Abstract
The two parts of the Caenorhabditis elegans reproductive system, the germ cells and the somatic reproductive tissues, each influence the life span of the animal. Removing the germ cells increases longevity, and this life span extension requires the somatic gonad. Here we show that the somatic gonad and the germ cells make distinct contributions to life span determination. The life span increase produced by loss of the germ cells requires the DAF-16/FOXO transcription factor. In response to germ-cell removal, DAF-16 accumulates in nuclei. We find that the somatic gonad is not required for DAF-16 nuclear accumulation or for the increased stress resistance that is produced by germ-cell removal. The somatic gonad is required, however, for expression of specific DAF-16 target genes. DAF-16 is known to be activated by reduced insulin/IGF-1 signaling in C. elegans. In certain insulin/IGF-1-pathway mutants, the somatic gonad is not required for germ-cell removal to extend life span. Our genetic experiments suggest that these mutations reduce insulin/IGF-1 signaling below a critical threshold level. At these low levels of insulin/IGF-1 signaling, factors normally provided by the somatic gonad are no longer needed for germ-cell removal to increase the expression of DAF-16 target genes.
THE reproductive system of Caenorhabditis elegans influences the animal's life span. When the germline precursor cells are removed at the time of hatching by laser microsurgery, life span is increased by ∼60% (Hsin and Kenyon 1999). This life span extension requires signals from the somatic reproductive tissues (somatic gonad), because it is not observed when both the germline and the somatic gonad are removed (Hsin and Kenyon 1999; Arantes-Oliveira et al. 2002). Removing the germ cells increases life span, at least in part, by influencing the FOXO-family transcription factor DAF-16, which is completely required for germline removal to extend life span (Hsin and Kenyon 1999). In animals lacking germ cells, DAF-16 accumulates in the nuclei of intestinal cells, and, to a lesser extent, those of other cell types (Lin et al. 2001). Intestinal DAF-16 activity appears to be important for life span extension, because in a daf-16(mu86) null mutant background, expressing daf-16 in the intestine is sufficient to rescue the entire life span extension produced by germ-cell removal (Libina et al. 2003). Germline removal appears to extend life span, at least in part, by activating a lipophilic signaling pathway (Hsin and Kenyon 1999; Broue et al. 2007; Gerisch et al. 2001, 2007) involving the intestinal adaptor protein KRI-1, which in turn mediates the nuclear localization of DAF-16 in the intestine (Berman and Kenyon 2006; Gerisch et al. 2007).
How the somatic reproductive tissues function to extend the life span of germline-less animals is not well understood. It is possible that the germ cells and the somatic tissues function in a purely linear pathway, with the somatic gonad sensing the absence of the germ cells and, in turn, sending life-span-extending signals to the other tissues. In this scenario, all of the effects of germline removal would require the presence of the somatic gonad. However, it is also possible that the germline and somatic gonad play qualitatively different roles in a more complex pathway that extends life span.
So far, the only gene implicated in the somatic-gonad signaling pathway is the insulin/IGF-1 receptor gene daf-2 (Hsin and Kenyon 1999). The insulin/IGF-1 signaling pathway is known to limit longevity in many organisms (Tatar et al. 2003; Kenyon 2005; Conover and Bale 2007; Selman et al. 2007; Taguchi et al. 2007). In normal animals with an intact reproductive system, daf-2 reduction-of-function mutations extend life span about two-fold, and this life span extension is daf-16 dependent (Kenyon et al. 1993; Larsen et al. 1995). In the wild type, DAF-2 activity is thought to shorten life span by activating the PI3-kinase AGE-1. The phosphorylated lipids generated by AGE-1 are predicted to activate several downstream kinases including PDK-1, AKT-1, AKT-2, and SGK-1 (Paradis and Ruvkun 1998; Paradis et al. 1999; Hertweck et al. 2004). Phosphorylation of DAF-16 by AKT-1, AKT-2, and SGK-1 prevents DAF-16 from accumulating in the nucleus (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001) and changing the expression of downstream genes whose expression more directly affects life span (Lee et al. 2003; McElwee et al. 2003; Murphy et al. 2003; Oh et al. 2006; Dong et al. 2007). By dephosphorylating the phospholipids generated by AGE-1, DAF-18, a lipid phosphatase, acts in opposition to AGE-1 (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999). Loss-of-function mutation of daf-18 prevents DAF-16 nuclear localization and shortens life span (Dorman et al. 1995; Larsen et al. 1995; Lin et al. 2001).
In animals carrying the daf-2(e1370) reduction-of-function mutation, which changes a residue in the intracellular tyrosine-kinase domain (Kimura et al. 1997), removing the germ cells extends life span even in the absence of the somatic gonad (Hsin and Kenyon 1999). This finding is consistent with the idea that the somatic gonad extends the life span of animals that lack germ cells by downregulating the insulin/IGF-1 pathway (Hsin and Kenyon 1999). Alternatively, the daf-2(e1370) mutation could activate a parallel pathway that compensates for the loss of the somatic gonad. Curiously, not all daf-2 mutations behave like daf-2(e1370). For example, in animals carrying the daf-2 ligand-binding domain mutation e1368 (Kimura et al. 1997), the additional life span extension produced by germline removal requires the somatic gonad, as in the wild type (Hsin and Kenyon 1999).
In this study, we address key questions about the role of the somatic gonad in the longevity of animals that lack the germline. First, to better understand how the germ cells and somatic tissues interact to affect longevity, we ask whether the somatic gonad is required for specific events that occur when the germline is removed. In addition, we ask why different daf-2 mutations have different effects on the reproductive signaling system.
MATERIALS AND METHODS
C. elegans strains:
All strains used in this study were maintained as described previously (Brenner 1974). The following strains were used:
N2 (wild type)
CF2049 akt-1(ok525) obtained from the CGC and outcrossed to our laboratory N2 three times.
CF2050 akt-2(ok393) obtained from the CGC and outcrossed to our laboratory N2 three times.
JT709 pdk-1(sa709)
CF1379 daf-2(mu150)
CF1934 daf-16(mu86); muIs109[Pdaf-16∷gfp∷daf-16cDNA + Podr-1∷rfp]
CF2688 daf-16(mu86); daf-2(e1368); muIs112[Pdaf-16∷gfp∷daf-16cDNA + Podr-1∷rfp]
CF1553 muIs84[Psod-3∷gfp]
CF1874 daf-16(mu86); muIs84[Psod-3∷gfp]
CF2533 daf-2(e1368); muIs84[Psod-3∷gfp]
CF1580 daf-2(e1370); muIs84[Psod-3∷gfp]
CF2683 daf-16(mu86); daf-2(e1368); muIs84[Psod-3∷gfp]
CF1588 daf-16(mu86); daf-2(e1370); muIs84[Psod-3∷gfp]
CF2630 sIs10314[Pdod-8∷gfp + pCeh361], obtained by outcrossing BC12544 to our laboratory N2 two times.
CF2676 daf-16(mu86); sIs10314[Pdod-8∷gfp + pCeh361]
CF2760 muEx405[Pdod-8∷rfpnls]
CF2922 daf-16(mu86); muEx405[Pdod-8∷rfpnls]
BC11128 dpy-5(e907); sEx11128[Pgpd-2∷gfp + pCeh361]
BC10466 dpy-5(e907); sEx10466[Pnnt-1∷gfp + pCeh361]
CF2923 daf-16(mu86); sEx10466[Pnnt-1∷gfp + pCeh361].
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health's National Center for Research Resources. The Pdod-8∷rfp transcriptional fusion was constructed using 1856 bp of DNA upstream of the predicted dod-8 translational start site. The Psod-3∷gfp strain was described previously (Libina et al. 2003). The daf-16∷gfp strain used was described previously (Berman and Kenyon 2006). All other DAF-16 target-gene reporter fusions were obtained from the Genome British Columbia C. elegans Gene Expression Consortium (McKay et al. 2003).
Laser ablation:
Laser ablations of germ-cell (Z2 and Z3) or somatic-gonad (Z1 and Z4) precursor cells in newly hatched L1 larvae were performed as described previously (Hsin and Kenyon 1999) using a VSL-337 nitrogen pumped dye laser (Laser Sciences). At adulthood, the absence of the gonad or germ cells was confirmed using a dissecting microscope. Intact controls were anesthetized and recovered from the same NaN3 agarose pads as experimental animals.
