Abstract
Foliar fungal pathogens from the genus Mycosphaerella affect eucalypts in natural forests and plantations worldwide. QTL analysis was conducted to dissect the genetic control of resistance in Eucalyptus globulus to a natural infection by Mycosphaerella leaf disease, using a clonally replicated outbred F2 family (112 genotypes) planted in a field trial. Two major QTL, with high LOD support (20.2 and 10.9) and high genomewide significance, explained a large proportion (52%) of the phenotypic variance in the severity of damage by Mycosphaerella cryptica, which may be indicative of oligogenic control. Both QTL were validated in a second F2 family and one was validated in a third F2 family. The mean values of different genotype classes at both major QTL argue for Mendelian inheritance with resistance dominant over susceptibility. There were strong correlations between the levels of Mycosphaerella damage in related genetic material planted in three widely separated locations in Tasmania. These findings together provide evidence that the genes controlling resistance to Mycosphaerella damage are stable in different genetic backgrounds and across different environments.
THE genus Eucalyptus (Myrtaceae) forms an integral part of the Australian flora, including >800 species that dominate most forest types, from coastal to subalpine habitats (Williams and Brooker 1997). Eucalyptus globulus (sensu Brooker 2000) is native to southeastern Australia, including Tasmania and the Bass Strait Islands (Williams and Potts 1996) where it is an important component of low-altitude forest ecosystems. Eucalypts are the major hardwood species grown in pulpwood plantations throughout the world (Cossalter and Pye-Smith 2003) and E. globulus is the main species grown in temperate regions (Potts et al. 2004).
Foliar diseases, mostly caused by fungi, are common on eucalypts (Park et al. 2000). One of the most prevalent is Mycosphaerella leaf disease (MLD), a foliar fungal disease affecting eucalypts in natural forests and plantations worldwide (Carnegie et al. 2001; Van Zyl et al. 2002; Mohammed et al. 2003). Over 30 species of Mycosphaerella have been detected on eucalypt leaves (Mohammed et al. 2003), of which Mycosphaerella cryptica and M. nubilosa are the most common in southern Australia (Carnegie et al. 1998; Park et al. 2000). M. cryptica appears to be a more generalized pathogen than M. nubilosa, as it infects both juvenile and adult foliage of many eucalypt species and is able to penetrate directly through the leaf cuticle. In contrast, M. nubilosa is largely confined to juvenile leaves of a limited range of closely related eucalypt species and can penetrate the leaf only via stomatal pores (Park et al. 2000). Mycosphaerella leaf disease causes leaf necrosis and defoliation that can be highly detrimental to growth (Carnegie and Ades 2002; Milgate et al. 2005b). Crown damage from MLD can range from <10% necrosis of leaves to extreme cases of complete defoliation and tree death. However, even relatively minor damage can cause a significant loss in growth (Carnegie and Ades 2002) because the disease adversely affects photosynthesis beyond a simple reduction in photosynthetic area (Pinkard and Mohammed 2006).
Mycosphaerella pathogens are hemibiotrophic fungi that must spend part of their life cycle living and reproducing on living plant tissue. These fungi infect foliage via wind-dispersed ascospores and/or splash-dispersed conidia (Park 1988). Ascospore development requires free water and is temperature dependent; consequently warm wet weather (i.e., summer rain) is conducive to disease epidemics (Park 1988; Carnegie et al. 1994). Furthermore, E. globulus like many eucalypt species is markedly heteroblastic and newly formed juvenile foliage is particularly susceptible to infection. As a result, significant leaf disease will occur only in areas where climatic conditions suitable for leaf infection, lesion development, and spore discharge coincide with the disease-prone stage of the host (Mohammed et al. 2003).
