼50% of the tillers had emerged from the flag leaf sheaths for approximately half of their length (stage 55 in the Zadoks scale; Zadoks et al. 1974). At maturity, PH was measured from the ground to the tip of the ear (excluding awns) on five main culms per plot.
Genetic map:
The SSR markers publicly available in GrainGenes (http://wheat.pw.usda.gov) were used to search for parental polymorphisms. The SSR probe sets used were BARC (Xbarc marker loci), CFA (Xcfa), CFD (Xcfd), CNL, KSUM, WMC (Xwmc), and WMS (Xgwm). A WMS primer set not publicly available was provided by Martin Ganal, TraitGenetics, Gatersleben, Germany. Preference was given to markers included in the wheat SSR consensus maps [the Ta-SSR-2004 (Somers et al. 2004), the Ta-Synthetic/Opata-BARC (Song et al. 2005), and the Wheat-Composite 2004 (see http://wheat.pw.usda.gov/)]. More than 800 markers were tested to detect polymorphisms between Kofa and Svevo. SSR profiles of the two parents were evaluated in high-resolution agarose gels (2% SeaKem LE agarose plus 1% MetaPhor agarose gels, both from Cambrex Bio Science Rockland, Rockland, ME), 6 mm in thickness, or in 5% polyacrylamide manual sequencing gels (45 cm long and 0.4 mm thick) and stained according to the silver-nitrate method. A unique thermocycling protocol was used for all probes: 94° for 3 min; 20 cycles of touchdown PCR including 94° for 45 sec, 61°/51° for 45 sec (−0.5° sec−1), and 72° for 60 sec; followed by 23 cycles including 94° for 45 sec, 51° for 45 sec, and 72° for 60 sec, with a final extension at 72° for 10 min. SSRs selected on the basis of base pair differences between the two parental alleles were profiled on the entire RIL population using either 3% agarose gel electrophoresis or the automated Li-Cor (Lincoln, NE) 4200 IR2 System with forward primers labeled with IR700/IR800 fluorochromes and 25-cm-long, 0.4-mm-thick polyacrylamide gels.
Only SSRs with a percentage of missing marker data points <8% were considered. After inspecting the grouping results obtained with LOD thresholds from 2 to 8, markers were grouped using a minimum conservative LOD threshold of 5.0. The majority of groups was then assigned to the A and B wheat chromosomes using the information from the publicly available wheat SSR maps and from an updated version of the WMS map (M. Ganal, unpublished results). Linkage groups assigned to each chromosome were rechecked for association using relaxed LOD thresholds (from 2 to 5) and then merged accordingly.
Haldane's mapping function was used to calculate map distances. Linkage groups were constructed with the linkage software JoinMap 3.0 on the basis of a regression mapping procedure with a weighted least-squares method that sequentially adds markers into the map (Stam 1993). Briefly, only data with recombination frequency <0.45 and LOD >1.0 were used; the “jump in goodness-of-fit” threshold for locus removal was set to 3.0; and the “ripple” command was used each time after adding a locus to the linkage group and three “mapping rounds” were performed for each linkage group. The graphical genotype of each RIL was then inspected for the presence of suspect data points (double-recombination events within short distances) and the original data were checked accordingly. Subsequently, a second round of mapping with JoinMap 3.0 was performed to obtain the final map distances. Marker order was also checked with the program “Carthagene,” using all the available computational options (Schiex and Gaspin 1997). In some cases with reliable linkage groups but high recombination frequencies among markers (e.g., chromosomes 2B, 3A, 5B, and 7A), the LOD threshold for mapping was lowered and the recombination threshold was increased. In some cases, the “fixed marker order” function of Joinmap was used to produce the map of each linkage group on the basis of the consensus SSR published data and of the most probable marker order output from Carthagene (I. Jurman et al., unpublished results). A total of 254 SSRs were profiled on the complete RIL set and grouped into 23 linkage groups. Cosegregating markers mapping within a 1-cM interval were excluded from the final map used to perform QTL analysis and only one marker for each cluster was retained. The final molecular map used for the QTL analysis was based on 232 SSRs distributed on 23 linkage groups, covering in total 2347 cM (Haldane's mapping function), with an average marker distance of 10.2 cM. On the basis of the Ta-SSR-2004 map (Somers et al. 2004) for the A and B genomes, we estimate that our map covers ∼70% of the durum genome. All SSR markers assayed in the remaining portion of the genome (mainly at the distal regions of chromosomes 1AS, 2AL, 2BS, 3AS, and 5BS) were found monomorphic; it is conceivable that the lack of polymorphism at these regions is prevailingly due to identity by descent. In this respect, it is interesting to note that a low marker density is observed in the most distal regions, where a high recombination rate is a general feature of the wheat chromosomes (Somers et al. 2004); the number of SSRs mapped in these distal and highly recombinogenic regions was relatively low, as compared to the hundreds of SSRs mapped in the proximal regions of the wheat chromosomes.
Statistical analysis of phenotypic data and QTL analysis:
Phenotypic data were analyzed by restricted maximum likelihood (REML) to fit a mixed model with checks as a fixed effect and rows, columns, and unreplicated entries as random effects (Littel et al. 1996). The REML model produced best linear unbiased predictors (BLUPs) for the phenotypic data of each genotype at each environment to be used in subsequent analyses. The analysis was performed using the MIXED procedure of the SAS statistical package (SAS 2001).
The heritability value (h2) was calculated for each trait across environments as
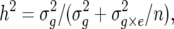 |
where n is the number of environments,
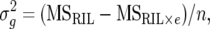 |
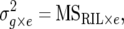 |
and MS is the mean square. Correlations between environmental factors and phenotypic traits as well as among traits and/or among environments for each trait and their significance levels were calculated as Pearson's correlation coefficient values using Minitab v. 14.0 (Minitab statistical software; Ryan et al. 1985).
Composite-interval mapping (CIM) (Zeng 1994) was used to search for QTL using the BLUP data of each trait separately for (i) each of the 16 environments, (ii) the average value across environments in each year (2004 and 2005), and (iii) the average value across all the environments. CIM analysis was performed in Windows QTL-Cartographer version 2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm; Basten et al. 2005; Wang et al. 2005). The parameter setup of “model 6 standard analysis” in QTL Cartographer was used: a walk speed of a 2-cM step, “forward” regression for the selection of the markers to control for the genetic background (control markers or cofactors), up to 10 control markers, and a blocked window size of 10 cM to exclude closely linked control markers at the testing site.
The threshold for declaring the presence of a significant QTL for each trait–environment combination was defined by 1000 permutations at P ≤ 0.10 (Churchill and Doerge 1994) and the minimum LOD score of 2.9 was chosen. Additionally, the QTL with a LOD score reaching a lower threshold of 2.5 have been reported as “suggested QTL”, provided that their peaks map approximately at the same position in at least two environments.
The QTL were identified adopting the nomenclature suggested by the catalog of gene symbols for wheat (http://wheat.pw.usda.gov/ggpages/wgc/98).
The value of the additive effect at each QTL was computed as half of the difference between the mean phenotypic values of the two groups of RILs, which, based on the information of the flanking markers, were assumed to be homozygous for one or the other of the parental alleles at that QTL region. In particular, the additive QTL effect (a) was defined as  (Svevo − Kofa); therefore a was positive when the Kofa allele showed the lower value. QTL detected in different environments were considered to be the same if the estimated map position of their peaks was positioned within 20 cM. To obtain more precise information on QTL effects and positions and to evaluate for the presence of digenic epistatic interactions across the QTL pairwise combinations, multiple-interval mapping (MIM) (Kao et al. 1999; Zeng et al. 1999), as implemented in WinQTLCart, was used by considering as initial QTL models the CIM results obtained for each trait and environment. Thus the initial CIM-derived QTL models were subjected to a refinement of the CIM-QTL positions and to a forward, stepwise-regression search for significant epistatic interactions among all pairwise combinations of QTL. Both main additive effects and their epistatic interactions were tested for significance using the Bayesian information criterion (BIC) with the penalty function c(n) = log(n), with n = 249 (Schwarz 1978; Zeng et al. 1999); the final main additive and epistatic QTL effects, the variance components, and the R2 values of the models were then estimated.
(Svevo − Kofa); therefore a was positive when the Kofa allele showed the lower value. QTL detected in different environments were considered to be the same if the estimated map position of their peaks was positioned within 20 cM. To obtain more precise information on QTL effects and positions and to evaluate for the presence of digenic epistatic interactions across the QTL pairwise combinations, multiple-interval mapping (MIM) (Kao et al. 1999; Zeng et al. 1999), as implemented in WinQTLCart, was used by considering as initial QTL models the CIM results obtained for each trait and environment. Thus the initial CIM-derived QTL models were subjected to a refinement of the CIM-QTL positions and to a forward, stepwise-regression search for significant epistatic interactions among all pairwise combinations of QTL. Both main additive effects and their epistatic interactions were tested for significance using the Bayesian information criterion (BIC) with the penalty function c(n) = log(n), with n = 249 (Schwarz 1978; Zeng et al. 1999); the final main additive and epistatic QTL effects, the variance components, and the R2 values of the models were then estimated.
Epistatic effects were also calculated according to Cheverud and Routman (1995). The two-locus genotypic values (Gijkl) are defined as the average phenotypic values of the RILs homozygous for the i (Svevo, S) or the j (Kofa, K) allele at the first marker locus (referred to as A) and the k (S) or the l (K) allele at the second marker locus (referred to as B). The single-locus genotypic values (Gij.. and G..kl) are defined as the unweighted average of the two genotypic classes at each locus irrespectively from the other locus; thus at locus A Gij.. = (GijSS + GijKK)/2 and at locus B G..kl = (GSSkl + GKKkl)/2. The additive (a) effects were calculated as follows: aA = (GSS.. − GKK..)/2 and aB = (G..SS − G..KK)/2. Assuming no interaction between the considered QTL, the nonepistatic two-locus genotypic values can be estimated as neijkl = G…. + aA + aB; the deviation of the two-locus genotypic values from the nonepistatic ones represents the epistatic effect: eijkl = Gijkl − neijkl.
Multiple-trait composite-interval mapping (Jiang and Zeng 1995) was used to test for the presence of QTL × environment (Q × E) interaction at the main chromosome regions affecting the target traits. The Q × E interaction was tested using a likelihood-ratio statistic developed for the null hypothesis that a1 = a2 = … = aj, where aj is the additive effect of a QTL in the jth environment and implemented in WinQTLCart. The significance of this statistic for a few chromosome regions harboring major QTL was obtained utilizing the 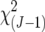 distribution. Tests were carried out by using the data from all the environments as well as from only those environments with significant additive effects at the main QTL regions.
distribution. Tests were carried out by using the data from all the environments as well as from only those environments with significant additive effects at the main QTL regions.
RESULTS
Variation among environments and RILs:
The mean values of the two parents and RILs across the 16 environments are shown in Table 1. The two parents showed a high and rather similar productivity, as expected in the case of elite cvs. well adapted to Mediterranean conditions. Their average performance was similar to that observed for cv. Vitron, chosen as common check in all trials (data not shown) due to its high yield potential and adaptation across Mediterranean environments (Pfeiffer et al. 2000). The broad variation among RILs (Figure 1) and their wide transgressive segregation are in keeping with the rather different pedigree/genetic background of the two parental cvs. Heritability values calculated across environments (Table 1) were similar to or higher than those reported in a similar experiment carried out in hexaploid wheat (Huang et al. 2006). However, it should be noted that in both cases the heritability values were obtained on the basis of an unreplicated experimental design and were thus likely overestimated because the adopted unreplicated experimental design does not allow for a proper estimate of the experimental error.