Life span analysis:
Life span analysis was performed at 20° as described previously (Kenyon et al. 1993; Arantes-Oliveira et al. 2003). Ablated animals were examined at day 1 of adulthood for the absence of germ cells or the whole gonad. Statview 4.5 software (Abacus) was used for statistical analysis.
Stress resistance assays:
To test oxidative stress resistance, animals were grown to day 2 of adulthood on standard agar plates and then placed in 300 mm paraquat dissolved in M9 media. Death, scored as an absence of movement, was assayed every hour. Statview 4.5 software (Abacus) was used for statistical analysis.
RNA mediated interference (RNAi):
RNAi by feeding was performed as described previously (Timmons et al. 2001). dsRNA production was induced by adding 100 μl of 0.1 m IPTG to bacterial lawns several hours to one day before adding worms. RNAi treatment was initiated shortly after the animals were ablated as young L1 larvae. For life span analysis, animals were moved to fresh lawns every 4 to 7 days. HT115 bacteria carrying the pAD48 construct described previously (Dillin et al. 2002) was used to knock down daf-2. HT115 bacteria carrying the backbone vector only construct pAD12 was used for the pdk-1(sa709) and daf-2(mu150) life spans described below. All other life spans were performed using OP50 bacteria.
GFP fluorescence microscopy and quantification:
On day 2 of adulthood, animals were anesthetized on agarose pads containing either 0.15 m NaN3 (DAF-16 target expression) or levamisole (DAF-16∷GFP localization). Whole worm images were taken using a Retiga EXi Fast1394 CCD digital camera (QImaging, Burnaby, British Columbia, Canada) using the 10× objective on a Zeiss Axioplan 2 compound microscope (Zeiss, Germany). Because expression of the various transgenes was primarily in the intestine, each image was taken so that the intestine was in focus. For an individual trial, exposure time was calibrated to minimize the number of saturated pixels for the set of animals. Openlab 4.0.2 software was used to quantify intensity of fluorescent worm images. For Psod-3∷gfp quantification, the vulval expression, which was very bright, was excluded, since this structure is not present in animals lacking the gonad. For all other GFP constructs, fluorescence of the entire animal was measured. None of the constructs had visible expression in embryos while retained in the adult prior to egg laying. Total fluorescence was calculated by the Open Lab program as measured by intensity of each pixel in the selected area of image (i.e., the worm). Image processing for figures was performed using Adobe Photoshop 7.0.
To assess differences in expression of DAF-16 target genes, we also attempted qRT–PCR. However, problems with normalization of gene expression between intact and germline-less animals confounded interpretation of the results. The qRT–PCR must be done during adulthood, when we observe changes in DAF-16 localization. However, at this stage, animals with intact gonads have approximately three times the number of cells as do animals missing the germline (adult hermaphrodites contain ∼2000 germ cells and 959 somatic cells). Thus, for example, normalization to a gene expressed in both the germline and soma would make the expression of a soma-specific gene appear to be increased in animals missing the germline. Moreover, methods to determine the level of gene expression in the germline are problematic: in situ hybridization in C. elegans is not straightforward, and transgene expression is often silenced in the germline.
RESULTS
The somatic gonad is not required for loss of the germline to stimulate DAF-16 nuclear localization:
To better understand the relationship between the germ cells and the somatic gonad in this longevity pathway, we asked whether the molecular events known to occur when the germline is removed require the presence of the somatic gonad. In wild-type animals with an intact reproductive system, a functional GFP-tagged DAF-16 protein is distributed diffusely throughout the cells of the animal. Laser ablation of the two germ-cell (germline) precursors, Z2 and Z3, in newly hatched animals causes DAF-16∷GFP to accumulate in the nuclei of intestinal cells, where it functions to extend life span when the animal reaches adulthood (Lin et al. 2001; Arantes-Oliveira et al. 2002; Libina et al. 2003). To determine whether the presence of the somatic gonad was required for this nuclear localization of DAF-16, we removed the whole gonad, that is, both the germline and the somatic gonad, by killing the cells Z1 and Z4. These two cells give rise to all of the somatic reproductive tissues, which in turn are required for the development of the germline. We found that in adult animals lacking the somatic gonad as well as the germ cells, DAF-16∷GFP was present in intestinal nuclei (Figure 1A). Thus, the somatic gonad is not required for DAF-16 nuclear accumulation in animals lacking a germline. This finding argues against the model that loss of the germline extends life span exclusively by derepressing a longevity function of the somatic gonad. Instead, it appears that the germ cells influence DAF-16 nuclear localization independently of the somatic gonad, and the somatic gonad plays another role that is required for longevity.
Figure 1.—
The effects of somatic-gonad removal on the pattern of DAF-16∷GFP. (A) The somatic gonad is not required for DAF-16∷GFP nuclear localization in animals that lack germ cells. Arrows indicate nuclear localization of DAF-16∷GFP in intestinal cells of day-2 adults lacking the germ cells (Z2 and Z3 ablated at hatching) or the germ cells as well as the somatic gonad (Z1 and Z4 ablated at hatching). Approximately 100 animals were examined in multiple trials, and the animals shown are representative. Nuclear localization of DAF-16∷GFP was observed in all of the animals lacking either germ cells or the whole gonad. (B) Somatic-gonad removal affects the level of DAF-16∷GFP. Removing the somatic gonad produced a modest but statistically significant decrease in the level of DAF-16∷GFP fluorescence. CF1934 intact control, n = 8, m = 1 ± 0.11; Z2/3, n = 14, m = 0.87 ± 0.037, P = 0.28; Z1/4, n = 6, m = 0.76 ± 0.028, P = 0.072, P′ = 0.039. Mean fluorescence intensity given is relative to intact control. P, the P-value (Student's t-test) compared to intact. P′, the P-value comparing germ-cell (Z2/Z3) ablation to whole-gonad (Z1/Z4) ablation.
The finding that nuclear-localized DAF-16 is not sufficient to extend life span is in keeping with previous findings. For example, when the AKT-phosphorylation sites on DAF-16 are mutated, the protein localizes to the nucleus constitutively but extends life span only modestly. When a daf-2 mutation is introduced, or the germline is removed, life span is greatly extended (Lin et al. 2001; Berman and Kenyon 2006). Thus, both daf-2 mutation and germline ablation must do more to extend life span than simply trigger DAF-16 nuclear localization.
Removal of the somatic gonad lowers the level of DAF-16:
Since the somatic gonad does not control DAF-16 nuclear localization, how does it contribute to longevity? When we quantified the amount of DAF-16∷GFP by measuring fluorescence intensity, we observed that animals lacking both the germline and the somatic gonad had somewhat lower levels of DAF-16∷GFP than did animals lacking only the germline (Figure 1B). The significance of this is not clear at this time, especially since we did not measure endogenous DAF-16 protein levels. However, this finding raises the possibility that the somatic gonad may promote longevity by elevating the level of DAF-16.
The somatic gonad affects the ability of DAF-16 to activate some of its target genes:
We next asked whether the somatic gonad influences DAF-16's ability to activate its target genes in animals lacking germ cells. Genes whose expression changes in a daf-16-dependent fashion in daf-2 mutants have been identified using microarray analysis and other methods (McElwee et al. 2003; Murphy et al. 2003; Oh et al. 2006; Dong et al. 2007). Thus, we began by asking whether any of these genes was also regulated by the reproductive system.
We attempted to analyze gene expression levels using quantitative RT–PCR, but this approach was confounded by the fact that the reproductive system comprises so much of the mass of the animal (see materials and methods). Instead, we obtained transgenic animals carrying GFP or RFP promoter fusions to a number of these genes to assess changes in expression in the animal by fluorescence intensity. We tested whether the expression of each was increased in response to germline ablation, and, if so, whether its upregulation required the somatic gonad. As described next, we identified four germline-regulated genes among these transgenic lines, and these genes fell into two classes.