In contrast to native forests, where epidemics may be restricted by the variable age structure and diversity of forest communities, eucalypt plantations are particularly susceptible to damage by MLD (Burgess and Wingfield 2002). This is especially notable in plantations on sites outside the normal range of the species, suggesting that MLD may influence the ecological niches of different eucalypt species and the suitability of different species for plantation development in certain areas. For example, in the northwest of Tasmania (a center for the expanding plantation industry on the island), E. globulus plantations have suffered repeatedly from outbreaks of MLD, prompting the forest industry to favor planting E. nitens in this area (Mohammed et al. 2003). Severe outbreaks of MLD have also affected eucalypt plantations outside Australia, in countries such as New Zealand, Chile, and South Africa; MLD was one of the factors that prevented the development of a commercial plantation industry for E. globulus in South Africa (Park et al. 2000). Although coevolution of eucalypts with their foliar parasites appears to maintain a balance between the hosts and their parasites in many native forests, the disturbance of natural forest communities can also promote disease epidemics (Park et al. 2000).
While the genetic control of disease resistance in crop plants has received considerable attention, relatively little is known about the inheritance of disease resistance in forest trees. In these long-lived organisms most of the existing knowledge comes from QTL studies of diseases such as rust in pines (Cronartium sp.; e.g., Devey et al. 1995; Li et al. 2006) and poplar (Melampsora sp.; e.g., Cervera et al. 1996; Lefèvre et al. 1998; Dowkiw and Bastien 2004; Jorge et al. 2005) and chestnut blight (Cryphonectria sp.; e.g., Kubisiak et al. 1997). In comparison, very few studies have addressed the issue of disease resistance in Eucalyptus and most have used a quantitative genetic approach (e.g., Carnegie et al. 1994; Dungey et al. 1997; Carnegie and Ades 2002; Milgate et al. 2005b). However, in the case of E. grandis, bulked segregation analysis has allowed the identification of a major gene influencing resistance to rust (Puccinia psidii; Junghans et al. 2003).
Quantitative genetic studies have reported genetic variation in the susceptibility of E. globulus to MLD, both within and between provenances (Carnegie et al. 1994; Dungey et al. 1997; Milgate et al. 2005a,b) as well as within segregating families (Milgate et al. 2005a). There is some evidence for a positive correlation between resistance to the disease and rainfall, consistent with more intense natural selection for disease resistance in high-rainfall areas (Carnegie et al. 1994; Dungey et al. 1997). In terms of genetic variability within the pathogen, in general, a broad range of host adaptation as evident in M. cryptica would suggest a lack of specialization in response to host species or resistance (Milgroom and Peever 2003). However, Milgate et al. (2005a) provided evidence for a close association between host and pathogen, finding specialization in the occurrence of two different biotypes of M. cryptica between resistant and susceptible E. globulus individuals. Specifically, RAPD analysis showed that 18 M. cryptica genotypes clustered into two biotypes, one of which was more frequent on resistant trees, whereas the other was more frequent on susceptible trees (Milgate et al. 2005a).
The genetic control of resistance to MLD remains poorly understood and consequently intraspecific variability in resistance has yet to be exploited at an operational level, either in breeding or in deployment programs. A thorough understanding of MLD resistance in E. globulus is necessary for effective management of the disease. This requires a detailed understanding of the genetic variation within both the host and the pathogens and the genetic control of the host–pathogen interactions (Keane et al. 2000; Burdon 2001).
Quantitative trait loci (QTL) analysis has been the main approach used to obtain insights into the number and location of loci affecting quantitative traits in forest trees (Sewell and Neale 2000). The moderate to high heritability previously reported for MLD in E. globulus (Dungey et al. 1997; Milgate et al. 2005b) suggests there are good prospects for finding QTL influencing MLD resistance. Such information would be a significant step toward understanding the inheritance of resistance to MLD in E. globulus and would be relevant to disease management in both breeding populations and natural forests. This study aims to determine the number and location of QTL underlying the variation in MLD resistance of E. globulus, using a clonally replicated F2 family. The QTL were validated in different F2 families and the stability of resistance was assessed in different environments.