The detailed statistics on a single-environment basis are reported in Table 2; the environments are listed on the basis of a decreasing order of GY values. The environments can be classified as high yielding (GY > 50 q ha−1 in Lbn-i05, Syr-i04, Itl-r04, Syr-i05, and Itl-r05), medium yielding (GY comprised between 25 and 50 q ha−1 in Syr-05, Syr-04, Spn1-r04, Mrc-i05, Tns-i04, Lbn-r05, and Lbn-i04), and low yielding (GY < 25 q ha−1 in Tns-r04, Lbn-r04, Spn2-r05, and Mrc-r05). The average PH of the RILs approached ∼90 cm in the most favorable conditions, while it was reduced to ∼55 cm in water-stressed conditions.
TABLE 2.
Mean phenotypic values of parents and RILs for each environment
| Grain yield (q ha−1)
|
Heading date (days)c: | Plant height (cm): | ||||
|---|---|---|---|---|---|---|
| Parentsa
|
RILsb
|
RILsb
|
||||
| Environment | Svevo | Kofa | Mean | Range | Mean | Mean |
| Lbn-i05 | 58.6 | 71.1 | 58.8 | 53.6–62.8 | 105.3 | 78.8 |
| Syr-i04 | 58.9 | 58.0 | 57.4 | 54.1–59.9 | 108.7 | 81.3 |
| Itl-r04 | 66.4 | 61.8 | 57.3 | 42.2–66.9 | 107.6 | 82.6 |
| Syr-i05 | 58.9 | 57.4 | 56.8 | 44.7–65.9 | 109.6 | 89.4 |
| Itl-r05 | 57.9 | 57.9 | 54.0 | 44.8–60.7 | 143.2 | 83.1 |
| Syr-r05 | 46.5 | 42.7 | 43.1 | 39.3–45.2 | 133.8 | 82.2 |
| Syr-r04 | 44.8 | 44.4 | 41.7 | 37.0–45.8 | 112.9 | 77.9 |
| Spn1-r04 | 42.0 | 44.5 | 34.8 | 24.7–45.1 | 135.4 | 84.9 |
| Mrc-i05 | 36.0 | 31.2 | 30.9 | 29.1–32.3 | 106.4 | 84.4 |
| Tns-i04 | 38.0 | 31.1 | 29.6 | 26.4–33.2 | 101.6 | 88.9 |
| Lbn-r05 | 29.6 | 31.4 | 28.1 | 26.1–30.5 | 105.7 | 67.4 |
| Lbn-i04 | 31.0 | 32.6 | 26.5 | 7.7–46.0 | 93.4 | 56.7 |
| Tns-r04 | 24.1 | 15.5 | 17.6 | 10.9–26.9 | 126.4 | 86.5 |
| Lbn-r04 | 17.5 | 21.2 | 16.7 | 4.2–30.3 | 94.6 | 53.4 |
| Spn2-r05 | 17.0 | 16.4 | 15.3 | 14.1–16.9 | 140.2 | 53.6 |
| Mrc-r05 | 7.0 | 6.4 | 5.6 | 1.1–16.0 | 100.8 | 64.0 |
| Mean | 39.6 | 39.0 | 35.9 | 30.9–38.7 | 115.3 | 75.9 |
Mean values of parents and RILs for grain yield, heading date, and plant height in the 16 field trials carried out in 2004 and 2005 are shown. Environments are listed according to the decreasing mean grain yield values of the RILs. Environments are indicated by acronyms where the first three letters indicate the locations, followed by the water regime indicated with either i (irrigated) or r (rainfed) and the year 04 (2003/2004) or 05 (2004/2005). Location: Itl, Cerignola, Italy; Lbn, Tel Amara, Lebanon; Mrc, Sidi El Aidi, Morocco; Spn1, Granada, Spain; Spn2, Lleida, Spain; Syr, Tel Hadya, Syria; Tns-i04, Koudiat, Tunisia; Tns-r04, Kef, Tunisia.
Mean values of 20 replicated plots per experiment, as in the augmented design experimental scheme adopted in this study.
Mean and range of the best linear unbiased predictor data (field experiments with unreplicated plots).
Days from emergence.
Relationships between phenotypic traits and environmental factors and among traits across environments:
The analysis of the relationships between phenotypic traits and environmental variables across environments showed the marked influence of the environmental conditions during the critical period from heading to harvest on GY and PH. Correlation coefficients between the mean phenotypic values of the RILs in each environment and the phenological and environmental factors are reported in supplemental Table 2 (supplemental data set 2) at http://www.genetics.org/supplemental/. Grain yield and PH showed a positive association with the total crop-cycle length (from emergence to harvest) and particularly with the duration of the phase from heading to harvest (r = 0.64 and 0.62, respectively; P ≤ 0.01); conversely, the duration of the emergence to HD was not associated with GY and PH. Thus, GY was mostly affected by environmental conditions around heading time and from heading to maturity.
Surprisingly, no significant correlation was observed between water input (including rainfall and irrigations) and the investigated traits; nonetheless, a weak, positive association was present between water input from heading to harvest and PH (r = 0.42, P = 0.10) but not with GY. Conversely, positive correlations were evidenced between average soil moisture at grain filling and GY (r = 0.50, P ≤ 0.05). Mean and maximum temperatures at heading and during grain filling showed no association with the investigated traits.
The correlations among phenotypic traits on a single-environment basis for the 249 RILs are reported in supplemental Table 3 (supplemental data set 2) at http://www.genetics.org/supplemental/. The correlation coefficients were often of low magnitude even when statistically significant. PH showed a positive correlation with GY in 13 of 16 environments (r from 0.27 to 0.56, P ≤ 0.001). The correlation between HD and GY was significant (P ≤ 0.001) in only six environments with the r values consistently negative (r from −0.25 in Lbn-r04 to −0.49 in Syr-i05).
QTL results:
Figure 2 reports the map position and main features of the QTL detected in this study. The QTL characterized by LOD scores >3.0 and R2 values >10% in at least one environment and also based on the mean of the 16 environments (Figure 2 and Table 5) are referred to as “major” QTL. Sixteen, 15, and 11 distinct QTL regions were detected for GY, HD, and PH, respectively (Table 3). As compared to GY, both HD and PH showed a considerably higher number of QTL with significant effects across more than three environments (two QTL for GY, five for HD, and six for PH), characterized by rather sizeable R2 values (up to 17.3, 27.5, and 15.8%, respectively).
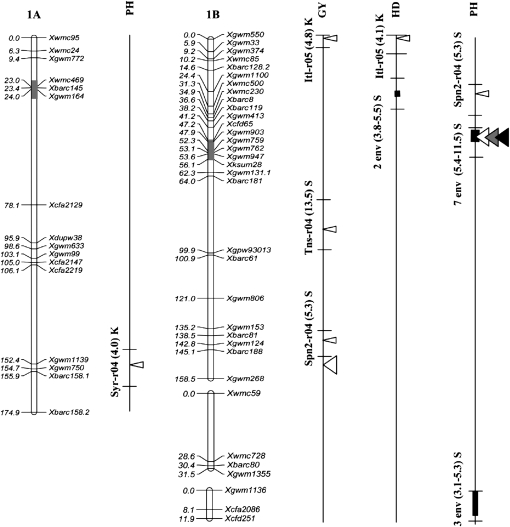
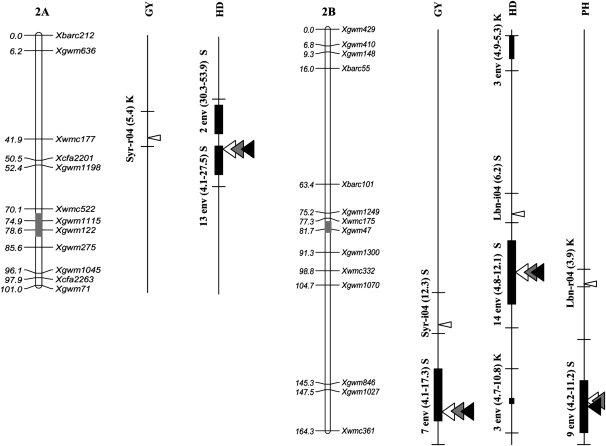
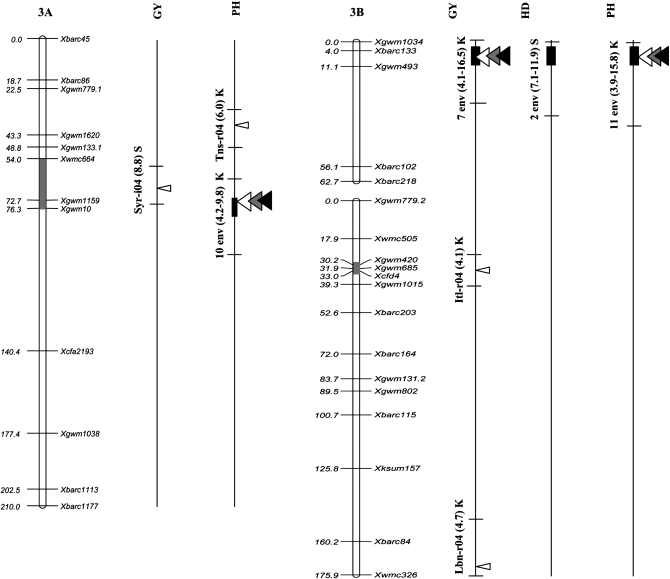
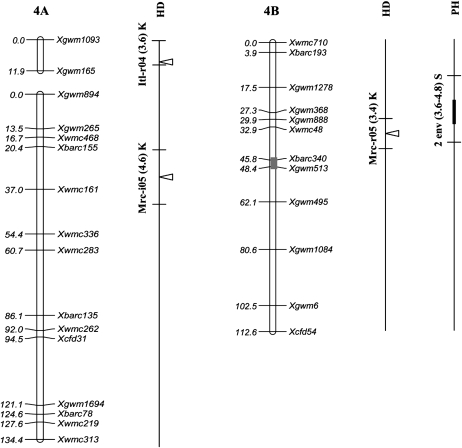
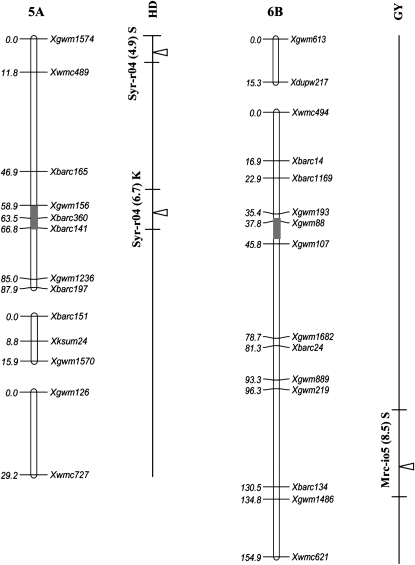
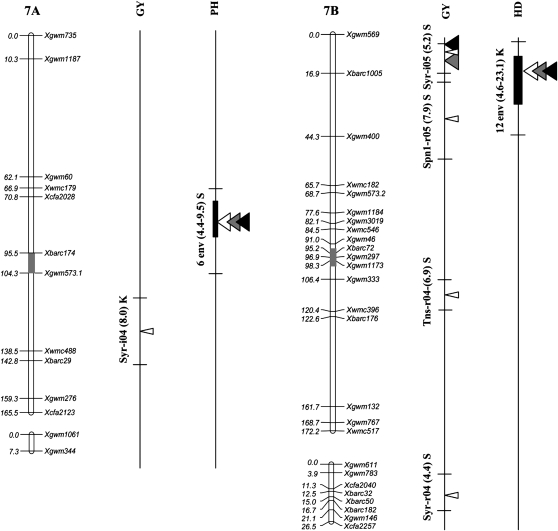
Figure 2.—
Position of QTL detected in the Kofa × Svevo RIL mapping population, tested in 16 Mediterranean environments. The QTL with R2 value >1% detected with composite-interval mapping (CIM) analysis are shown. QTL peak positions are reported (from left to right) for grain yield (GY), heading date (HD), and plant height (PH). QTL identified in a single environment are indicated with narrow triangles, and QTL identified on the basis of the mean values of 2004, 2005, and both years (i.e., all 16 environments) are indicated with wide triangles (open, shaded, and solid triangles, respectively). Horizontal black bars indicate the (LOD − 1) supporting intervals of QTL. QTL found in two or more environments with LOD peaks within a 20-cM interval are indicated with vertical bars of three different thicknesses based on their R2 values. For QTL detected in single environments the environment code, the R2 value, and the parent contributing the increasing allele (Kofa, K; Svevo, S) are reported, while for QTL found in two or more environments, the number of significant environments, the range of R2 values, and the parent contributing the increasing allele are indicated.