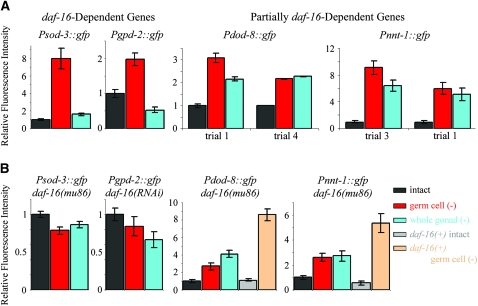
The sod-3 (Mn++ superoxide dismutase) promoter contains multiple canonical DAF-16-binding elements [T(G/A)TTTAC] and binds DAF-16 directly (Honda and Honda 1999; Furuyama et al. 2000; Oh et al. 2006). Previously, we constructed a transcriptional Psod-3∷gfp fusion gene, and found that its expression was increased in many tissues in response to daf-2 mutation in a daf-16 dependent fashion (Libina et al. 2003), as predicted by previous findings (Honda and Honda 1999). We found that this Psod-3∷gfp transgene was also upregulated in response to germline removal, in a daf-16-dependent fashion (Figure 2B; Table 2). Next, we asked whether the upregulation of Psod-3∷gfp in response to germline removal required the activity of the somatic gonad. We found that when we removed the whole gonad, the expression of sod-3 decreased dramatically relative to germ-cell-ablated animals, to a level only slightly higher than in animals with an intact gonad. We observed this substantial decrease in expression in each of three experiments (Figure 2A; Table 1).
Figure 2.—
The somatic gonad is required for expression of some DAF-16-regulated genes in animals that lack germ cells. (A) Expression of GFP reporters in animals lacking germ cells (Z2 and Z3 ablated) or the somatic gonad and germ cells (Z1 and Z4 ablated). Values for histograms are given in Table 1. Psod-3∷gfp and Pgpd-2∷gfp expression requires the presence of the somatic gonad in germ-cell ablated animals. In five of nine trials, there was a statistically significant decrease in expression of dod-8 in whole-gonad ablated animals. Trial 1, shown here, is representative of this observation. In four of nine trials (such as trial 4, displayed here) there was no significant decrease in dod-8 expression upon whole gonad ablation. In one of four trials (trial 3, shown here) there was a statistically significant decrease in expression of nnt-1 when the somatic gonad as well as the germ cells were removed (Z1 and Z4 ablated). In three of four trials (such as trial 1, shown here) there was no significant decrease in nnt-1 expression upon whole gonad ablation. (B) Expression of GFP reporters in daf-16(mu86) mutants lacking either the germ cells (Z2 and Z3 ablated) or the germ cells and the somatic gonad (Z1 and Z4 ablated). Thus, the increase in Psod-3∷gfp and Pgpd-2∷gfp expression produced by germline removal requires daf-16 and the somatic gonad. In contrast, the increase in Pdod-8∷gfp and Pnnt-1∷gfp expression is partially daf-16 and somatic-gonad independent. For complete data set, see Table 2.
TABLE 2.
daf-16 dependence of gene expression in animals lacking germ cells
| Construct | Construct no. | Strain | Trial | daf-16 genotype | Intact | n | P | Germ cell (−) | n | P | Whole gonad (−) | n | P | P′ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sod-3∷gfp | muIs84 | CF1874 | 1 | daf-16(mu86) | 1 ± 0.044 | 29 | — | 0.79 ± 0.044 | 26 | 0.001 | 0.87 ± 0.042 | 24 | 0.034 | 0.17 |
| gpd-2∷gfp | sEx11128 | BC11128 | 1 | daf-16(RNAi) | 1 ± 0.087 | 20 | — | 0.84 ± 0.13 | 19 | 0.301 | 0.66 ± 0.11 | 18 | 0.019 | 0.29 |
| BC11128 | daf-16(+) | 0.9 ± 0.077 | 19 | 0.42 | 1.5 ± 0.16 | 17 | 0.005 | |||||||
| BC11128 | 2 | daf-16(RNAi) | 1 ± 0.087 | 25 | — | 0.55 ± 0.11 | 16 | <0.001 | 0.24 ± 0.062 | 27 | <0.001 | 0.14 | ||
| BC11128 | daf-16(+) | 0.63 ± 0.068 | 25 | 0.002 | 1.8 ± 0.12 | 21 | <0.001 | |||||||
| dod-8∷gfp | sIs10314 | CF2676 | 1 | daf-16(mu86) | 1 ± 0.077 | 24 | — | 1.8 ± 0.14 | 17 | <0.001 | 2.3 ± 0.15 | 27 | <0.001 | 0.016 |
| CF2630 | daf-16(+) | 1.3 ± 0.077 | 30 | 0.006 | ||||||||||
| CF2676 | 2 | daf-16(mu86) | 1 ± 0.19 | 21 | — | 2.7 ± 0.44 | 18 | 0.002 | 4.1 ± 0.39 | 9 | <0.001 | 0.023 | ||
| CF2630 | daf-16(+) | 1.1 ± 0.15 | 24 | 0.67 | 8.6 ± 0.69 | 25 | <0.001 | |||||||
| dod-8∷rfp | muEx405 | CF2922 | 1 | daf-16(mu86) | 1 ± 0.14 | 17 | — | 2.3 ± 0.29 | 24 | <0.001 | 1.6 ± 0.19 | 28 | 0.011 | 0.07 |
| CF2760 | daf-16(+)a | 0.84 ± 0.13 | 20 | 0.42 | 4.0 ± 0.33 | 18 | <0.001 | 2.4 ± 0.33 | 17 | <0.001 | ||||
| CF2922 | 2 | daf-16(mu86) | 1 ± 0.12 | 12 | — | 2.0 ± 0.30 | 13 | 0.009 | 2.6 ± 0.42 | 17 | 0.001 | 0.19 | ||
| CF2760 | daf-16(+)b | 1.7 ± 0.23 | 13 | 0.016 | 3.6 ± 0.32 | 16 | <0.001 | 3.7 ± 0.38 | 12 | <0.001 | ||||
| nnt-1∷gfp | sEx10466 | BC10466 | 1 | daf-16(RNAi) | 1 ± 0.21 | 19 | — | 1.5 ± 0.23 | 28 | 0.125 | 2.2 ± 0.26 | 40 | <0.001 | 0.03 |
| BC10466 | daf-16(+) | 1.2 ± 0.24 | 16 | 0.48 | 2.8 ± 0.41 | 17 | <0.001 | |||||||
| CF2923 | 2 | daf-16(mu86) | 1 ± 0.17 | 35 | — | 2.6 ± 0.32 | 39 | <0.001 | 2.7 ± 0.40 | 33 | <0.001 | 0.88 | ||
| BC10466 | daf-16(+)c | 0.56 ± 0.15 | 31 | 0.05 | 5.4 ± 0.787 | 23 | <0.001 | 6.2 ± 0.96 | 25 | <0.001 |
Values represent mean fluorescence intensity relative to intact daf-16(−) controls. Fluorescence was measured on day 2 of adulthood. P represents the P-value (Student's t-test) compared to intact control, and P′ represents the P-value compared to animals lacking germ cells (Z2 and Z3 ablated).
Same data labeled trial 4 for dod-8∷rfp in Table 1.
Same data labeled trial 5 for dod-8∷rfp in Table 1.
Same data labeled trial 4 for nnt-1∷gfp in Table 1.
TABLE 1.