MATERIALS AND METHODS
Genetic material, trial design, and disease assessment:
To study genetic variation in E. globulus susceptibility to MLD, three large outcross F2 families (families 1–3) were generated, with grandparents originating from King Island (KI) in Bass Strait and Taranna (T) in the southeast of Tasmania (Figure 1). Family 1 (T7/KI157//T144/KI5) was used to construct a linkage map and perform QTL analysis, while the two other controlled-cross families [families 2 (T7/KI157//KI5/KI157) and 3 (KI2/KI161//KI5/KI157)] were used for QTL validation. The F2 families were designed to have varying levels of resistance to MLD. Family 1 was constructed from crossing unrelated F1's each derived from grandparents predicted by Dungey et al. (1997) to have highly divergent breeding values for MLD resistance. This maximized the possibility of segregation for disease resistance in the F2. Family 3 was designed to create a relatively resistant family on the basis of these breeding values. Family 1 contained 240 genotypes, of which 160 were replicated clonally (one ramet and one ortet), resulting in two trees per genotype. Families 2 and 3 contained 240 and 238 genotypes, respectively, of which 160 and 100 were replicated clonally. Clonal replication allowed an improved estimate of genetic variation in each family, by accounting for some of the environmental heterogeneity in the field trial. For example, clonal replication would reduce the chance of both individuals representing a single genotype having low disease severity solely due to disease escape, as opposed to genetically governed resistance.
Figure 1.—
Map of Tasmania, showing the provenances of the Eucalyptus globulus genetic material used (King Island and Taranna) and the locations of field trial sites (Woolnorth, Weilangta, and Ridgley).
The three controlled-cross families were planted in a field trial at Woolnorth in northwest Tasmania (Figure 1) in May 1998, as part of an experiment also containing open-pollinated families collected from four of the grandparents (KI2, KI5, T7, T5; see Milgate et al. 2005a for a full description of the trial design). The trial featured an incomplete block design, consisting of two replicates, each with 20 incomplete blocks of 30 or 36 trees. The ortet and the ramet of each cloned genotype were assigned to separate replicates at random. The same and closely related genetic material was planted at two other sites in Tasmania, Weilangta in the southeast and Ridgley in the central north of the state (Figure 1), which allowed us to assess the environmental stability of MLD resistance. The genetic material planted at Ridgley in 1990 comprised an (inter- and intraprovenance) F1 factorial mating design with parents from King Island and Taranna (see Dungey et al. 1997 for a full description of the trial design). The F2 families grown at Woolnorth and Weilangta were derived from crossing between the F1 families grown at Ridgley. The Weilangta field trial was planted in 1999 and also contained 14 open-pollinated families, including 7 families from King Island and 7 from Taranna, 6 of which were derived from the grandparents of the F2 families (1–3), in addition to the same F2 families as planted at Woolnorth. Each family contained between 18 and 56 trees at the time of disease assessment. All families were planted in a randomized single-tree plot, in 20–51 replicates.
All field trials became naturally infected with MLD [M. cryptica at Woolnorth, identified by Milgate et al. (2005a), and both M. cryptica and M. nubilosa at Ridgley (Carnegie and Ades 2002) and possibly also at Weilangta (Pinkard and Mohammed 2006)] <1 year after planting. Both species of Mycosphaerella are capable of causing severe damage in isolation (Milgate et al. 2005a,b), although they often cooccur in diseased trees. When they cooccur, such as at Ridgley, it is not feasible to separately assess the damage by each pathogen species; hence following previous studies (Carnegie et al. 1994; Dungey et al. 1997) only total disease severity was assessed. Disease severity was recorded after 12 months at Woolnorth, 15 months at Weilangta, and 18 months at Ridgley. Disease severity (myco) was quantified by estimating the percentage area of necrotic lesions present on each tree, on a 10-point scale (see Milgate et al. 2005a).
Analysis:
The myco trait values used for QTL analysis in family 1 were least-square means calculated for each of the 112 genotypes with map information, by fitting to the logn-transformed data a mixed model including the effects of replicate (fixed), genotype (fixed), incomplete block within replicate (random), and random error. All families in the field trial were included and analysis was undertaken with PROC MIXED of SAS V9.1 (SAS Institute, Cary, NC). The distribution of genotype least-square means was examined for myco (by the Kolmogorov–Smirnov test using PROC UNIVARIATE of SAS), to test the assumption of a normal distribution for interval and multiple-QTL model (MQM) mapping. Even with transformation, the distribution of least-squares means departed from normality (P < 0.01).