TABLE 5.
Features of QTL for grain yield, heading date, and plant height based on the averaged data of 16 environments
| CIM QTL analysis
|
MIM QTL analysis
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | QTL | Flanking markers | LOD | Peak position (cM) | R2 (%) | Effecta | LOD-1 support interval (cM) | Peak position (cM) | R2 (%) | Effect |
| Grain yield | QYld.idw-2Bb | Xgwm1027–Xwmc361 | 8.9 | 155 | 15.6 | 0.55 | 147–161 | 155 | 21.5 | 0.69 |
| QYld.idw-3Bb | Xbarc133–Xgwm493 | 10.0 | 8 | 15.3 | −0.55 | 2–13 | 8 | 13.8 | −0.52 | |
| QYld.idw-7B | Xgwm569–Xbarc1005 | 3.0 | 0 | 4.1 | 0.29 | 0–10 | 0 | 3.2 | 0.26 | |
| Q.idw-2B × Q.idw-3B | 14.0 | 0.57 | ||||||||
| Multiple R2 | 52.5 | |||||||||
| Heading date | QHd.idw-2A.2b | Xwmc177–Xcfa2201 | 21.4 | 46 | 32.2 | 0.9 | 44–48 | 45 | 32.7 | 0.9 |
| QHd.idw-2B.2c | Xgwm1300–Xwmc332 | 7.3 | 95 | 9.2 | 0.5 | 91–103 | 93 | 11.5 | 0.5 | |
| QHd.idw-7Bb | Xgwm569–Xbarc1005 | 9.9 | 25 | 17.8 | −0.7 | 19–33 | 25 | 17.8 | −0.7 | |
| Multiple R2 | 62.0 | |||||||||
| Plant height | QPht.idw-1B.1c | Xbarc119–Xgwm413 | 7.1 | 41 | 8.7 | 0.9 | 40–45 | 42 | 9.1 | 0.9 |
| QPht.idw-2Bb | Xgwm1027–Xwmc361 | 5.4 | 149 | 7.4 | 0.9 | 136–157 | 148 | 10.6 | 1.1 | |
| QPht.idw-3Ac | Xgwm1159–Xgwm10 | 4.8 | 73 | 5.6 | −0.7 | 68–88 | 74 | 5.2 | −0.8 | |
| QPht.idw-3Bb | Xbarc133–Xgwm493 | 10.3 | 10 | 14.2 | −1.2 | 8–17 | 10 | 15.7 | −1.3 | |
| QPht.idw-7Ac | Xcfa2028–Xbarc174 | 4.4 | 78 | 7.3 | 0.9 | 68–90 | 78 | 6.6 | 0.8 | |
| Q.idw-2B × Q.idw-3B | 10.0 | 1.1 | ||||||||
| Multiple R2 | 57.3 | |||||||||
QTL for grain yield, heading date, and plant height were identified from the averaged data of 16 environments using the CIM (LOD > 2.5) and MIM analyses. For each QTL, the flanking markers, the peak LOD score, the chromosome position of the LOD peak, the R2 value, the additive effect, and the (LOD-1) QTL supporting interval from the CIM analysis are reported. For each QTL, the chromosome position of the LOD peak, the R2 value, and the additive effect from the MIM analysis are also reported. The significant epistatic interactions between QTL and the corresponding additive × additive effect as estimated in the MIM analysis are indicated in italics.
Additive effect for grain yield (q ha−1), heading date (days), and plant height (cm) computed as half of the difference between the mean phenotypic values of the RILs homozygous for the Svevo and Kofa alleles [(Svevo − Kofa)/2].
QTL influencing more than one trait in a range of environments.
Trait-specific QTL present in a range of environments.
TABLE 3.
QTL number and R2 range in the 16 environments
| Unique QTL
|
QTL common to different environments
|
|||||
|---|---|---|---|---|---|---|
| Trait | One environment (no.) | R2 (range) (%) | Two environments (no.) | R2 (range) (%) | Three or more environments (no.) | R2 (range) (%) |
| Grain yield | 14 | 4.1–13.5 | 0 | — | 2 | 4.1–17.3 |
| Heading date | 7 | 3.4–6.7 | 3 | 3.8–53.9 | 5 | 4.1–27.5 |
| Plant height | 4 | 3.9–6.0 | 1 | 3.6–4.8 | 6 | 3.1–15.8 |
The number and R2 range of QTL identified in the 16 environments for grain yield, heading date, and plant height are shown. The number of QTL identified in a single environment, in two environments, and across three or more environments is summarized. Results from the CIM (LOD > 2.5) are reported. Only QTL with the R2 value >1% were considered. QTL were considered common to different environments when the corresponding chromosome peak positions were located within 20 cM.
The total number of QTL detected for each trait and environment, together with the corresponding R2 values and their significant digenic epistatic interactions (calculated in the CIM and/or MIM analysis), is reported in Table 4. In general, on a single-environment basis, both the total number of significant QTL identified and the multiple-R2 value (comprising the significant additive QTL effects and their significant epistatic interactions) were higher in environments characterized by medium- to high-GY values, compared to those with low yield (i.e., <25 q ha−1).
TABLE 4.
Features of QTL for grain yield, heading date, and plant height on a single-environment basis
| Lbn-i05a | Syr-i04 | Itl-r04 | Syr-i05 | Itl-r05 | Syr-r05 | Syr-r04 | Spn1-r04 | Mrc-i05 | Tns-i04 | Lbn-r05 | Lbn-i04 | Tns-r04 | Lbn-r04 | Spn2-r05 | Mrc-r05 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 58.8b | 57.4 | 57.3 | 56.8 | 54.0 | 43.1 | 41.8 | 34.8 | 30.9 | 29.6 | 28.1 | 26.5 | 17.6 | 16.7 | 15.3 | 5.6 | |
| a. Grain yield | ||||||||||||||||
| Total QTL no. | 2 | 3 | 3 | 3 | 3 | 0 | 3 | 3 | 1 | 2 | 0 | 0 | 3 | 1 | 2 | 0 |
| QYld.idw-2B | ||||||||||||||||
| LOD (CIM) | 4.1 | 6.3 | 3.2 | 3.5 | 3.2 | 7.5 | 6.9 | (2.6) | ||||||||
| R2 (MIM) | 9.9 | 12.4 | 8.7 | 7.2 | 5.3 | 11.3 | 11.7 | 3.5 | ||||||||
| ac (MIM) | 0.52 | 1.70 | 1.28 | 0.72 | 0.39 | 1.55 | 0.38 | 0.48 | ||||||||
| a (%)d | 0.8 | 2.9 | 2.3 | 1.3 | 0.9 | 4.4 | 1.3 | 2.7 | ||||||||
| QYld.idw-3B | ||||||||||||||||
| LOD | 3.1 | 7.5 | 9.6 | 3.4 | 3.5 | (2.5) | 3.3 | |||||||||
| R2 | 4.8 | 12.3 | 18.1 | 5.4 | 7.5 | 5.1 | 7.5 | |||||||||
| a | −0.35 | −1.62 | −1.73 | −0.35 | −1.24 | −0.24 | −0.13 | |||||||||
| a (%) | 5.9 | 2.8 | 3.0 | 0.9 | 3.6 | 0.8 | 0.9 | |||||||||
| QYld.idw-2B × QYld.idw-3B | ||||||||||||||||
| R2 | 6.6 | 16.7 | 7.6 | 2.4 | 5.7 | |||||||||||
| a × ae | 0.43 | 1.91 | 1.22 | 0.78 | 0.28 | |||||||||||
| a × a (%) | 7.3 | 3.3 | 2.1 | 2..2 | 1.0 | |||||||||||
| Multiple R2 (MIM) | 21.3 | 31.1 | 44.7 | 37.9 | 17.0 | — | 14.8 | 26.2 | 6.5 | 22.5 | — | — | 11.7 | 4.7 | 13.7 | |
| b. Heading date | ||||||||||||||||
| Total QTL no. | 3 | 2 | 4 | 2 | 3 | 4 | 5 | 4 | 4 | 3 | 4 | 6 | 3 | 3 | 4 | 4 |
| QHd.idw-1B | ||||||||||||||||
| LOD (CIM) | 4.7 | 3.3 | ||||||||||||||
| R2 (MIM) | 5.4 | 4.1 | ||||||||||||||
| ac (MIM) | 0.4 | 0.3 | ||||||||||||||
| QHd.idw-2A.1 | ||||||||||||||||
| LOD | 13.8 | 8.7 | ||||||||||||||
| R2 | 35.3 | 21.9 | ||||||||||||||
| a | 2.7 | 1.4 | ||||||||||||||
| QHd.idw-2A.2 | ||||||||||||||||
| LOD | 10.9 | 11.5 | 18.7 | 9.5 | 15.5 | 6.1 | 19.2 | 15.2 | 8.6 | (2.8) | 17.5 | 3.2 | 13.8 | |||
| R2 | 18.1 | 17.8 | 34.7 | 14.7 | 24.2 | 10.0 | 28.4 | 22.8 | 12.5 | 3.7 | 22.4 | 5.3 | 17.4 | |||
| a | 0.7 | 0.8 | 1.3 | 0.7 | 1.1 | 0.5 | 1.0 | 1.1 | 0.6 | 0.3 | 1.1 | 0.3 | 0.6 | |||
| QHd.idw-2B.1 | ||||||||||||||||
| LOD | 3.7 | 4.1 | 3.3 | |||||||||||||
| R2 | 3.2 | 4.7 | 5.1 | |||||||||||||
| a | −0.5 | −0.4 | −0.3 | |||||||||||||
| QHd.idw-2B.2 | ||||||||||||||||
| LOD | 8.5 | 5.5 | 4.2 | 3.5 | 7.5 | 6.9 | 3.6 | 5.5 | 7.3 | 3.4 | 5.5 | 6.2 | 7.5 | 3.1 | ||
| R2 | 12.3 | 8.7 | 7.0 | 6.3 | 11.3 | 8.7 | 6.3 | 8.2 | 13.0 | 1.8 | 5.5 | 12.8 | 10.2 | 4.2 | ||
| a | 0.5 | 0.6 | 0.5 | 0.5 | 0.5 | 0.6 | 0.6 | 1.2 | 0.6 | 0.2 | 0.6 | 0.5 | 0.5 | 0.6 | ||
| QHd.idw-2B.3 | ||||||||||||||||
| LOD | 7.7 | 4.2 | 3.2 | |||||||||||||
| R2 | 15.2 | 6.5 | 4.1 | |||||||||||||
| a | −0.6 | −0.4 | −0.3 | |||||||||||||
| QHd.idw-3B | ||||||||||||||||
| LOD | 7.3 | 4.3 | ||||||||||||||
| R2 | 13.3 | 9.3 | ||||||||||||||
| a | 0.5 | 0.4 | ||||||||||||||
| QHd.idw-7B | ||||||||||||||||
| LOD | 4.6 | 5.3 | 8.5 | 5.1 | 5.0 | 5.0 | 6.9 | 4.4 | 5.4 | 7.5 | 11.0 | 7.8 | ||||
| R2 | 13.9 | 18.6 | 16.7 | 11.4 | 13.7 | 12.5 | 14.3 | 6.5 | 14.2 | 16.5 | 24.3 | 12.8 | ||||
| a | −0.6 | −0.9 | −0.7 | −0.7 | −0.6 | −0.7 | −0.9 | −1.1 | −0.6 | −0.9 | −0.7 | −1.1 | ||||
| QHd.idw-2A.2 × QHd.idw-7B | ||||||||||||||||
| R2 | 4.5 | 2.7 | ||||||||||||||
| a × ae | 0.5 | 0.4 | ||||||||||||||
| QHd.idw-2B.3 × QHd.idw-3B | ||||||||||||||||
| R2 | 12.8 | 11.5 | ||||||||||||||
| a × a | 0.5 | 0.5 | ||||||||||||||
| QHd.idw-2A.2 × QHd.idw-3B | ||||||||||||||||
| R2 | 2.1 | |||||||||||||||
| a × a | 0.3 | |||||||||||||||
| Multiple R2 (MIM) | 44.4 | 26.5 | 64.9 | 41.3 | 35.3 | 47.2 | 45.8 | 55.0 | 47.1 | 50.0 | 44.4 | 46.0 | 47.1 | 22.2 | 56.1 | 41.6 |
| c. Plant height | ||||||||||||||||
| Total QTL no. | 6 | 0 | 4 | 6 | 4 | 3 | 4 | 7 | 0 | 6 | 0 | 3 | 3 | 4 | 1 | 1 |
| QPht.idw-1B.1 | ||||||||||||||||
| LOD (CIM) | 3.8 | 7.1 | 8.1 | 5.5 | 4.0 | 5.8 | 3.4 | |||||||||
| R2 (MIM) | 4.7 | 9.9 | 12.0 | 11.3 | 5.9 | 7.8 | 6.3 | |||||||||
| ac (MIM) | 0.9 | 2.3 | 2.3 | 2.7 | 1.4 | 1.4 | 0.3 | |||||||||
| QPht.idw-1B.2 | ||||||||||||||||
| LOD | 3.5 | (2.8) | (2.5) | |||||||||||||
| R2 | 5.8 | 5.7 | 4.1 | |||||||||||||
| a | 1.1 | 1.7 | 0.9 | |||||||||||||
| QPht.idw-2B | ||||||||||||||||
| LOD | 3.7 | 5.7 | 5.2 | (2.7) | 3.3 | 6.2 | 4.5 | 8.4 | (2.5) | |||||||
| R2 | 10.7 | 12.8 | 8.7 | 7.5 | 6.2 | 12.0 | 6.9 | 14.8 | 4.4 | |||||||
| a | 1.5 | 2.1 | 2.1 | 1.3 | 1.7 | 1.9 | 1.1 | 2.4 | 0.4 | |||||||
| QPht.idw-3A | ||||||||||||||||