The somatic gonad is required for DAF-16 to activate some of its target genes in animals that lack germ cells
| Construct | Construct no. | Strain | Trial | Intact | n | Germ cell (−) | n | P | Whole gonad (−) | n | P | P′ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sod-3∷gfpa | muIs84 | CF1553 | 1 | 1 ± 0.048 | 34 | 8.0 ± 1.17 | 26 | <0.001 | 1.6 ± 0.12 | 32 | <0.001 | <0.001 |
| gpd-2∷gfp | sEx11128 | BC11128 | 1 | 1 ± 0.12 | 19 | 2.0 ± 0.185 | 17 | <0.001 | 0.53 ± 0.073 | 16 | 0.002 | <0.001 |
| 2 | 1 ± 0.076 | 28 | 0.91 ± 0.12 | 26 | <0.001 | 0.60 ± 0.042 | 32 | <0.001 | 0.031 | |||
| 3 | 1 ± 0.12 | 19 | 1.47 ± 0.18 | 13 | 0.04 | 0.76 ± 0.14 | 18 | 0.21 | 0.004 | |||
| dod-8∷gfp | sIs10314 | CF2630 | 1 | 1 ± 0.007 | 28 | 2.17 ± 0.026 | 38 | <0.001 | 2.3 ± 0.017 | 32 | <0.001 | 0.60 |
| 2 | 1 ± 0.11 | 36 | 15.9 ± 1.5 | 35 | <0.001 | 9.9 ± 0.49 | 46 | <0.001 | <0.001 | |||
| 3 | 1 ± 0.10 | 30 | 3.3 ± 0.35 | 27 | <0.001 | 2.5 ± 0.15 | 33 | <0.001 | 0.033 | |||
| 4 | 1 ± 0.086 | 31 | 3.1 ± 0.22 | 34 | <0.001 | 2.2 ± 0.091 | 35 | <0.001 | <0.001 | |||
| dod-8∷rfp | muEx405 | CF2760 | 1 | 1 ± 0.13 | 27 | 2.8 ± 0.19 | 26 | <0.001 | 1.9 ± 0.14 | 38 | <0.001 | <0.001 |
| 2 | 1 ± 0.12 | 34 | 4.2 ± 0.40 | 37 | <0.001 | 4.2 ± 0.35 | 43 | <0.001 | 0.92 | |||
| 3 | 1 ± 0.17 | 28 | 4.8 ± 0.34 | 35 | <0.001 | 4.0 ± 0.44 | 21 | <0.001 | 0.14 | |||
| 4b | 1 ± 0.16 | 20 | 4.8 ± 0.40 | 18 | <0.001 | 2.9 ± 0.40 | 17 | <0.001 | 0.002 | |||
| 5c | 1 ± 0.13 | 13 | 2.2 ± 0.19 | 16 | <0.001 | 2.2 ± 0.23 | 12 | <0.001 | 0.93 | |||
| nnt-1∷gfp | sEx10466 | BC10466 | 1 | 1 ± 0.25 | 23 | 6.0 ± 0.92 | 23 | <0.001 | 5.1 ± 0.92 | 23 | 0.002 | 0.57 |
| 2 | 1 ± 0.23 | 34 | 3.5 ± 0.43 | 29 | <0.001 | 4.2 ± 0.47 | 35 | <0.001 | 0.25 | |||
| 3 | 1 ± 0.14 | 30 | 9.1 ± 1.0 | 32 | <0.001 | 6.4 ± 0.80 | 29 | <0.001 | 0.04 | |||
| 4d | 1 ± 0.27 | 31 | 9.6 ± 1.4 | 23 | <0.001 | 11 ± 1.7 | 25 | <0.001 | 0.51 |
Values represent mean fluorescence intensity relative to intact controls. Fluorescence was measured on day 2 of adulthood. P represents the P-value (Student's t-test) compared to intact control, and P′ represents the P-value compared to animals lacking germ cells (Z2 and Z3 ablated).
Changes in Psod-3∷gfp expression were observed twice by eye and quantification was performed on the third trial.
Same data labeled trial 1 of dod-8∷rfp in Table 2.
Same data labeled trial 2 of dod-8∷rfp in Table 2.
Same data labeled trial 2 of nnt-1∷gfp in Table 2.
The glyceraldehyde 3-phosphate dehydrogenase gene gpd-2 was identified as a DAF-16-regulated gene through microarray analysis (Murphy et al. 2003). The promoter fragment of gpd-2 used to construct the gfp transgene contains one canonical DAF-16 binding site, but whether this gene (or the other two genes we examined) is a direct target of DAF-16 is not known. We found that, like Psod-3∷gfp, Pgpd-2∷gfp was regulated by the germline in a daf-16-dependent fashion (Figure 2B; Table 2). We observed increased expression in germ-cell-ablated animals in four of five trials, and we saw no induction of expression when daf-16 was reduced by RNAi (Figure 2B; Table 2). As with sod-3, we also saw a consistent decrease in transgene expression relative to that seen in germline-ablated animals upon whole-gonad ablation (Figure 2A; Table 1).
The remaining two genes proved to be regulated in a different fashion. The putative steroid dehydrogenase gene dod-8 (also called stdh-1) is upregulated in daf-2 mutants in a daf-16-dependent fashion (Murphy et al. 2003). We utilized two transcriptional fusions to analyze dod-8 expression, a Pdod-8∷gfp fusion containing 0.5 kb of upstream promoter sequence and a Pdod-8∷rfp fusion containing a 1.8 kb promoter fragment. Both of these promoter sequences contained the sequence CTTATCA, which is overrepresented among DAF-16-regulated genes (Murphy et al. 2003), but only the long form contained two canonical DAF-16 binding sites. The two constructs behaved similarly. We found increased expression of both transgenes in response to germline removal. Interestingly, this increase was only partially DAF-16 independent. Although we observed decreased transgene expression in germline-less daf-16(mu86) mutant animals relative to germline-less daf-16(+) animals, the level of expression of both transgenes remained higher than in intact daf-16(mu86) animals (Figure 2B; Table 2). To determine whether the increase in expression produced by loss of the germ cells required the somatic gonad, we removed the entire gonad. We found that dod-8 expression was invariably increased relative to intact controls. In four of the nine trials we saw a similar level of induction in animals lacking either the germ cells or the whole gonad (Figure 2A; Table 1). However, in five of nine trials, the level of expression of both transgenes was somewhat lower in animals missing the whole gonad than it was in animals missing germ cells alone. We do not know the source of this variability, which was observed with both constructs.
Finally, we examined the nicotinamide nucleotide transhydrogenase gene nnt-1. The promoter fusion we examined contains three canonical DAF-16 binding sites. As with dod-8, we observed an increase in expression upon germ-cell ablation that was partially daf-16 independent (Figure 2B; Table 2). In three of four trials, somatic-gonad removal had no effect on the increase in Pnnt-1∷gfp expression produced by loss of the germ cells. However, in one trial, there was a significant decrease relative to germ-cell removal alone (Figure 2A; Table 1). Thus, nnt-1 expression was similar to that of dod-8, described above.
Our results suggest there are at least two classes of genes whose expression is regulated by DAF-16 and the germline. Expression of genes in the first class, including sod-3 and gpd-2, increases in germline-less animals in a completely daf-16-dependent fashion. This increase also requires, to a large extent, the somatic gonad. In contrast, the increase seen in the second class, including dod-8/stdh-1 and nnt-1, only partially depends on daf-16 and is largely or completely independent of the somatic gonad. Together, these results suggest that the somatic gonad is required for the activation of a subset of daf-16-regulated genes in animals lacking a germline. We did not observe an obvious correlation between the expression of these transgenes and the presence of potential DAF-16 binding sites; however, since only sod-3 is known to be a direct DAF-16 target, the significance of this is not clear.
The somatic gonad is not required for the oxidative stress resistance of animals that lack germ cells:
Another consequence of germline ablation is an increase in the animals' resistance to heat and oxidative stress (Arantes-Oliveira et al. 2002). To ask whether this increased stress resistance requires the somatic gonad, we measured resistance to the oxidative stressor paraquat of animals in which both the somatic gonad and the germ cells had been removed. We found that the stress resistance of animals lacking germ cells was not diminished by the removal of the somatic gonad (Table 3). This finding strengthens the argument that the somatic gonad is not required for all of the events triggered by removing the germ cells.
TABLE 3.
The somatic gonad is not required for germ-cell removal to increase oxidative stress resistance
| Trial | Strain | Intact (hr) | n | Germ cell (−) | n | P | Whole gonad (−) | n | P | P′ |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Wild type | 3.46 ± 0.16 | 70 | 4.40 ± 0.25 | 53 | <0.001 | 4.34 ± 0.22 | 66 | <0.001 | 0.94 |
| 2 | Wild type | 3.28 ± 0.14 | 79 | 4.10 ± 0.19 | 72 | <0.001 | 4.65 ± 0.20 | 79 | <0.001 | 0.065 |
| 3 | daf-16(mu86) | 2.90 ± 0.11 | 28 | 4.04 ± 0.17 | 38 | <0.001 | 3.90 ± 0.15 | 32 | <0.001 | 0.63 |
| Wild type | 3.55 ± 0.13 | 36 | 4.43 ± 0.18 | 35 | <0.001 | |||||
| 4 | daf-16(mu86) | 3.12 ± 0.14 | 30 | 4.03 ± 0.14 | 27 | <0.001 | 4.25 ± 0.13 | 33 | <0.001 | 0.29 |
| Wild type | 3.22 ± 0.14 | 31 | 4.22 ± 0.19 | 34 | <0.001 |
Animals were subjected to 300 mm paraquat as day-2 adults. Mean survival time is given in hours. P represents the P-value (Logrank Mantel/Cox) compared to intact control, and P′ represents the P-value compared to animals lacking germ cells (Z2 and Z3 ablated).