In the case of the analysis examining the stability of susceptibility to MLD across different environments, Pearson's correlation coefficients were calculated between the parental breeding values and the myco least-square means in open-pollinated and controlled-cross families grown at Woolnorth and Weilangta. The least-square means for the Woolnorth families were calculated by fitting a mixed model with replicate and families as fixed effects and incomplete blocks as random effects. In the case of Weilangta, replicate was treated as random and family as a fixed effect. Breeding values were from Dungey et al.'s (1997) analysis of a factorial mating design at Ridgley and the performance of the three F2 families was predicted assuming complete additivity.
QTL analysis was conducted with MAPQTL 4.0 (Van Ooijen et al. 2002), using the consensus linkage map constructed in family 1 (described in detail in Freeman et al. 2006). Putative QTL were declared at two different levels, significant (genomewide type I error <0.05) and suggestive (chromosomewide type I error rate <0.05). A relaxed type I error rate (i.e., presenting QTL at the suggestive level) was adopted to facilitate comparative QTL mapping (Beavis 1998; Van Ooijen 1999). Comparative QTL mapping will be particularly useful in a genus such as Eucalyptus, in which QTL mapping is in its infancy. The inclusion of numerous transferable microsatellite markers in the linkage map used for QTL analysis in this study (Freeman et al. 2006) will facilitate comparative mapping. The LOD threshold for genomewide significance was determined by permutation testing (1000 replications; Churchill and Doerge 1994). An average chromosomewide LOD threshold (LOD 3) for suggestive QTL was determined by empirical simulations (Van Ooijen 1999). Interval mapping was initially performed to scan the genome for map intervals significantly associated with myco, using the default parameters of MAPQTL 4.0 (Van Ooijen et al. 2002). The suggestive threshold was used to select putative QTL as cofactors for the MQM procedure. Where map intervals exceeding this threshold existed, a single marker closest to the peak LOD was chosen as a cofactor for the MQM (Jansen 1993, 1994; Jansen and Stam 1994). MQM mapping removes the residual variance attributable to the selected cofactors, thereby increasing the power in subsequent scans for multiple QTL. An iterative approach combining forward and backward selection of cofactors was used for QTL detection, as suggested by Van Ooijen et al. (2002). All methods of QTL discovery have their biases, so a combination of approaches can increase confidence in the detection of putative QTL (Kearsey and Farquhar 1998; Asins 2002). Because myco distribution departed from normality, single-marker Kruskal–Wallis tests were also used for QTL detection. Where the closest marker to the QTL peak segregated from the opposite parent, the significance of the closest marker segregating from the same parent as the QTL was tested. This analytical method is robust to departures from normality (Van Ooijen et al. 2002). Where more than one unlinked marker significantly affected myco (genomewide type I error <0.05 in interval mapping), they were tested for epistasis by testing for an interaction effect using PROC GLM of SAS.
Validation of the two major putative QTL for MLD severity was attempted in two other F2 families (families 2 and 3, described above), using a selective genotyping strategy. Although each of the F2 families was interrelated (see above), in each case where they shared grandparents, a different individual of the F1 generation was used for crossing. The myco scores were logn transformed to optimize residual normality and least-square means were calculated from two clones of each genotype, as described above for family 1. The extremes in genotype least-square means (25 and 20 genotypes from each extreme in families 2 and 3, respectively) were assayed using the closest polymorphic SSR marker(s) to each of the putative QTL. A putative QTL was considered validated if the marker genotypes significantly affected MLD severity (P < 0.05) in a Kruskal–Wallis test. The Kruskal–Wallis test allowed an assessment of the effects of different allelic combinations on the mean trait value, for the major QTL in each F2 family.