| LOD | 4.8 | 4.5 | 3.0 | 3.0 | 3.2 | 3.0 | 3.8 | 5.5 | 4.7 | 4.8 | ||||||
| R2 | 7.3 | 5.6 | 3.5 | 3.9 | 3.2 | 3.8 | 4.5 | 6.2 | 7.0 | 10.6 | ||||||
| a | −1.3 | −1.2 | −1.7 | −1.4 | −0.8 | −1.3 | −1.3 | −0.9 | −0.4 | −0.4 | ||||||
| QPht.idw-3B | ||||||||||||||||
| LOD | 4.9 | 4.2 | 11.6 | 6.9 | (2.5) | 4.3 | 6.5 | 5.7 | 4.5 | 9.3 | 4.5 | |||||
| R2 | 7.3 | 11.2 | 14.1 | 10.9 | 4.3 | 7.0 | 10.1 | 7.9 | 8.5 | 16.5 | 4.9 | |||||
| a | −1.2 | −1.8 | −2.8 | −2.2 | −1.7 | −1.2 | −2.1 | −1.5 | −1.2 | −2.4 | −0.3 | |||||
| QPht.idw-4B | ||||||||||||||||
| LOD | 2.8 | (2.8) | ||||||||||||||
| R2 | 4.1 | 2.7 | ||||||||||||||
| a | 0.9 | 1.1 | ||||||||||||||
| QPht.idw-7A | ||||||||||||||||
| LOD | 3.1 | 5.9 | 3.1 | 3.1 | (2.6) | 3.4 | ||||||||||
| R2 | 3.7 | 10.3 | 5.7 | 7.1 | 6.1 | 4.9 | ||||||||||
| a | 0.8 | 2.4 | 1.7 | 2.1 | 1.6 | 1.1 | ||||||||||
| QPht.idw-2B × QPht.idw-3B | ||||||||||||||||
| R2 | 4.8 | 15.2 | 5.4 | 6.5 | 3.1 | 9.6 | 7.1 | 8.2 | ||||||||
| a × ae | 1.0 | 2.3 | 1.9 | 1.2 | 1.3 | 1.8 | 1.1 | 1.8 | ||||||||
| QPht.idw-2B × QPht.idw-7A | ||||||||||||||||
| R2 | 4.7 | 2.1 | ||||||||||||||
| a × a | 1.0 | 1.2 | ||||||||||||||
| QPht.idw-3B × QPht.idw-7A | ||||||||||||||||
| R2 | 1.4 | 1.7 | ||||||||||||||
| a × a | 1.2 | 1.1 | ||||||||||||||
| Multiple R2 (MIM) | 47.4 | — | 50.6 | 60.0 | 34.2 | 22.7 | 26.8 | 41.6 | — | 50.8 | — | 28.6 | 44.2 | 22.2 | 10.6 | 6.3 |
Main features of the QTL for grain yield, heading date, and plant height detected in the 16 environments are listed in decreasing order according to the mean RIL grain yield values. The total number of QTL found in each environment (CIM, LOD > 2.5) and the corresponding multiple R2 obtained in the MIM model are reported. For each QTL with significant effect in at least two environments (QTL peaks located within 20 cM) the corresponding LOD value (from CIM analysis) is reported. The R2 and the additive and epistatic effects (as from the MIM analysis) are also reported. The LOD values of QTL with LOD peaks between 2.5 and 2.9 are included in parentheses.
Environments listed according to the decreasing mean yield value of the RILs.
Mean grain yield data (q ha−1) of each environment.
Additive effect for grain yield (q ha−1), heading date (days), and plant height (cm) computed as half of the difference between the mean phenotypic values of the RILs homozygous for the Svevo and Kofa alleles [(Svevo − Kofa)/2] in the multiple-interval mapping analysis.
Additive effect expressed as percentage of the mean grain yield of the RILs in the specific environment.
Additive-by-additive epistatic effect as computed in the multiple-interval-mapping analysis.
QTL for grain yield:
A total of 16 different QTL were detected, 14 of which were found to be specific for a single environment (Figure 2; Table 3), with additive effects and R2 values ranging from 0.1 to 1.1 q ha−1 and from 4.1 to 13.5%, respectively. Both parental cultivars contributed the favorable alleles at these QTL (6 and 10 by Kofa and Svevo, respectively). The LOD profile was always <2.5 in Syr-r05, Lbn-r05, Lbn-i04, and Mrc-r05; thus no significant GY QTL was evidenced in these environments, all of which were characterized by medium- to low-GY values (from 5.6 q ha−1 in Mrc-r05 to 43.1 q ha−1 in Syr-r05).
Two major GY QTL (QYld.idw-2B and QYld.idw-3B) with multiple R2 values (including also their significant epistatic interaction) up to 44.7% (Itl-r04) were identified across several environments showing a broad range of mean productivity (from 15.3 to 58.8 q ha−1; Tables 4a and 5). It is interesting to underline that both QTL reached a LOD > 3 in five environments as well as when considering the mean GY values across environments.
QYld.idw-2B, located in the distal region of chromosome (chr.) 2BL, had a LOD value >2.5 in eight environments with LOD peaks positioned within a 19-cM interval between Xgwm846/Xgwm1027 and Xwmc361, R2 values from 3.5 to 12.4%, and additive effect from 0.38 to 1.70 q ha−1 (MIM analysis; Table 4a). This QTL was thus detected across environments with an approximately threefold range in yield values (from 17.6 to 58.8 q ha−1). Additionally, in the same chromosome region, peaks of LOD score for GY comprised between 2.0 and 2.5 (indicative of the presence of a putative QTL effect) were observed in two additional environments (Syr-r05 and Lbn-i04, data not reported). In all environments, including those with LOD peaks between 2.0 and 2.5, Svevo contributed the favorable allele.
The second major GY QTL (QYld.idw-3B), located on the distal region of chr. 3BS and flanked by Xbarc133 and Xgwm493, was detected in seven environments with R2 values ranging from 4.8 to 18.1% (Table 4a). In addition, the putative presence of this QTL for GY was detected in the Syr-r05 trial (LOD peak of 2.0). In all environments, the increasing allele was consistently contributed by Kofa (additive effects from −0.13 to −1.73 q ha−1).
QTL analysis on the mean GY values of 2004 (eight environments), 2005 (eight environments), and across both years (Figure 2 and Table 5) evidenced significant effects at both QYld.idw-2B and QYld.idw-3B. A third chr. region (QYld.idw-7B between Xgwm569 and Xbarc1005) was detected when using 2005 and all the 16-environment mean data; however, this QTL showed LOD, R2, and additive values lower than those of QYld.idw-2B and QYld.idw-3B.
Considering the results of the CIM analysis carried out using the GY mean values across the 16 environments (Table 5), the chr. 2BL and 3BS major QTL (QYld.idw-2B and QYld.idw-3B, respectively) showed, respectively, peak LOD values of 8.9 and 10.0, R2 values of 15.6 and 15.3%, and additive effects of similar magnitude (0.55 q ha−1), although of opposite sign. MIM-based QTL analysis confirmed the QTL peak positions, evidenced additive effects of 0.69 (chr. 2BL) and −0.52 (chr. 3BS) q ha−1, and revealed a strong, significant epistatic interaction between these two QTL (Table 5).
QTL for heading date:
Although this trait showed a transgressive segregation in both directions (Figure 1), ∼50% of the RILs headed within 2 days. This result indicates that no major gene with large effects for HD segregated in the Kofa × Svevo population, an important feature when evaluating different genotypes in Mediterranean environments where variation in HD usually shows, on an adaptive basis, significant and negative association with GY in both bread and durum wheat (Richards 1996; Araus et al. 1998, 2003a,b; Del Moral et al. 2003), thus requiring its adoption as a covariate if the objective is the identification of QTL for GY on a per se (i.e., constitutive) basis, rather than on a more adaptive basis.
Three to four QTL per environment (Table 4b) were most frequently observed (3.6, on average), with multiple R2 values ranging from 22.2 to 64.9%. Major HD QTL with significant effects in several (six or more) environments as well as on the mean values across environments were detected on chr. 2AS (QHd.idw-2A.2), chr. 2BL (QHd.idw-2B.2), and chr. 7BS (QHd.idw-7B). Interestingly, these three major QTL for HD mapped in positions (Figure 2; Tables 4b and 5) other than those influencing GY. QHd.idw-2A.2, located on the proximal region of chr. 2AS, was detected in 13 of 16 environments, with R2 values from 3.7 to 34.7% (Table 4b), as well as when considering the mean values of 2004, 2005, and across all 16 environments (Figure 2), where this QTL accounted for 32.2% of the phenotypic variation among RILs, with a very narrow LOD − 1 supporting interval (4 cM) located between Xwmc177 and Xcfa2201 (Table 5, CIM analysis). On a single-environment basis, LOD profiles for GY on the chromosome region underlying QHd.idw-2A.2 did not support the presence, even putatively, of a QTL for GY. A second QTL (QHd.idw-2A.1) that maps near QHd.idw-2A.2 reached the significance threshold in two environments. On chr. 2BS, in the homeologous position corresponding to QHd.idw-2A.2, a QTL (QHd.idw-2B.1, flanked by Xgwm429 and Xgwm148) with significant effect on HD was detected in three environments (LOD from 3.3 to 4.1).
A second major QTL for HD (QHd.idw-2B.2) was located on the proximal region of the 2BL chr. within the Xgwm1300–Xwmc332 interval. Although this QTL was detected in almost all environments (14 of 16), the associated R2 values and additive effects were lower than those of QHd.idw-2A.2 (Table 4b). QHd.idw-2B.2 showed no concomitant effect on GY and PH. In the Lbn-i04 trial, the LOD and R2 values of the two major QTL for HD (QHd.idw-2A.2 and QHd.idw-2B.2) were considerably lower than the average and in the Syr-i05 trial the LOD peaks were <2.0.