The finding that whole-gonad-ablated animals are paraquat resistant but not long lived indicates that mechanisms that increase paraquat resistance are not sufficient to increase life span. Consistent with this, we previously found that removing the germline increased the heat resistance of daf-16(−) mutants in spite of the fact that it did not increase their life span (Libina et al. 2003). We asked whether this was the case for paraquat resistance as well, and found that it was: daf-16(mu86) mutants lacking either the germline or whole gonad survived longer in paraquat than did daf-16(mu86) mutants with an intact gonad (Table 3). Thus, a daf-16-independent mechanism increases heat and oxidative stress resistance (but not longevity) following loss of the germ cells or the entire reproductive system.
Low levels of insulin-like signaling eliminate the requirement for the somatic gonad:
How does the somatic gonad communicate with other tissues? Previous studies have suggested that the somatic gonad may promote longevity in animals missing the germ cells by modulating the activity of the insulin-like receptor DAF-2 (Hsin and Kenyon 1999). In the daf-2(e1370) mutant, which carries a mutation affecting the DAF-2 tyrosine-kinase domain, the somatic gonad is not required for germline removal to extend life span (Hsin and Kenyon 1999). Curiously, animals carrying the daf-2(e1368) mutation, which changes a residue in the DAF-2 ligand-binding domain, respond like wild type to germline and whole-gonad ablation. Germline ablation further extends life span, and this life span extension requires the somatic gonad. We found that the same was true for animals carrying daf-2(mu150) (Figure 3B), another daf-2 mutation (Garigan et al. 2002) that affects the ligand-binding domain (D. Gems, personal communication). This difference was unexpected, because all of these daf-2 mutations extend the life span of intact animals.
Figure 3.—
The reproductive system and insulin-like signaling. (A) The somatic gonad is required for the life span extension produced by germline removal in weak insulin/IGF-1-pathway mutants. daf-2(RNAi) intact control, n = 128/113 (uncensored/total analyzed), m = 36.5 ± 1.7 (days); Z2/3(−), n = 90/73, m = 72.8 ± 2.5, P < 0.001; Z1/4(−), n = 93/80, m = 58.2 ± 2.2, P < 0.0001, P′ < 0.0001. pdk-1(sa709) intact control, n = 64/34, m = 26.6 ± 1.2; Z2/3(−), n = 44/37, m = 33.7 ± 2.0, P = 0.0026; Z1/4(−), n = 48/45, m = 28.0 ± 0.98, P = 0.54, P′ = 0.0015. akt-1(ok525) intact control, n = 90/71, m = 21.3 ± 0.73; Z2/3(−), n = 94/46, m = 28.0 ± 1.6, P < 0.0001; Z1/4(−), n = 88/85, m = 23.9 ± 1.1, P = 0.015, P′ = 0.048; akt-1(+), n = 90/72, m = 16.7 ± 0.53, P < 0.0001. akt-2(ok393) intact control, n = 90/64, m = 14.0 ± 0.61; Z2/3(−), n = 88/57, m = 19.8 ± 1.3, P < 0.0001; Z1/4(−), n = 104/96, m = 17.3 ± 0.54, P = 0.0001, P′ = 0.0363; akt-2(+), n = 81/61, m = 14.9 ± 0.55, P = 0.48. P refers to the P-value compared to intact. P′ refers to the P-value comparing germ-cell (Z2/Z3) ablation to whole-gonad (Z1/Z4) ablation. (B) Reducing daf-2 levels in daf-2(mu150) mutants with RNAi allows germline removal to extend life span independently of the somatic gonad. daf-2(mu150) fed HT115 bacteria carrying the pAD12 vector only control plasmid: intact control, n = 79/61, m = 27.0 ± 0.862; Z2/3(−), n = 48/34, m = 44.0 ± 2.7, P <0.001; Z1/4(−), n = 52/51, m = 28.8 ± 1.0, P = 0.12, P′ < 0.0001. daf-2(mu150, RNAi) fed HT115 bacteria carrying the pAD43 (daf-2 RNAi) plasmid: intact control, n = 77/61, m = 49.1 ± 1.9; Z2/3(−), n = 46/39, m = 92.3 ± 4.5, P < 0.0001; Z1/4(−), n = 51/45, m = 102.4 ± 3.4, P < 0.0001, P′ = 0.27. (C) Quantitative model to explain the difference in life span seen with whole gonad ablation in various daf-2 mutant backgrounds. Decreasing the amount of insulin-like signaling below a certain threshold eliminates the requirement for the somatic gonad.
One way to explain this difference is by postulating that the DAF-2 receptor can bind to two different ligands (Hsin and Kenyon 1999). To make this idea clear, we will describe a simple version of this model. Here, one ligand activates the DAF-2 receptor in normal, intact animals. All of the daf-2 mutants we examined lack the ability to respond to this ligand, so they are all long lived. A second ligand is produced by the somatic gonad, and it inactivates the DAF-2 receptor. In this model, the two ligand-binding domain mutants, daf-2(e1368) and daf-2(mu150), can still bind to this second ligand, so they respond normally to the loss of the somatic gonad. In contrast, the daf-2(e1370) mutant DAF-2 protein cannot respond to either ligand, so daf-2(e1370) mutants do not respond to loss of the somatic gonad.
Alternatively, different daf-2 mutants could respond differently to the somatic gonad because they reduce insulin/IGF-1 signaling to different extents. In this quantitative model, a modest reduction in DAF-2 activity would not allow animals lacking germ cells to live long in the absence of the somatic gonad, whereas a more severe reduction would. Consistent with this model, the daf-2(mu150) allele is likely to be weaker than daf-2(e1370), since daf-2(mu150) produces a smaller life span extension in otherwise normal animals. Likewise, in some (Hsin and Kenyon 1999) but not all (Gems et al. 1998) studies, e1368 mutants have been found to have shorter lives than e1370 mutants. We note that these two models are not mutually exclusive. Specifically, one could imagine that a somatic-gonad-dependent ligand has a higher affinity for the DAF-2 receptor than does the classical DAF-2 ligand. However, the quantitative model admits many more mechanistic possibilities.
To test the quantitative model, we asked whether modest reductions in insulin/IGF-1 signaling could produce phenotypes similar to those produced by the daf-2 ligand-binding domain mutations. We began by lowering the level of wild-type DAF-2 protein using RNAi. daf-2(RNAi) animals probably have higher residual levels of daf-2 activity than do e1370 mutants, because they have more modest life span extensions. We found that the further life span extension produced by germline ablation in daf-2(RNAi) animals was partially dependent on the somatic gonad (Figure 3A). Thus, one does not require ligand-binding domain mutations to produce a daf-2(−) animal that lives long but responds at least partially normally to germline and whole-gonad ablation.
The quantitative model also predicts that a modest reduction in the activity of a downstream gene in the insulin/IGF-1 pathway could also produce a phenotype similar to that produced by daf-2(mu150) or daf-2(e1368). We found that this was the case for the relatively weak pdk-1 allele sa709 (Paradis et al. 1999), which extended the life span of intact animals, but did not allow germline removal to further extend life span independently of the somatic gonad. In addition, the somatic gonad was partially required for germline removal to extend the life spans of akt-1(ok525) and akt-2(ok393) null mutations (Figure 3A). In these pdk-1, akt-1, and akt-2 mutants, the DAF-2 receptor is wild type and should be able to bind to any ligand.
Finally, the quantitative model predicts that further reduction of DAF-2 activity in an animal carrying a weak allele of daf-2 such as daf-2(mu150) will behave like a strong allele such as daf-2(e1370). Indeed, in daf-2(mu150) animals subjected to daf-2 RNAi, removal of the somatic gonad no longer suppressed the longevity seen with germ-cell ablation alone (Figure 3B). So, whereas both daf-2(mu150) and daf-2(RNAi) animals retain the requirement of the somatic gonad for germ-cell-ablated animals to live long, the combination of the two, daf-2(mu150, RNAi) removes this requirement. Consistent with this, we found previously that subjecting daf-2(e1368) mutants to daf-2(RNAi) allowed whole-gonad ablation to further extend life span (Arantes-Oliveira et al. 2003).