RESULTS
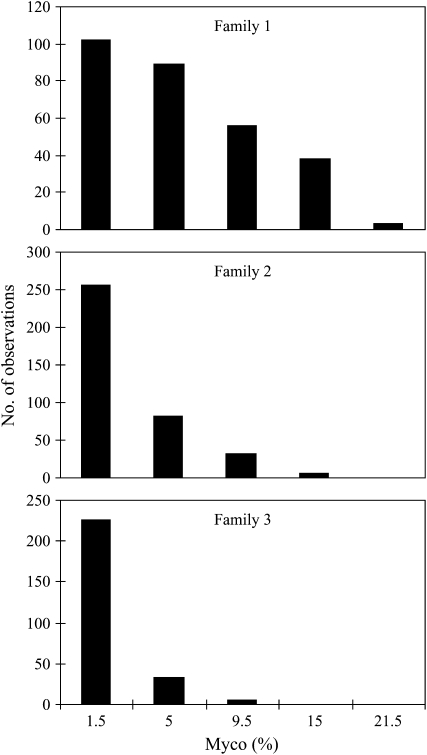
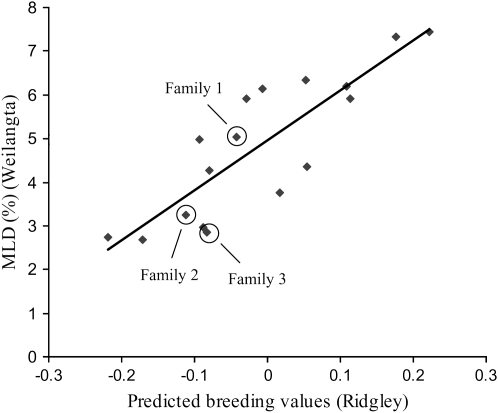
The frequency distribution of myco phenotypes at Woolnorth departed significantly from normality in each family (Figure 2). Each family exhibited a right-skewed distribution (i.e., skewed toward resistance), which may reflect some plants escaping the disease. However, escapees must have been a minority, since the effects of both replicate (P = 0.16) and incomplete block (P = 0.26) were not significant, suggesting infection was uniform across the field trial. Furthermore, the within-family clonal repeatability was high (H2 = 0.59 across all families), indicating a genetic basis to variation in disease severity. Family 1 exhibited the greatest variance followed by family 2 and then family 3, consistent with the proportion of King Island and Taranna in the pedigrees of each family. Disease severity was strongly positively correlated in the same families planted at Woolnorth and Weilangta (d.f. = 6, r = 0.98, P = 0.0001). Furthermore, breeding values calculated from the field trial at Ridgley by Dungey et al. (1997) provided a good prediction of the disease severity in families planted at Woolnorth (d.f. = 6, r = 0.95, P = 0.001) and Weilangta (d.f. = 16, r = 0.81, P < 0.0001; Figure 3).
Figure 2.—
Frequency distributions for the severity of Mycosphaerella cryptica damage (myco %) for individual Eucalyptus globulus trees in each F2 family at the Woolnorth field trial, expressed as the midpoint of each class. All surviving trees in each family were used.
Figure 3.—
Scatter plot of observed Mycosphaerella leaf disease (MLD %) in open-pollinated and controlled-cross families of Eucalyptus globulus at Weilangta, vs. predicted breeding values for MLD damage from related genetic material at Ridgley. Breeding values are centered on a population mean of zero. The three controlled-cross F2 families are circled.