A third major QTL for HD (QHd.idw-7B), located on chr. arm 7BS (flanked by Xgwm569 and Xbarc1005), was detected in 12 environments with R2 values ranging from 6.5 to 24.3%. This QTL was not detected in Syr-i04, Syr-i05, Lbn-r04, and Lbn-i04.
The two major GY QTL on chrs. 2BL and 3BS significantly affected HD in only three and two environments, respectively (QHd.idw-2B.3 in Syr-i05, Lbn-i04, and Lbn-r04 and QHd.idw-3B in Syr-i05 and Lbn-i04), with Svevo contributing the allele for earliness at the first chr. region and Kofa at the second (Table 4b).
QTL for plant height:
The number of QTL detected per environment (Table 4c) ranged from zero to seven, with three to four QTL most frequently identified and multiple R2 values up to 60.0% (Syr-i05).
As compared to GY and HD, PH showed the lowest number of QTL detected in one environment only (14, 7, and 4 for GY, HD, and PH, respectively; see Figure 2 and Table 3). Conversely, PH was the trait influenced by the highest number of major QTL (5 in total, Table 4c) that were consistently detected also on the basis of the mean values of 2004, 2005, and across both years (Figure 2 and Table 5).
Five major QTL for PH were detected on chr. 1BS (proximal region, QPht.idw-1B.1 in 7 environments), chr. 2BL (distal region, QPht.idw-2B in 9 environments), chr. 3AL (proximal, QPht.idw-3A in 10 environments), chr. 3BS (distal region, QPht.idw-3B in 11 environments), and chr. 7AS (proximal region, QPht.idw-7A in 6 environments). At each QTL, the sign of the additive effect was consistent across all the significant environments and both parents contributed QTL alleles increasing PH.
It is interesting to note that QPht.idw-2B was located in the same chr. region (between Xgwm1027 and Xwmc361) where the chr. 2BL GY major QTL mapped; for both traits, the plus allele was contributed by Svevo. On a single-environment basis, QPht.idw-2B showed R2 values from 4.4 to 14.8% and additive effect from 0.4 to 2.4 cm (Table 4c).
Another major QTL for PH (QPht.idw-3B; chr. 3BS region between Xbarc133 and Xgwm493) was coincident with the chr. 3BS major QTL for GY. QPht.idw-3B was identified in all seven environments where significant effects for GY were evidenced plus four additional ones, Itl-r05, P7r05, Lbn-i04, and Tns-r04, with the last two trials characterized by rather low GY (26.5 and 17.6 q ha−1, respectively). As for GY, the increasing allele was consistently contributed by Kofa (additive effects from −0.3 to −2.8 cm).
Excluding the chr. 2BL and 3BS QTL, none of the three remaining major QTL for PH overlapped with QTL for GY.
Considering the results of the MIM analysis on the mean values of the 16 environments, QPht.idw-3B had the highest R2 value (15.7%) and additive effect (−1.3 cm); the other four major QTL showed similar R2 values that ranged from 5.2 to 10.6% (Table 5).
Epistasis and QTL × environment interaction at the two major QTL on chrs. 2BL and 3BS:
The epistatic interactions detected by MIM together with their average effects and contributions to genetic variances (R2 values) are reported in Tables 4 and 5. Most of the epistatic effects detected with the MIM procedure and retained in the MIM models were due to the interaction between the chr. 2BL and 3BS major QTL. In this case, significant epistatic interactions with a magnitude of R2 values and effects comparable to those estimated for the additive QTL effects were detected for both GY and PH across 13 of the 14 trait–environment combinations where both QTL were significant and also when considering the mean values across all 16 environments (see Tables 4, a and c, and 5). In the latter case, the R2 values and the additive effects of the epistatic interaction between the chr. 2BL and 3BS QTL were, respectively, 14.0% and 0.57 q ha−1 for GY and 10.0% and 1.1 cm for PH.
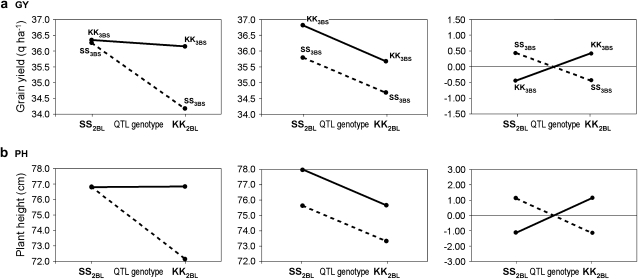
The additive and additive × additive interaction effects computed according to a linear two-marker model using the marker more highly associated to each QTL peak are reported together with the phenotypic values of the parental and recombinant genotypic classes in Table 6a and Figure 3.
TABLE 6.
Epistatic interactions between QTL
| Lbn-i05a | Itl-r04 | Syr-i05 | Syr-r04 | Spn1-r04 | Tns-i04 | Lbn-i04 | Tns-r04 | Mean | |
|---|---|---|---|---|---|---|---|---|---|
| Epistatic effects | 58.8b | 57.3 | 56.8 | 41.8 | 34.8 | 29.6 | 26.5 | 17.6 | 35.9 |
| a. Epistatic interactions between the two major QTL for grain yield on chromosomes 2BL and 3BS | |||||||||
| Grain yield (q ha−1) | |||||||||
| QYld.idw-2B × QYld.idw-3B | |||||||||
| a (Xgwm1027) | 0.48 | 1.54 | 1.14 | — | 1.52 | 0.35 | — | — | 0.57 |
| a (Xbarc133) | −0.36 | −1.49 | −1.48 | — | −1.11 | −0.23 | — | — | −0.52 |
| a × a parental genotypesc | 0.39 | 1.57 | 1.00 | — | 0.77 | 0.16 | — | — | 0.47 |
| a × a recombinant genotypesd | −0.39 | −1.57 | −1.00 | — | −0.77 | −0.16 | — | — | −0.47 |
| Heading date (days) | |||||||||
| QHd.idw-2B.1 × QHd.idw-3B | |||||||||
| a (Xgwm1027) | — | — | −0.5 | — | — | — | −0.4 | — | — |
| a (Xbarc133) | — | — | 0.5 | — | — | — | 0.5 | — | — |
| a × a parental genotypes | — | — | −0.5 | — | — | −0.5 | — | — | |
| a × a recombinant genotypes | — | — | 0.5 | — | — | — | 0.5 | — | — |
| Plant height (cm) | |||||||||
| QPht.idw-2B × QPht.idw-3B | |||||||||
| a (Xgwm1027) | 1.3 | 1.7 | 2.9 | 1.3 | 1.8 | 2.1 | 1.0 | 2.2 | 1.3 |
| a (Xbarc133) | −1.2 | −1.7 | −2.9 | −1.1 | −1.9 | −1.6 | −0.9 | −2.1 | −1.2 |
| a × a parental genotypes | 1.4 | 1.8 | 2.4 | 1.2 | 1.4 | 1.9 | 1.1 | 1.8 | 1.2 |
| a × a recombinant genotypes | −1.4 | −1.8 | −2.4 | −1.2 | −1.4 | −1.9 | −1.1 | −1.8 | −1.2 |
| Lbn-i05a | Itl-r04 | Syr-i05 | Itl-r05 | Syr-r05 | Tns-r04 | |
| Epistatic effects | 58.8b | 57.3 | 56.8 | 54.0 | 43.1 | 17.6 |
| b. Epistatic interactions between other major QTL | ||||||
| Heading date (days) | ||||||
| QHd.idw-2A.2 × QHd.idw-7B | ||||||
| a (Xgwm1198) | — | 1.0 | — | — | — | 1.2 |
| a (Xbarc1005) | — | −0.6 | — | — | — | −0.6 |
| a × a parental genotypesc | — | −0.2 | — | — | — | −0.4 |
| a × a recombinant genotypesd | — | 0.2 | — | — | — | 0.4 |
| QHd.idw-2A.2 × QHd.idw-2B.1 | ||||||
| a (Xwmc177) | — | — | — | — | 0.8 | — |
| a (Xgwm429) | — | — | — | — | −0.4 | — |
| a × a parental genotypes | — | — | — | — | −0.4 | — |
| a × a recombinant genotypes | — | — | — | — | 0.4 | — |
| Plant height (cm) | ||||||
| QPht.idw-2B × QPht.idw-7A | ||||||
| a (gwm1027) | 1.0 | — | 1.8 | — | — | — |
| a (cfa2028) | 0.9 | — | 2.3 | — | — | — |
| a × a parental genotypes | −0.6 | — | −1.0 | — | — | — |
| a × a recombinant genotypes | 0.6 | — | 1.0 | — | — | — |
| QPht.idw-3B × QPht.idw-7A | ||||||
| a (Xbarc133) | — | — | −2.5 | −2.0 | — | — |
| a (Xcfa2028) | — | — | 2.1 | 1.2 | — | — |
| a × a parental genotypes | — | — | 0.5 | 0.2 | — | — |
| a × a recombinant genotypes | — | — | −0.5 | −0.2 | — | — |
Additive and epistatic effects for the QTL with a significant epistatic interaction are shown. For each environment with a significant epistatic effect and for the mean data across all environments, the additive (regular font) and epistatic (italic font) values were calculated according to a linear two-marker model using the marker most associated to each QTL peak.
Environments are listed according to the decreasing mean grain yield value of the RILs.
Mean grain yield (q ha−1) of each environment.
Parental genotypes: KK1stQTLKK2ndQTL and SS1stQTLSS2ndQTL.
Recombinant genotypes: KK1stQTLSS2ndQTL and SS1stQTLKK2ndQTL.
Figure 3.—
Additive and epistatic effects for grain yield (GY) (a) and plant height (PH) (b) calculated on the data averaged across 16 environments. A two-marker model including Xgwm1027 for the QTL on chr. 2BL and Xbarc133 for the QTL on chr. 3BS was adopted. For each trait, from left to right the first diagram shows the mean phenotypic values observed at the four genotypic classes, the diagram in the center shows the phenotypic values expected under the additive model only, while the third diagram depicts the epistatic effects, which are either positive or negative, but equal in magnitude, under the unweighted model.
For both GY and PH, the epistatic interaction between the chr. 2BL and the 3BS QTL was positive for the two parental (nonrecombinant) genotypic classes (i.e., KK2BLKK3BS and SS2BLSS3BS), hence increasing GY and PH, and negative for the recombinant classes (KK2BLSS3BS and SS2BLKK3BS), as reported in Table 6a. Syr-i05 and Lbn-i04 were the only two environments where both QTL significantly affected HD on a single-QTL basis (Table 4b) and also on an epistatic basis (additive × additive interaction effect of 0.5 days in both environments; Table 6a). In both environments, the parental genotypes were characterized by an earlier heading and the recombinant genotypes by a delayed heading.
The significant epistatic interactions observed for HD and PH between other QTL pairs are reported in Table 6b. In general, both their magnitude and the number of environments showing these interactions were lower than those observed between the two chr. 2BL and 3BS regions. For PH, each of the two major QTL on chrs. 2BS and 3BS showed a significant interaction with QPht.idw-7A, a major QTL for PH only, in two environments.