Strong daf-2 mutations render sod-3 expression independent of the somatic gonad:
In the wild type, the somatic gonad is required for germline removal to increase expression of the DAF-16 target gene sod-3. Because strong daf-2 mutations allow germline-less animals to live long independently of the somatic gonad, we wondered whether strong daf-2 mutations would also allow germline-less animals to upregulate sod-3 expression independently of the somatic gonad. To address this question, we examined sod-3∷gfp levels in a daf-2(e1370) mutant, in which either germline or whole-gonad removal extends life span. daf-2 mutations are known to elevate sod-3 levels relative to wild type (Honda and Honda 1999; Libina et al. 2003; McElwee et al. 2003; Murphy et al. 2003). We found that the level of sod-3∷gfp expression was even higher in daf-2(e1370) mutants lacking the germ cells (Figure 4; Table 4). However, unlike in wild type, this increased expression did not require the somatic gonad. In fact, removing the somatic gonad as well as the germline in daf-2(e1370) mutants produced a level of sod-3 expression that was slightly higher than the level produced by removing only the germline. This is consistent with previous findings that whole-gonad ablation can increase the life span of strong daf-2 mutants even more than does germline ablation (Hsin and Kenyon 1999) (Table 4). daf-2(e1368) mutants treated with daf-2 RNAi respond like daf-2(e1370) mutants to germline and whole-gonad ablation (Table 4) (Arantes-Oliveira et al. 2003), and we found that their sod-3 expression profiles under the same conditions were similar to those of e1370 mutants (Table 4).
Figure 4.—
Expression of the DAF-16 target gene sod-3 generally correlates with life span in daf-2 mutants lacking either the germ cells or the somatic gonad. Fluorescence intensity of GFP from the whole animal, excluding vulval regions, was measured and calculated relative to intact controls (see materials and methods). Values for histograms of sod-3∷gfp levels are given in Table 4.
TABLE 4.
Relative levels of Psod-3:gfp expression in daf-2 mutants subjected to germline or whole-gonad ablation
| Strain | Genotype | Psod-3∷gfp expression relative to intact | n | P | P′ | Mean life span (days) | n | P | P′ | |
|---|---|---|---|---|---|---|---|---|---|---|
| CF1580 | daf-2(e1370) | Intact | 1 ± 0.042 | 23 | — | 43.2 ± 0.93a | 245 | — | ||
| Germ cell (−) | 1.52 ± 0.063 | 17 | <0.001 | 64.5 ± 3.5a | 59 | <0.001 | ||||
| Whole gonad (−) | 1.98 ± 0.083 | 17 | <0.001 | <0.001 | 69.5 ± 6.0a | 31 | <0.001 | 0.33 | ||
| CF2533 | daf-2(e1368,RNAi) | Intact | 1 ± 0.19 | 14 | — | 51.0 ± 1.9b | 68 | — | ||
| Germ cell (−) | 1.17 ± 0.13 | 11 | 0.456 | 87.90 ± 4.4 | 37 | <0.001 | ||||
| Whole gonad (−) | 1.76 ± 0.18 | 15 | 0.007 | 0.014 | 124.1 ± 5.9b | 39 | <0.001 | <0.001 | ||
| CF1553 | daf-2(RNAi) | Intact | 1 ± 0.19 | 18 | — | 36.5 ± 1.7 | 113 | — | ||
| Germ cell (−) | 5.15 ± 0.60 | 16 | <0.001 | 72.8 ± 2.5 | 73 | <0.001 | ||||
| Whole gonad (−) | 3.09 ± 0.60 | 21 | 0.003 | 0.020 | 58.2 ± 2.2 | 80 | <0.001 | <0.001 | ||
| CF2533 | daf-2(e1368) | Intact | 1 ± 0.034 | 37 | — | 34.3 ± 1.0a | 110 | |||
| Germ cell (−) | 3.74 ± 0.19 | 35 | <0.001 | 71.0 ± 2.3a | 45 | <0.001 | ||||
| Whole gonad (−) | 3.48 ± 0.19 | 40 | <0.001 | 0.37 | 41.1 ± 1.5a | 45 | <0.001 | <0.001 | ||
| CF1588 | daf-16(mu86); daf-2(e1370) | Intact | 1 ± 0.044 | 25 | — | |||||
| Germ cell (−) | 0.74 ± 0.029 | 28 | <0.001 | |||||||
| Whole gonad (−) | 0.93 ± 0.042 | 25 | 0.26 | <0.001 | ||||||
| CF2683 | daf-16(mu86); daf-2(e1368) | Intact | 1 ± 0.067 | 19 | — | |||||
| Germ cell (−) | 0.85 ± 0.064 | 15 | 0.11 | |||||||
| Whole gonad (−) | 0.88 ± 0.044 | 11 | 0.14 | 0.69 |
Psod-3∷gfp expression values represent mean fluorescence intensity relative to intact controls. P represents the P-value (Student's t-test for Psod-3∷gfp expression and Logrank Mantel/Cox for life span data) compared to intact control, and P′ represents the P-value compared to animals lacking germ cells (Z2 and Z3 ablated).
Life span data previously published in Hsin and Kenyon (1999), and included here for comparison.
Life span data previously published in Arantes-Oliveira et al. (2003) and included here for comparison.
Next, we examined Psod-3∷gfp expression in weaker daf-2 mutants, in which the somatic gonad is required for germline ablation to increase life span. We found that the somatic gonad was partially required for increased sod-3 expression produced by germline loss in daf-2(RNAi) animals, consistent with the fact that the somatic gonad is partially required for the increased life span produced by germline loss in these animals. Unexpectedly, this was not the case for daf-2(e1368) mutants. In these animals, the somatic gonad was not required for germline ablation to further increase sod-3∷gfp expression, in spite of the fact that the somatic gonad was required for germline ablation to further extend life span (Figure 4; Table 4). Thus, overall, we observed a general correlation between life span and sod-3 expression in these experiments, but the correlation was not perfect.
In both daf-2 mutants and germline-less animals, increased sod-3 expression requires daf-16. Likewise, we found that the very high levels of sod-3 expression observed in daf-2 mutants lacking the germ cells or the whole gonad were completely dependent on daf-16. This finding suggests that the mechanisms that produce the very long life spans of these animals [which are also daf-16 dependent (Figure 4; Table 4)] are likely to involve increases in the levels of DAF-16-dependent gene expression.
DISCUSSION
The somatic gonad and germ cells act in different ways to control the life span of the animal:
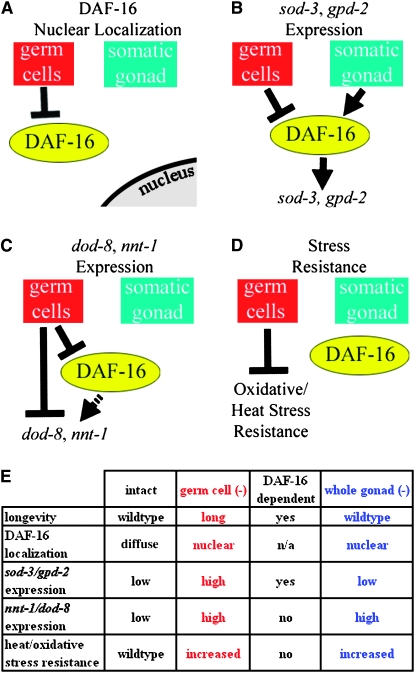
The life span extension produced by removing the germline of C. elegans depends on the presence of the somatic reproductive tissues (Hsin and Kenyon 1999). In principle, one could imagine that the germ cells and the somatic gonad function in a strictly linear pathway to affect life span. In this scenario, the presence of the germline inhibits a life-span-extending activity of the somatic gonad. Removal of the germline relieves this inhibition, thereby increasing longevity. If this were the case, then all of the effects of germline removal should be reversed by also removing the somatic gonad. However, we found that removing the entire reproductive system does not prevent the nuclear localization of DAF-16 that is triggered by germline removal, nor does it prevent the increase in the animal's stress resistance. These findings suggest that instead of acting in a strictly linear pathway, the germ cells and the somatic gonad each send signals to the rest of the animal that modulate its physiology (Figure 5).