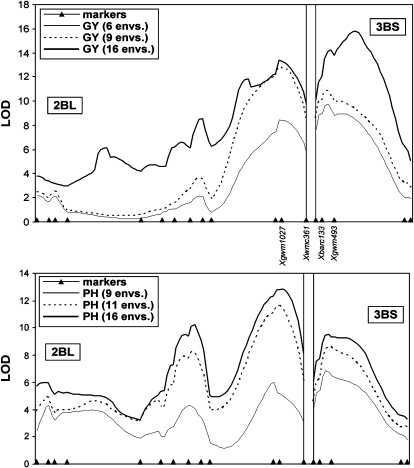
Notable LOD peaks for QTL × environment (Q × E) interaction were detected for GY and PH at the two major QTL on chrs. 2BL and 3BS (Figure 4). In both regions, Q × E LOD peaks for GY and PH, estimated over all 16 environments as well as on subsets of environments with significant QTL additive effects at these chromosome regions, were highly significant and were located close to those of the additive QTL effects. For GY, Q × E LOD peaks ranged from a minimum of 8.5 (chr. 2BL; Q × E computed on the subset of 6 environments with LOD > 2.5 at both chr. 2BL and 3BS QTL) to a maximum of 15.8 (chr. 3BS; Q × E computed across all environments). For PH, Q × E LOD peaks ranged from 6.0 (chr. 2BL; subset of nine environments with LOD > 2.5 in at least one of the two QTL) to 12.8 (chr. 2BL; all environments) (Figure 4). The Q × E interaction effects associated with the two major QTL were sizeable (data not reported) when evaluated across all environments and on the selected subset of environments. Importantly, it should be noted that in all cases the Q × E interactions were determined by fluctuations in the magnitude of the effects rather than by their direction, as shown by the sign of the additive effects that were always consistent across environments (Tables 4a and 4c).
Figure 4.—
LOD score plots of the QTL × environment interaction (G × E) for grain yield (GY) and plant height (PH) in the chromosome regions harboring the two major QTL clusters (chrs. 2BL and 3BS). A G × E significance test was carried out for the combined analysis of the subset of environments showing significant QTL effects (LOD > 2.5) at both chromosome regions (thin line), of the subset of environments showing QTL effect (LOD > 2.5) in at least one of the two chromosome regions or in both of them (dashed line), and of all environments (thick line). The chromosome positions of the markers flanking the QTL peaks are shown.
DISCUSSION
The detection of few major QTL with relatively large effect and acting together with a plethora of minor QTL highly affected by environmental conditions has emerged as a common feature of quantitative traits in plants and animals (Tanksley 1993; Yano and Sasaki 1997; Mackay 2004; Holland 2007). Additionally, the comparative analysis of QTL data for different traits and from different crops/genetic backgrounds indicates the presence of QTL clusters, particularly for complex traits that affect plant growth, response to environmental conditions, and yield (Khavkin and Coe 1997; Cai and Morishima 2002; Tuberosa et al. 2002b; Sawkins et al. 2004). Our results highlight the presence of this QTL type on chrs. 2BL and 3BS in the Kofa × Svevo durum population, where LOD peaks for GY and PH obtained from the mean data of the 2 years were located in rather short intervals (<10 cM).
The evaluation of this RIL population for additional morpho-physiological traits relevant for wheat productivity on a per se and/or adaptive basis (e.g., root architecture, osmotic adjustment, photosynthetic capacity, assimilate relocation from the stem, etc.; Blum 1988, 2005; Ludlow and Muchow 1990; Tuberosa 2004; Sanguineti et al. 2007) will provide important clues on the functional basis of the effects that the chr. 2BS and 3BL QTL exert during the crop phenology stages more critical for final yield (e.g., early growth, tiller proliferation, spike development, anthesis, and particularly, grain filling).
Comparative analysis with the known major QTL for yield in wheat:
A survey of the literature reporting QTL for GY and related agronomic traits in both tetraploid and hexaploid wheat (Li et al. 2002; Peng et al. 2003; Elouafi and Nachit 2004; Huang et al. 2004, 2006; McCartney et al. 2005; Quarrie et al. 2005, 2007; Marza et al. 2006; Narasimhamoorthy et al. 2006; Kuchel et al. 2007; Kumar et al. 2007; Ma et al. 2007) revealed rather different scenarios for the two major GY QTL identified in our study.
The presence of QTL clusters for GY and related traits has been reported in (i) the International Triticeae Mapping Initiative (ITMI) population mainly on chr. groups 2 (short arms), 4, 5, and 6 (Börner et al. 2002); (ii) the cross Chinese Spring × SQ1 on all chr. groups, with particularly strong effects on chrs. 7AL and 7BL (Quarrie et al. 2005, 2007); and (iii) crosses among Canadian wheats on chr. groups 4, 5, and 7 (McCartney et al. 2005; Huang et al. 2006).
Our comparative analysis was carried out by relating the (LOD − 1) supporting interval of the QTL to the Ta-SSR-2004 consensus map (Somers et al. 2004). For the Xgwm846-Xgwm1027 interval on chr. 2BL, shown to harbor a major QTL for GY in our study, no QTL for GY was previously reported. QTL for GY and PH have been assigned to the proximal region of chr. 2BL (Börner et al. 2002; Marza et al. 2006), and QTL for GY, kernel weight, and number of kernels per spike have been reported on chr. 2AL (Börner et al. 2002; Peng et al. 2003; Huang et al. 2004; McCartney et al. 2005), but not in the distal region. Conversely, major QTL for GY have been identified in two different genetic backgrounds (Börner et al. 2002; Campbell et al. 2003) on the short distal regions of chr. group 3. One study (Campbell et al. 2003; Dilbirligi et al. 2006) dissected the genetic basis of GY and yield components using a set of recombinant inbred chromosome lines (RICLs) obtained from the introgression of chr. 3A segments from Wichita into the genetic background of Cheyenne (CNN(WI3A)). Among the four major QTL on chr. 3AS that have been shown to influence GY and yield components, one was precisely located in the distal region homeologous to that herein reported for GY and PH on chr. 3BS (Xbarc133–Xgwm493). In the ITMI mapping population (Börner et al. 2002), a QTL specific for PH but with no effect on GY or yield components was found in the distal portion of chr. 3BS.
The chr. 3BS QTL for GY evidenced in our study and also by Campbell et al. (2003) and Börner et al. (2002) is located on the deletion bin 3BS8 0.78-1.00 (Sourdille et al. 2004; Liu et al. 2006), one of the regions with the highest density of mapped ESTs (gene-rich region 3S0.9 in the consensus physical map of chr. group 3) among those defined by deletion bins (Munkvold et al. 2004; Dilbirligi et al. 2006). In both ITMI and RICLs–(CNN(WI3A)) populations, an additional major QTL for GY and PH was located on a proximal region of chr. 3AS that in Kofa × Svevo underlined a major QTL for PH (QPht.idw-3A) with a high and consistent effect across environments. In our case no concurrent effect on GY was observed at this chr. region.
QTL for heading date:
HD has long been considered as a major adaptive trait with a particularly relevant role also in domestication (Snape et al. 2001; Peng et al. 2003; Laurie et al. 2004; Hanocq et al. 2006). Adaptation of wheat to a range of environments with very different photoperiod conditions and winter temperatures is mainly accomplished by combinations of natural alleles at major genes for vernalization requirements and photoperiod sensitivity (Worland 1996). As compared to hexaploid wheat, the major elite durum wheat gene pools show no major vernalization requirements (spring wheat), while functionally variant alleles are present at main loci for the photoperiod-sensitive response (Clarke et al. 1998). Nonetheless, durum types with various vernalization requirements have been described (Marque et al. 2004; Motzo and Giunta 2007).
In Mediterranean environments, early and medium-early, photoperiod-insensitive genotypes have been selected by breeders to escape the combined burden of terminal drought and heat stress that frequently occurs in the cultivation areas of durum wheat; terminal heat stress is also typical of the southwestern United States where Kofa was selected. However, this feature cannot be generalized to all environments suitable for durum wheat cultivation.
Considering the major homeologous gene series controlling the photoperiod response in tetraploid and hexaploid wheats (Ppd-A1 on chr. 2AS and Ppd-B1 on chr. 2BS), the position of a major QTL for HD (QHd.IDW-2A.2; Xwmc177-Xgwm1198) detected in our population under field conditions across most environments suggests Ppd-A1 as a feasible candidate for this QTL. However, due to the relatively lower relevance in bread wheat germplasm of Ppd-A1 compared to both Ppd-B1 and Ppd-D1 (Hanocq et al. 2006), its position on chr. 2AS has not yet been clearly defined. The map position of QHd.IDW-2A.2 coincides with the most probable position of Ppd-A1 on the basis of published observations and the homeologous relationships with Ppd-B1 and Ppd-D1 (Sourdille et al. 2003; Hanocq et al. 2004; Mohler et al. 2004; Kuchel et al. 2006).
It is worth noting that one of the major QTL detected in our study on chr. 2BS (QHd.IDW-2B.1; Xgwm429-Xgwm148) is located on the same chromosome region where Ppd-B1 was originally mapped (near Xgwm148; Hanocq et al. 2004; Mohler et al. 2004; Kuchel et al. 2006). The role of this QTL in our study appears rather marginal, as shown by the fact that its effect was significant only in Syria and Lebanon (Syr-r05, Lbn-i04, and Lbn-r05).
On the same chromosome harboring Ppd-B1, a major effect on HD was detected in the proximal region of the long arm of chr. 2BL (QHd.idw-2B.2; Xgwm47-Xgwm1070); this region did not overlap with any major QTL for HD previously described in tetraploid and hexaploid wheat as well as in barley. However, earliness per se genes have been mapped on chrs. 2BL and 2DL (Scarth and Law 1983; Worland 1996) and a vernalization QTL was positioned near the QTL region detected in our population (Hanocq et al. 2006).
In general, the QTL for HD showed a weak association with GY and PH. This finding suggests that a nearly optimum balance between earliness and yield potential has been attained in this particular genetic background. Possible cause–effect relationships between HD and GY were evidenced for QHd.idw-7B and the two main QTL clusters on chrs. 2BL and 3BS in a few environments only; in these cases, earliness was positively associated with GY as expected in Mediterranean environments (Araus et al. 2003a,b). On the basis of the definition of escape mechanisms (Blum 1988; Ludlow and Muchow 1990), constitutive earliness in cereals adapted to Mediterranean environments can be considered as one of the best-studied examples of effective escape from drought constraints (Zaharieva et al. 2001). However, excessive earliness in the elite materials has often been correlated with constitutively low biomass and low GY potential (Reynolds et al. 2007). This is not the case for the Kofa × Svevo population as shown by the lack of correlation of HD with GY and PH in the highest-yielding environments. Therefore, in this population the overall role of escape in supporting grain productivity in environments subjected to terminal stress was limited, though significant in some cases (Lbn-i04, Lbn-r04, Syr-i05, Syr-r05, Itl-r05, and Spn2-r05).
Epistasis at the main QTL on chrs. 2BL and 3BS:
Epistasis has great relevance for the outcome of marker-assisted breeding (Zeng et al. 1999; Li et al. 2003; Blanc et al. 2006). Nevertheless, notable epistatic effects in wheat have been validated and analyzed in detail only for vernalization requirement (Yan et al. 2006; Szucs et al. 2007), a trait with a genetic basis less complex than that of GY. Overall, negative epistatic effects prevailed in the RILs in comparison to the average performance of the two parental cvs. The presence of negative epistatic interactions in the nonparental genotypes is a common feature for GY, particularly in mapping populations derived from inbred elite materials (Li et al. 1997, 2001; Luo et al. 2001; Melchinger et al. 2003). In the Kofa × Svevo population, the two major QTL for GY and PH on chrs. 2BL and 3BS showed a significant and sizeable epistatic interaction in the five environments where both QTL were concomitantly detected. This result differs in part from those reported in rice (Li et al. 1997; Mei et al. 2005, 2006) and maize (Blanc et al. 2006), where numerous digenic interactions with rather limited effects and not coincident with the locations of the major QTL effects were evidenced.
Conclusions:
To the best of our knowledge, this is the most extensive study reporting QTL results for grain yield in durum wheat grown in a range of environments broadly different for the amount of water available to the crop. The approach followed in our study allows for the detection of QTL with a consistent effect across different environments as shown for the major QTL on chrs. 2BL and 3BS. Of these two QTL, the one on chr. 2BL has not been previously described in wheat, while the one on chr. 3BS confirms the importance of this region as reported by other studies. These results warrant additional work to fine map these two major QTL and to elucidate their functional basis. Isogenization of QTL is an important prerequisite toward their fine mapping and, eventually, positional cloning (Salvi and Tuberosa 2005, 2007). In our case, because both QTL also affected plant height, their fine mapping and positional cloning will be greatly facilitated if based on the measurement of plant height rather than grain yield, with the assumption being that pleiotropy and not linkage is the genetic cause of the concurrent effects of these QTL on plant height and grain yield. Additionally, the fine mapping of the two QTL will allow more markers to be precisely mapped in the two underlying chromosome regions, thus enabling a more detailed investigation of the haplotype variation present in wheat at these QTL regions (He et al. 2007; Mackay and Powell 2007). We have also identified a number of QTL highly interactive with the environments explored in our study. These QTL could provide insight into understanding the factors crucial for the adaptation of durum wheat to specific environmental conditions.
Acknowledgments
The authors thank Sandra Stefanelli, Stefano Vecchi, and Marco Mantovani for technical assistance throughout improving the durum wheat for water use efficiency and yield stability through physiological and molecular approaches (IDuWUE) project. The financial contribution of the European Union [IDuWUE project; International Scientific Cooperation project (INCO) contract no. ICA3-CT-2002-10 028] is gratefully acknowledged.
References
- Araus, J. L., T. Amaro, J. Casadeus, A. Asbati and M. M. Nachit, 1998. Relationships between ash content, carbon isotope discrimination and yield in durum wheat. Aust. J. Plant Physiol. 25: 835–842. [Google Scholar]
- Araus, J. L., G. A. Slafer, M. P. Reynolds and C. Royo, 2002. Plant breeding and drought in C-3 cereals: What should we breed for? Ann. Bot. 89: 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus, J. L., J. Bort, P. Steduto, D. Villegas and C. Royo, 2003. a Breeding cereals for Mediterranean conditions: ecophysiological clues for biotechnology application. Ann. Appl. Biol. 142: 129–141. [Google Scholar]
- Araus, J. L., D. Villegas, N. Aparicio, L. F. Del Moral, S. El Hani et al., 2003. b Environmental factors determining carbon isotope discrimination and yield in durum wheat under Mediterranean conditions. Crop Sci. 43: 170–180. [Google Scholar]
- Basten, C., B. S. Weir and Z.-B. Zeng, 2005. QTL Cartographer. North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/.
- Blanc, G., A. Charcosset, B. Mangin, A. Gallais and L. Moreau, 2006. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor. Appl. Genet. 113: 206–224. [DOI] [PubMed] [Google Scholar]
- Blanco, A., R. Simeone, A. Cenci, A. Gadaleta, O. A. Tanzarella et al., 2004. Extension of the Messapia x dicoccoides linkage map of Triticum turgidum (L.) Thell. Cell Mol. Biol. Lett. 9: 529–541. [PubMed] [Google Scholar]
- Blum, A., 1988. Breeding for Stress Environments. CRC Press, Boca Raton, FL.
- Blum, A., 2005. Drought resistance, water-use efficiency, and yield potential: Are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 56: 1159–1168. [Google Scholar]
- Borevitz, J. O., and J. Chory, 2004. Genomics tools for QTL analysis and gene discovery. Curr. Opin. Plant Biol. 7: 132–136. [DOI] [PubMed] [Google Scholar]
- Börner, A., E. Schumann, A. Fürste, H. Cöster, B. Leithold et al., 2002. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 105: 921–936. [DOI] [PubMed] [Google Scholar]
- Bortiri, E., D. Jackson and S. Hake, 2006. Advances in maize genomics: the emergence of positional cloning. Curr. Opin. Plant Biol. 9: 164–171. [DOI] [PubMed] [Google Scholar]
- Cai, H. W., and H. Morishima, 2002. QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Campbell, B. T., P. S. Baezinger, K. S. Gill, K. M. Eskridge, H. Budak et al., 2003. Identification of QTLs and environmental interactions associated with agronomic traits on chromosome 3A of wheat. Crop Sci. 43: 1493–1505. [Google Scholar]
- Cattivelli, L., P. Baldi, C. Crosatti, N. Di Fonzo, P. Faccioli et al., 2002. Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Mol. Biol. 48: 649–665. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., and E. J. Routman, 1995. Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J. M., R. M. Depauw and L. L. Thiessen, 1998. Registration of wheat genetic stocks near-isogenic for photoperiod sensitivity. Crop Sci. 38: 282. [Google Scholar]
- Condon, A. G., R. A. Richards, G. J. Rebetzke and G. D. Farquhar, 2004. Breeding for high water-use efficiency. J. Exp. Bot. 55: 2447–2460. [DOI] [PubMed] [Google Scholar]
- Del Moral, L. G., Y. Rharrabti, D. Villegas and C. Royo, 2003. Evaluation of grain yield and its components in durum wheat under Mediterranean conditions: an ontogenic approach. Agron. J. 95: 266–274. [Google Scholar]
- De Vita, P., O. Li Destri Nicosia, F. Nigro, C. Platani, C. Riefolo et al., 2007. Breeding progress in morpho-physiological, agronomical and qualitative traits of durum wheat cultivars released in Italy during the 20th century. Eur. J. Agron. 26: 39–53. [Google Scholar]
- Dilbirligi, M., M. Erayman, B. T. Campbell, H. S. Randhawa, P. S. Baezinger et al., 2006. High-density mapping and comparative analysis of agronomically important traits on wheat chromosome 3A. Genomics 88: 74–87. [DOI] [PubMed] [Google Scholar]
- Dunkeloh, A., and J. Jacobeit, 2003. Circulation dynamics of Mediterranean precipitation variability 1948–98. Int. J. Climatol. 23: 1843–1866. [Google Scholar]
- Elouafi, I., and M. M. Nachit, 2004. A genetic linkage map of the Durum x Triticum dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits. Theor. Appl. Genet. 108: 401–413. [DOI] [PubMed] [Google Scholar]
- Gallagher, J. N., 1979. Field studies of cereal leaf growth: I. Initiation and expansion in relation to temperature and ontogeny. J. Exp. Bot. 117: 625–636. [Google Scholar]
- Gupta, P. K., R. K. Varshney, P. C. Sharma and B. Ramesh, 1999. Molecular markers and their applications in wheat breeding. Plant Breed. 118: 369–390. [Google Scholar]
- Gupta, P. K., S. Rustgi and N. Kumar, 2006. Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 49: 565–571. [DOI] [PubMed] [Google Scholar]
- Hanocq, E., M. Niarquin, E. Heumez, M. Rousset and J. Le Gouis, 2004. Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor. Appl. Genet. 110: 106–115. [DOI] [PubMed] [Google Scholar]
- Hanocq, E., A. Laperche, O. Jaminon, A. L. Lainé and J. Le Gouis, 2006. Most significant genome regions involved in the control of earliness traits in bread wheat, as revealed by QTL meta-analysis. Theor. Appl. Genet. 114: 569–584. [DOI] [PubMed] [Google Scholar]
- He, G., X. Luo, F. Tian, K. Li, Z. Zhu et al., 2007. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 16: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J. B., 2007. Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Huang, X. Q., H. Kempf, M. W. Ganal and M. S. Röder, 2004. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 933–943. [DOI] [PubMed] [Google Scholar]
- Huang, X. Q., S. Cloutier, L. Lycar, N. Radovanovic, D. G. Humphreys et al., 2006. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor. Appl. Genet. 113: 753–766. [DOI] [PubMed] [Google Scholar]
- Jiang, C., and Z. B. Zeng, 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.-H., Z.-B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavkin, E., and E. Coe, 1997. Mapped genomic locations for developmental functions and QTLs reflect concerted groups in maize (Zea mays L.). Theor. Appl. Genet. 95: 343–352. [Google Scholar]
- Kuchel, H., G. Hollamby, P. Langridge, K. Williams and S. P. Jefferies, 2006. Identification of genetic loci associated with ear-emergence in bread wheat. Theor. Appl. Genet. 113: 1103–1112. [DOI] [PubMed] [Google Scholar]
- Kuchel, H., K. J. Williams, P. Langridge, H. A. Eagles and S. P. Jefferies, 2007. Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theor. Appl. Genet. 115: 1029–1041. [DOI] [PubMed] [Google Scholar]
- Kumar, N., P. L. Kulwal, H. S. Balyan and P. K. Gupta, 2007. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 19: 163–177. [Google Scholar]
- Kusterer, B., J. Muminovic, H. F. Utz, H. P. Piepho, S. Barth et al., 2007. Analysis of a triple testcross design with recombinant inbred lines reveals a significant role of epistasis in heterosis for biomass-related traits in Arabidopsis. Genetics 175: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceras, J. C., G. Pantuwan, B. Jongdee and T. Toojinda, 2004. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 135: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi, P., M. C. Sanguineti, C. Liu, Y. Li, T. Y. Wang et al., 2007. Root-ABA1 QTL affects root lodging, grain yield, and other agronomic traits in maize grown under well-watered and water-stressed conditions. J. Exp. Bot. 58: 319–326. [DOI] [PubMed] [Google Scholar]
- Laurie, D. A., S. Griffiths, R. P. Dunford, V. Christodoulou, S. A. Taylor et al., 2004. Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crop Res. 90: 87–99. [Google Scholar]
- Leemans, R., and W. Cramer, 1991. The IIASA database for mean monthly values of temperature, precipitation and cloudiness on a global terrestrial grid. Pub. RR-91–18, International Institute of Applied Systems Analyses, Laxenburg, Austria.
- Li, W. L., J. C. Nelson, C. Y. Chu, L. H. Shi, S. H. Huang et al., 2002. Chromosomal locations and genetic relationships of tiller and spike characters in wheat. Euphytica 125: 357–366. [Google Scholar]
- Li, Z. K., S. R. Pinson, W. D. Park, A. H. Paterson and J. W. Stansel, 1997. Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. K., J. L. Luo, H. W. Mei, D. L. Wang, Q. Y. Shu et al., 2001. Overdominant epistatic loci are the primary genetic basis for inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158: 1737–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. K., B. Yu, H. R. Lafitte, N. Huang, B. Courtois et al., 2003. QTL x environment interactions in rice. I. Heading date and plant height. Theor. Appl. Genet. 108: 141–153. [DOI] [PubMed] [Google Scholar]
- Littel, R. C., G. A. Milliken, W. W. Stroup and R. D. Wollfinger, 1996. SAS System for Mixed Models. SAS Institute, Cary, NC.