Figure 5.—
The somatic reproductive tissues are required for some, but not all, of the processes triggered by germline removal. (A) Loss of the germ cells triggers DAF-16 nuclear localization independently of the somatic gonad. (B) The somatic gonad is required for the increased expression of sod-3 and gpd-2 that occurs when the germ cells are removed. (C) The somatic gonad is not required for the increased expression of dod-8 and nnt-1 that occurs when the germ cells are removed. Moreover, DAF-16 is only partially required for this upregulation. (D) Neither DAF-16 nor the somatic gonad is required for the increased stress resistance that occurs when the germ cells are removed. (E) Specific findings demonstrating that the somatic gonad is required for some, but not all processes triggered by germ-cell removal.
Although the somatic gonad does not regulate the subcellular localization of DAF-16, it is required for the proper expression of a subset of DAF-16-regulated genes in animals lacking the germline. The somatic gonad may affect DAF-16 transcriptional activity in a number of ways, for example, by controlling a covalent modification of the DAF-16 protein or by activating a cofactor. Because removing the somatic gonad appeared to decrease the amount of DAF-16∷GFP protein, the somatic gonad could also influence DAF-16 activity by affecting DAF-16 levels. As the transcription of some target genes may be more sensitive than others to the level of DAF-16, a change in DAF-16 protein levels could conceivably affect the transcription of some genes more than others. In our experiments, the somatic gonad was consistently required for sod-3 and gpd-1 expression but not for dod-8 and nnt-1 expression in animals that lack germ cells. Thus one could imagine that these genes differ in their affinity for DAF-16 protein.
We did not observe a correlation between the presence of any potential DAF-16 binding sites in the promoters of the genes we examined and their behavior in our assay. However, only sod-3 is known to be a direct target of DAF-16. Thus, it is difficult to speculate about mechanism at this point.
The somatic gonad is not required for loss of the germline to increase paraquat resistance:
In addition to analyzing the regulation of individual genes, we also examined the somatic-gonad dependence of a process that would seem likely to involve changes in expression of many genes: stress resistance. Like many long-lived mutants, animals lacking germ cells are resistant to heat and oxidative stress. Because DAF-16 is required for the longevity produced by germ-cell removal, one would expect DAF-16 to be required for the increased stress resistance produced by germ-cell removal. However, we previously found that this was not the case for heat resistance (Libina et al. 2003), and in this study we found that DAF-16 was not required for paraquat resistance either. We also found that germline-less animals lacking the somatic gonad, which are not long lived, are stress resistant. Together these findings indicate that increased stress resistance is not sufficient for the longevity of germline-less animals, since in both types of experiments, we obtained stress-resistant animals that were not long lived.
These data also show that increased sod-3 activity is not required for increased stress resistance. Wild-type animals lacking the whole gonad, as well as daf-16 mutants lacking the germ cells, are stress resistant but have relatively low levels of sod-3 expression. Perhaps other anti-oxidant and/or stress-tolerance genes are responsible for the increased oxidative and heat-stress resistance of these animals.
Is stress resistance required for longevity? Previously, we found that whereas intestinal daf-16 completely rescues the longevity of daf-16(−) mutants lacking a germline, it only partially rescues thermotolerance (Libina et al. 2003). Thus, it is not clear to what extent mechanisms that increase thermotolerance are required for the longevity of animals that lack a germline. Together these findings increase the list of cases in which increased resistance to specific types of environmental stressors has been uncoupled from longevity (Libina et al. 2003; Van Remmen et al. 2003; Fujii et al. 2004; Henderson et al. 2006; Wolff and Dillin 2006; Wolff et al. 2006).
The finding that the somatic gonad is not required for increased stress resistance solidifies the notion that not all of the effects of germline removal are mediated through the somatic gonad. Stress resistance is a fundamental physiological change that probably requires a substantial shift in gene expression patterns. In this context, it seems significant that the increased expression of two of the genes we examined, dod-8 and nnt-1, was partially daf-16-independent (and somatic-gonad independent). Possibly genes regulated in this fashion underlie the increased stress resistance produced by loss of the germline.
The somatic gonad may influence life span independently of the daf-2 pathway:
How does the somatic gonad signal to the rest of the animal? Because sharply reducing the activity of daf-2 allows germline removal to extend life span independently of the somatic gonad, it is possible that the somatic gonad affects life span by regulating insulin/IGF-1 signaling (Hsin and Kenyon 1999). However, it is also possible that the insulin/IGF-1 pathway and the somatic gonad act in parallel to affect life span. In this case, in animals lacking a germline, strong daf-2 mutations would trigger events that duplicate, or compensate for, the function of the somatic gonad.
These two models can be evaluated by monitoring events that are known to be controlled by the DAF-2 pathway. One such event is DAF-16 nuclear localization. If the somatic gonad extends life span in animals lacking a germline by inhibiting insulin/IGF-1 signaling, then one would expect DAF-2-pathway activity to increase upon removal of the somatic gonad. This increase would further activate the AKT-1, AKT-2, and SGK-1 kinases, which in turn would phosphorylate DAF-16, inhibiting DAF-16's nuclear localization. Thus, in animals lacking the somatic gonad as well as the germline, we would expect to see at least some cytoplasmic DAF-16 protein. This is the case if insulin/IGF-1 signaling is increased in a germline-deficient animal by mutating the PTEN phosphatase gene daf-18. In germline-less daf-18 mutants, which are not long lived, DAF-16 is excluded from nuclei (Berman and Kenyon 2006). In contrast, when the somatic gonad was removed from animals lacking a germline, we did not observe any change in the subcellular localization of DAF-16. This finding argues against the idea that the somatic gonad increases life span by downregulating the insulin/IGF-1 pathway. However, because of the complexity of insulin/IGF-1 signaling, we cannot rule out this possibility altogether. In fact, the DAF-2 pathway is known to have outputs that can affect longevity independently of DAF-16 localization, since the life span of animals containing a constitutively nuclear AKT-site mutant DAF-16 protein is much longer in a daf-2(−) background than in a wild-type background (Lin et al. 2001; Berman and Kenyon 2006). In addition, the activity of mammalian FOXO6 protein is regulated by AKT independently of nuclear localization (Jacobs et al. 2003; Van Der Heide et al. 2005).
Thresholds and the insulin/IGF-1 pathway:
One goal of this study was to try to understand why different daf-2 mutations produce different effects on the reproductive signaling system. For example, the daf-2(e1370) mutation, which affects the DAF-2 tyrosine-kinase domain, allows germline removal to further extend life span independently of the somatic gonad, whereas the daf-2(e1368) and mu150 mutations, which affect the DAF-2 ligand-binding domain, do not (Hsin and Kenyon 1999; and this study). Here, we showed that one can mimic the effect of the ligand-binding-domain mutations by reducing the level of wild-type DAF-2 protein with RNAi, or by reducing the level of downstream signaling components such as PDK-1 or AKT-1/2 in animals that have a wild-type DAF-2 receptor. In addition, further reducing the level of the DAF-2 ligand-binding-domain mutant protein with RNAi allows germline removal to extend life span independently of the somatic gonad. Together, these findings support a quantitative model in which a modest reduction in insulin/IGF-1 signaling does not remove the requirement for the somatic gonad, but a more extensive reduction does remove this requirement (Figure 3C).
All of the daf-2-pathway mutants we analyzed were long lived, but only the strongest affected somatic-gonad signaling. Thus, a higher level of DAF-2 activity is required to prevent intact animals from living longer than is required for the reproductive signaling system to regulate longevity normally (Figure 4C). This finding suggests that the daf-2-regulated processes that trigger life span extension in intact animals are not entirely coincident with the daf-2-regulated processes that influence signaling from the reproductive system.
The genetics of extreme longevity:
daf-2 mutants that lack germ cells live much longer than intact daf-2 mutants or wild-type animals that lack germ cells (Hsin and Kenyon 1999). What mechanisms produce the extreme longevity of these animals? Because this entire life span increase is daf-16 dependent (Hsin and Kenyon 1999), one possibility is that the same set of life-span-extending genes that are upregulated in daf-2 mutants are upregulated even more when the germline is removed. Consistent with this, we observed a further increase in expression of sod-3 in these very long-lived animals, and this increase is daf-16 dependent. This finding is important because it indicates, for the first time, that the expression of longevity genes that are upregulated in daf-2 mutants can be increased even more by conditions that further increase life span. In the future, it will be interesting to measure global gene expression profiles in these very long-lived animals and to test the significance of individual gene activities with RNAi. In particular, it is possible that new genes, not previously identified, will make an important contribution to extreme longevity.
In general, we found that sod-3 expression levels correlated with life span extension in extremely long-lived animals. For example, in daf-2(RNAi) animals, sod-3 expression increased further in response to germline ablation, and this additional expression, like life span increase, partially required the somatic gonad. In strong daf-2(e1370) mutants, sod-3 expression increased more upon loss of the germ cells, and this increase was independent of the somatic gonad. In fact, in these animals, loss of the somatic gonad further increased sod-3 expression, just as it further increased life span. These findings suggest that daf-2 mutations affect the requirement for somatic-gonad signaling by affecting the expression of genes like sod-3. We note that the correlation we observed was not perfect. In daf-2(e1368) mutants, sod-3 levels rose when either the germ cells or the whole gonad was removed, but life span was only extended upon germline removal. To explain this, we suggest that when the system is operating near a threshold level, one will observe variation at the level of individual gene expression that will not always reflect the aggregate behavior of the system as a whole.
Conclusion:
Together these studies have helped to clarify the role of the somatic gonad in the regulation of life span by the reproductive system. They indicate that the somatic gonad is required for some, but not all, of the events that are triggered when the germ cells are removed. In particular, the somatic gonad is required for the proper regulation of a subset of DAF-16 target genes. It is not yet clear how the somatic gonad exerts its effect on gene expression in other tissues. Because loss of the somatic gonad does not produce the same spectrum of phenotypes produced by inhibition of the insulin/IGF-1 pathway, it is possible that the somatic gonad acts through a new, as yet unidentified signaling pathway.
Acknowledgments
Arjumand Ghazi discovered that germline ablation increases expression of Pnnt-1∷gfp, and Lisa Watson discovered that germline ablation increases expression of Pdod-8∷gfp. We thank these and other members of the Kenyon Lab for helpful comments and advice, especially M. Gaglia, A. Ghazi, M. Hansen, S. Lee, and L. Mitic. N.A.-O. was supported by the Gulbenkian Foundation, J.R.B. by a Howard Hughes predoctoral fellowship, and P.Z. by the Larry Hillblom Foundation. This work was supported by National Institutes of Health grant no. AG-023903 to C.K., who is an American Cancer Society Research Professor, the director of the University of California, San Francisco's Hillblom Center for the Biology of Aging, and a founder and director of Elixir Pharmaceuticals.
References
- Arantes-Oliveira, N., J. Apfeld, A. Dillin and C. Kenyon, 2002. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295: 502–505. [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira, N., J. R. Berman and C. Kenyon, 2003. Healthy animals with extreme longevity. Science 302: 611. [DOI] [PubMed] [Google Scholar]
- Berman, J. R., and C. Kenyon, 2006. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124: 1055–1068. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broue, F., P. Liere, C. Kenyon and E. E. Baulieu, 2007. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell 6: 87–94. [DOI] [PubMed] [Google Scholar]
- Conover, C. A., and L. K. Bale, 2007. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell 6: 727–729. [DOI] [PubMed] [Google Scholar]
- Dillin, A., D. K. Crawford and C. Kenyon, 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834. [DOI] [PubMed] [Google Scholar]
- Dong, M.-Q., J. D. Venable, N. Au, T. Xu, S. K. Park et al., 2007. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317: 660–663. [DOI] [PubMed] [Google Scholar]
- Dorman, J. B., B. Albinder, T. Shroyer and C. Kenyon, 1995. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141: 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, M., Y. Matsumoto, N. Tanaka, K. Miki, T. Suzuki et al., 2004. Mutations in chemosensory cilia cause resistance to paraquat in nematode Caenorhabditis elegans. J. Biol. Chem. 279: 20277–20282. [DOI] [PubMed] [Google Scholar]
- Furuyama, T., T. Nakazawa, I. Nakano and N. Mori, 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan, D., A. L. Hsu, A. G. Fraser, R. S. Kamath, J. Ahringer et al., 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems, D., A. J. Sutton, M. L. Sundermeyer, P. S. Albert, K. V. King et al., 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction, and longevity in Caenorhabditis elegans. Genetics 150: 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch, B., C. Weitzel, C. Kober-Eisermann, V. Rottiers and A. Antebi, 2001. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1: 841–851. [DOI] [PubMed] [Google Scholar]
- Gerisch, B., V. Rottiers, D. Li, D. L. Motola, C. L. Cummins et al., 2007. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc. Natl. Acad. Sci. USA 104: 5014–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, E. B., E. Malone Link, L. X. Liu, C. D. Johnson and J. A. Lees, 1999. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. USA 96: 2925–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, S. T., and T. E. Johnson, 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11: 1975–1980. [DOI] [PubMed] [Google Scholar]
- Henderson, S. T., M. Bonafe and T. E. Johnson, 2006. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A Biol. Sci. Med. Sci. 61: 444–460. [DOI] [PubMed] [Google Scholar]
- Hertweck, M., C. Gobel and R. Baumeister, 2004. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6: 577–588. [DOI] [PubMed] [Google Scholar]
- Honda, Y., and S. Honda, 1999. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13: 1385–1393. [PubMed] [Google Scholar]
- Hsin, H., and C. Kenyon, 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. [DOI] [PubMed] [Google Scholar]
- Jacobs, F. M., L. P. van der Heide, P. J. Wijchers, J. P. Burbach, M. F. Hoekman et al., 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278: 35959–35967. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., 2005. The plasticity of aging: insights from long-lived mutants. Cell 120: 449–460. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudner and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Larsen, P. L., P. S. Albert and D. L. Riddle, 1995. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139: 1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. Y., J. Hench and G. Ruvkun, 2001. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 11: 1950–1957. [DOI] [PubMed] [Google Scholar]
- Lee, S. S., S. Kennedy, A. C. Tolonen and G. Ruvkun, 2003. DAF-16 target genes that control C. elegans life-span and metabolism. Science 300: 644–647. [DOI] [PubMed] [Google Scholar]
- Libina, N., J. R. Berman and C. Kenyon, 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. [DOI] [PubMed] [Google Scholar]
- Lin, K., H. Hsin, N. Libina and C. Kenyon, 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28: 139–145. [DOI] [PubMed] [Google Scholar]
- McElwee, J., K. Bubb and J. H. Thomas, 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121. [DOI] [PubMed] [Google Scholar]
- McKay, S. J., R. Johnsen, J. Khattra, J. Asano, D. L. Baillie et al., 2003. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harbor Symp. Quant. Biol. 68: 159–169. [DOI] [PubMed] [Google Scholar]
- Mihaylova, V. T., C. Z. Borland, L. Manjarrez, M. J. Stern and H. Sun, 1999. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. USA 96: 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath et al., 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Ogg, S., and G. Ruvkun, 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell. 2: 887–893. [DOI] [PubMed] [Google Scholar]
- Oh, S. W., A. Mukhopadhyay, B. L. Dixit, T. Raha, M. R. Green et al., 2006. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38: 251–257. [DOI] [PubMed] [Google Scholar]
- Paradis, S., and G. Ruvkun, 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, S., M. Ailion, A. Toker, J. H. Thomas and G. Ruvkun, 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, J. P., P. E. Kuwabara, O. M. Sinilnikova, L. Duret, D. Thierry-Mieg et al., 1999. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr. Biol. 9: 329–332. [DOI] [PubMed] [Google Scholar]
- Selman, C., S. Lingard, A. I. Choudhury, R. L. Batterham, M. Claret et al., 2007. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. (in press). [DOI] [PubMed]
- Taguchi, A., L. M. Wartschow and M. F. White, 2007. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science 317: 369–372. [DOI] [PubMed] [Google Scholar]
- Tatar, M., A. Bartke and A. Antebi, 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351. [DOI] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- van der Heide, L. P., F. M. Jacobs, J. P. Burbach, M. F. Hoekman and M. P. Smidt, 2005. FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo-cytoplasmic shuttling. Biochem. J. 391: 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen, H., Y. Ikeno, M. Hamilton, M. Pahlavani, N. Wolf et al., 2003. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics 16: 29–37. [DOI] [PubMed] [Google Scholar]
- Wolff, S., and A. Dillin, 2006. The trifecta of aging in Caenorhabditis elegans. Exp. Gerontol. 41: 894–903. [DOI] [PubMed] [Google Scholar]
- Wolff, S., H. Ma, D. Burch, G. A. Maciel, T. Hunter et al., 2006. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell 124: 1039–1053. [DOI] [PubMed] [Google Scholar]