- Liu, S., X. Zhang, M. O. Pumphrey, R. W. Stack, B. S. Gill et al., 2006. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genomics 6: 83–89. [DOI] [PubMed] [Google Scholar]
- Loss, S. P., and K. H. M. Siddique, 1994. Morphological and physiological traits associated with wheat yield increases in Mediterranean environments. Adv. Agron. 52: 229–276. [Google Scholar]
- Ludlow, M. M., and R. C. Muchow, 1990. A critical evaluation of traits for improving crop yields in water-limited environments. Adv. Agron. 43: 107–153. [Google Scholar]
- Luo, L. J., Z. K. Li, H. W. Mei, Q. Y. Shu, R. Tabien et al., 2001. Overdominant epistatic loci are the primary genetic basis for inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158: 1755–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z., D. Zhao, C. Zhang, Z. Zhang, S. Xue et al., 2007. Molecular genetic analysis of five spike-related traits in wheat using RIL and immortalized F2 populations. Mol. Genet. Genomics 277: 31–42. [DOI] [PubMed] [Google Scholar]
- Maccaferri, M., M. C. Sanguineti, E. Natoli, J. L. Araus-Ortega, M. Bensalem et al., 2006. A panel of elite accessions of durum wheat (Triticum durum Desf.) suitable for association mapping studies. Plant Genet. Resour. 4: 79–85. [Google Scholar]
- Mackay, I., and W. Powell, 2007. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 12: 57–63. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14: 253–257. [DOI] [PubMed] [Google Scholar]
- Malmberg, R. L., and R. Mauricio, 2005. QTL-based evidence for the role of epistasis in evolution. Genet. Res. 86: 89–95. [DOI] [PubMed] [Google Scholar]
- Marque, V., A. K. Fritz, T. J. Martin and G. M. Paulsen, 2004. Agronomic and quality attributes of winter durum wheat in the central great plains. Crop Sci. 44: 878–883. [Google Scholar]
- Marza, F., G. H. Bai, B. F. Carver and W. C. Zhou, 2006. Quantitative trait loci for yield and related traits in the wheat population Ning7840 x Clark. Theor. Appl. Genet. 112: 688–698. [DOI] [PubMed] [Google Scholar]
- McCartney, C. A., D. J. Somers, D. G. Humphreys, O. Lukow, N. Ames et al., 2005. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 x ‘AC Domain’. Genome 48: 870–883. [DOI] [PubMed] [Google Scholar]
- Mei, H. W., Z. K. Li, Q. Y. Shu, L. B. Guo, Y. P. Wang et al., 2005. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 110: 649–659. [DOI] [PubMed] [Google Scholar]
- Mei, H. W., J. L. Xu, Z. K. Li, X. Q. Yu, L. B. Guo et al., 2006. QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor. Appl. Genet. 112: 648–656. [DOI] [PubMed] [Google Scholar]
- Melchinger, A. E., H. H. Geiger, H. F. Utz and F. W. Schnell, 2003. Effect of recombinant in the parent populations on the means and combining ability variances in hybrid populations of maize (Zea mays L.). Theor. Appl. Genet. 106: 332–340. [DOI] [PubMed] [Google Scholar]
- Mohler, V., R. Lukman, S. Ortiz-Islas, M. William, A. J. Worland et al., 2004. Genetic and physical mapping of photoperiod insensitive gene Ppd-B1 in common wheat. Euphytica 138: 33–40. [Google Scholar]
- Motzo, R., and F. Giunta, 2007. The effect of breeding on the phenology of Italian durum wheats: from landraces to modern cultivars. Eur. J. Agron. 26: 462–470. [Google Scholar]
- Munkvold, J. D., R. A. Greene, C. E. Bermudez-Kandianis, C. M. La Rota, H. Edwards et al., 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachit, M. M., I. Elouafi, M. A. Pagnotta, A. El Saleh, E. Iacono et al., 2001. Molecular linkage map for an intraspecific recombinant inbred population of durum wheat (Triticum turgidum L. var. durum). Theor. Appl. Genet. 102: 177–186. [Google Scholar]
- Narasimhamoorthy, B., B. S. Gill, A. K. Fritz, J. C. Nelson and G. L. Brown-Guedira, 2006. Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theor. Appl. Genet. 112: 787–796. [DOI] [PubMed] [Google Scholar]
- Peng, J., Y. Ronin, T. Fahima, M. S. Röder, Y. Li et al., 2003. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 100: 2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, W. H., K. D. Sayre and M. P. Reynolds, 2000. Enhancing genetic grain yield potential and yield stability in durum wheat, pp. 83–93 in Options Mediterraneennes, Serie A, Seminaires Mediterraneens, Vol. 40, edited by C. Royo, M. M. Nachit, N. di Fonzo and J. L. Araus. IAMZ, Zaragoza, Spain.
- Qi, L. L., B. Echalier, S. Chao, G. R. Lazo, G. E. Butler et al., 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie, S. A., A. Steed, C. Calestani, A. Semikhodskii, C. Lebreton et al., 2005. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 110: 865–880. [DOI] [PubMed] [Google Scholar]
- Quarrie, S. A., S. Pekic-Quarrie, R. Radosevic, D. Rancic, A. Kaminska et al., 2007. Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J. Exp. Bot. 57: 2627–2637. [DOI] [PubMed] [Google Scholar]
- Reynolds, M., D. Calderini, A. Condon and M. Vargas, 2007. Association of source/sink traits with yield, biomass and radiation use efficiency among random sister lines from three wheat crosses in a high-yield environment. J. Agric. Sci. 145: 3–16. [Google Scholar]
- Richards, R. A., 1996. Defining selection criteria to improve yield under drought. Plant Growth Reg. 20: 157–166. [Google Scholar]
- Richards, R. A., 2000. Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 51: 447–458. [DOI] [PubMed] [Google Scholar]
- Röder, M. S., V. Korzun, K. Wendehake, J. Plaschke, M. H. Tixier et al., 1998. A microsatellite map of wheat. Genetics 149: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, B. F., B. L. Joiner and T. A. Ryan, Jr., 1985. Minitab Handbook, Ed. 2. PWS Publishers, Boston.
- Salvi, S., and R. Tuberosa, 2005. To clone or not to clone plant QTLs: present and future challenge. Trends Plant Sci. 10: 297–304. [DOI] [PubMed] [Google Scholar]
- Salvi, S., and R. Tuberosa, 2007. Cloning plant QTLs, pp. 207–224 in Genomics Platforms and Approaches (Genomics-Assisted Crop Improvement, Vol. 1), edited by R. K. Varshney and R. Tuberosa. Springer, Dordrecht, The Netherlands.
- Sanguineti, M. C., S. Li, M. Maccaferri, S. Corneti, F. Rotondo et al., 2007. Dissection of genetic variability for root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 151: 291–305. [Google Scholar]
- SAS Institute, 2001. An introduction to the analysis of linear mixed models using SAS version 8.2. SAS Institute, Cary, NC.
- Sawkins, M. C., A. D. Farmer, D. Hoisington, J. Sullivan, A. Tolopko et al., 2004. Comparative map and trait viewer (CMTV): an integrated bioinformatic tool to construct consensus maps and compare QTL and functional genomics data across genomes and experiments. Plant Mol. Biol. 56: 465–480. [DOI] [PubMed] [Google Scholar]
- Scarth, R., and C. N. Law, 1983. The location of the photoperiod gene Ppd2 and an additional genetic factor for ear emergence time on chromosome 2B of wheat. Heredity 51: 607–619. [Google Scholar]
- Schaeffer, M., P. Byrne and E. H. Coe, 2006. Consensus quantitative trait maps in maize: a database strategy. Maydica 51: 357–367. [Google Scholar]
- Schiex, T., and C. Gaspin, 1997. CARTHAGENE: constructing and joining maximum likelihood gene maps. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 258–267. [PubMed] [Google Scholar]
- Schön, C. C., H. F. Utz, S. Groh, B. Truberg, S. Openshaw et al., 2004. Quantitative trait locus mapping based on resampling in a vast maize testcross experiment and its relevance to quantitative genetics for complex traits. Genetics 167: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, G., 1978. Estimating the dimension of a model. Ann. Stat. 6: 461–464. [Google Scholar]
- Slafer, G. A., and F. H. Andrade, 1993. Physiological attributes to the generation of grain yield in bread wheat cultivars released at different eras. Field Crops Res. 31: 351–367. [Google Scholar]
- Slafer, G. A., and J. L. Araus, 2007. Physiological traits for improving wheat yield under a wide range of environments, pp. 147–156 in Scale and Complexity in Plant Systems Research (Wageningen UR Frontis Series, Vol. 21), edited by J. H. J. Spiertz, P. C. Struik and H. H. van Laar. Springer, Dordrecht, The Netherlands.
- Slafer, G. A., D. F. Calderini and D. J. Miralles, 1996. Yield components and compensation in wheat: opportunities for further increasing yield potential, pp. 101–134 in Increasing Yield Potential in Wheat: Breaking the Barriers, edited by M. P. Reynolds, S. Rajaram and A. McNab. CIMMYT, El-Batan, Mexico.
- Snape, J. W., K. Butterworth, E. Whitechurch and A. J. Worland, 2001. Waiting for fine times: genetics of flowering time in wheat. Euphytica 119: 185–190. [Google Scholar]
- Somers, D. J., P. Isaac and K. Edwards, 2004. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Song, Q. J., J. R. Shi, S. Singh, E. W. Fickus, J. M. Costa et al., 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110: 550–560. [DOI] [PubMed] [Google Scholar]
- Song, X. J., W. Huang, M. Shi, M. Z. Zhu and H. X. Lin, 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Sourdille, P., T. Cadalen, H. Guyomarc'h, J. W. Snape, M. R. Perretant et al., 2003. An update of the Courtot x Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 106: 530–538. [DOI] [PubMed] [Google Scholar]
- Sourdille, P., S. Sukhwinder, T. Cadalen, G. Brown-Guedira, G. Gay et al., 2004. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 4: 12–25. [DOI] [PubMed] [Google Scholar]
- Spielmeyer, W., J. Hyles, P. Joaquim, F. Azanza, D. Bonnet et al., 2007. A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor. Appl. Genet. 115: 59–66. [DOI] [PubMed] [Google Scholar]
- Stam, P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3: 739–744. [Google Scholar]
- Stich, B., J. M. Yu, A. E. Melchinger, H. P. Piepho, H. F. Utz et al., 2007. Power to detect higher-order epistatic interactions in a metabolic pathway using a new mapping strategy. Genetics 176: 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szucs, P., J. S. Skinner, I. Karsai, A. Cuesta-Marcos, K. G. Haggard et al., 2007. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol. Genet. Genomics 277: 249–261. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., 1993. Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Tuberosa, R., 2004. Molecular approaches to unravel the genetic basis of water use efficiency, pp. 228–301 in Water Use Efficiency in Plant Biology, edited by M. A. Bacon. Blackwell, Oxford.
- Tuberosa, R., and S. Salvi, 2006. Genomics approaches to improve drought tolerance in crops. Trends Plant Sci. 11: 405–412. [DOI] [PubMed] [Google Scholar]
- Tuberosa, R., B. S. Gill and S. Quarrie, 2002. a Cereal genomics: ushering in a brave new world. Plant Mol. Biol. 48: 445–449. [DOI] [PubMed] [Google Scholar]
- Tuberosa, R., S. Salvi, M. C. Sanguineti, P. Landi, M. Maccaferri et al., 2002. b Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann. Bot. 89: 941–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, X. Y., J. M. Wan, J. F. Weng, L. Jiang, J. C. Bi et al., 2005. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 110: 1334–1346. [DOI] [PubMed] [Google Scholar]
- Wan, X. Y., J. M. Wan, L. Jiang, J. K. Wang, H. Q. Zhai et al., 2006. QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor. Appl. Genet. 112: 1258–1270. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten, P. Gaffney and Z. B. Zeng, 2005. Windows QTL Cartographer 2.5. North Carolina State University, Raleigh, NC.
- Worland, A., 1996. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89: 49–57. [Google Scholar]
- Yadav, R. S., C. T. Hash, F. R. Bidinger, G. P. Cavan and C. J. Howarth, 2002. Quantitative trait loci associated with traits determining grain and stover yield in pearl millet under terminal drought-stress conditions. Theor. Appl. Genet. 104: 67–83. [DOI] [PubMed] [Google Scholar]
- Yan, L., D. Fu, C. Li, A. Blechl, G. Tranquilli et al., 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., and T. Sasaki, 1997. Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35: 145–153. [PubMed] [Google Scholar]
- Zadoks, J. C., T. T. Chang and C. F. Konzak, 1974. A decimal code for the growth stage of cereals. Weed Res. 14: 415–421. [Google Scholar]
- Zaharieva, M., E. Gaulin, M. Havaux, E. Acevedo and P. Monneveux, 2001. Drought and heat responses in the wild wheat relative Aegilops geniculata Roth: potential interest for wheat improvement. Crop Sci. 41: 1321–1329. [Google Scholar]
- Zeng, H., Y. Zhong and L. Luo, 2006. Drought tolerance genes in rice. Funct. Integr. Genomics 6: 338–341. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 2005. QTL mapping and the genetic basis of adaptation: recent developments. Genetica 123: 25–37. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., C. H. Kao and C. J. Basten, 1999. Estimating the genetic architecture of quantitative traits. Genetics 74: 279–289. [DOI] [PubMed] [Google Scholar]