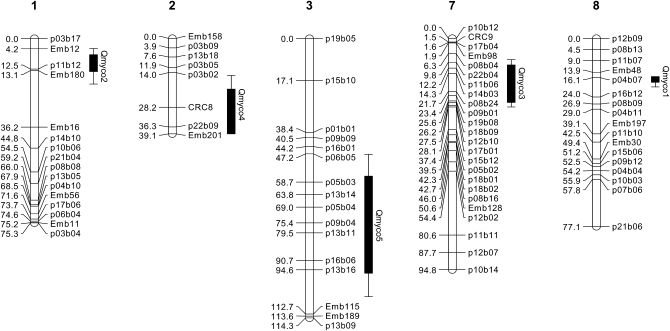
Three putative QTL with significant (genomewide type I error <0.05) effect on myco were located by MQM analysis on linkage groups 1, 7, and 8 (Qmyco2, Qmyco3, and Qmyco1, respectively; Table 1; Figure 4). The two most significant of these (Qmyco1 and Qmyco2) also exceeded the genomewide significance threshold from interval mapping and were confirmed by single-marker Kruskal–Wallis tests. Two additional putative QTL regions were located at the suggestive level (chromosomewide type I error <0.05), on linkage groups 2 and 3 (Qmyco4 and Qmyco5, LOD 4.0 and 3.5, respectively; Table 1). The closest markers to four of the five putative QTL were AFLPs, and for the remaining QTL it was a microsatellite (CRC8). Importantly, the two most significant QTL regions were located close to microsatellite markers. For example, while Qmyco1 peaked (LOD 20.2) closest to AFLP marker p04b07, it was very close to microsatellite marker Emb48 (Figure 4). Similarly, Qmyco2 peaked (LOD 10.9) closest to AFLP marker p11b12, but was closely flanked by microsatellites Emb12 and Emb180 (Figure 4).
TABLE 1.
Putative QTL for the severity of foliar damage caused by Mycosphaerella cryptica in Eucalyptus globulus family 1, identified by MQM mapping and single-marker Kruskal–Wallis tests
| QTL | Linkage group | Segregationa | Adjacent marker | Map position (cM) | LODb | % expd | Kruskal– Wallis P |
|---|---|---|---|---|---|---|---|
| Qmyco2 | 1 | F | p11b12 | 12.5 | 10.9*** | 15.4 | *** |
| Qmyco4 | 2 | F | CRC8 | 28.2 | 4.0c | 5.8 | * |
| Qmyco5 | 3 | M | p09b04 | 75.4 | 3.5c | 6.2 | |
| Qmyco3 | 7 | F | p08b24 | 21.7 | 6.0* | 9.0 | |
| Qmyco1 | 8 | F | p04b07 | 16.1 | 20.2*** | 36.3 | *** |
The parent from which the QTL effect segregates: M, male; F, female.
Peak LOD score for each QTL. Genomewide significance (P) is indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Suggestive QTL based on a chromosomewide significance P < 0.05.
The percentage of phenotypic variation between genotype least-square means explained by each QTL peak.
Figure 4.—
The location of each putative QTL for Mycosphaerella cryptica resistance of Eucalyptus globulus exceeding the chromosomewide significance level from MQM mapping. Linkage groups and loci are as described by Freeman et al. (2006) with microsatellite markers named starting with Emb or CRC. Solid bars and lines represent 1- and 2-LOD support intervals, respectively. The 2-LOD support interval corresponds to an ∼95% confidence interval (Lander and Botstein 1989).
In combination, the two major putative QTL explained 51.7% of the phenotypic variance (i.e., the variation in genotype least-square means) in myco. All putative QTL for myco damage segregated on the female side, except Qmyco5 (Table 1). The interaction between the two major QTL was nonsignificant (F = 0.20; P = 0.9), suggesting that there is no epistasis between them. Assuming strictly additive effects, the presence of the positive alleles at both of the major QTL in family 1 (the a allele for Emb12 and the f allele for Emb48) would lead to 7.4% less damage (myco), compared to the negative alleles (the b allele for Emb12 and the e allele for Emb48; Table 2).
TABLE 2.
Genotype means for fully informative microsatellite markers adjacent to the major QTL for severity of Mycosphaerella cryptica damage in each F2 family of Eucalyptus globulus
| Family | LGa | QTL | Markerb | Parental genotypesc | Genotype means (% damage) | Family means (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Qmyco2 | Emb12*** | ab × cd | ac | ad | bc | bd | 4.7 |
| (rs) (ss) | (rs) | (rs) | (ss) | (ss) | |||||
| 4.4 | 3.1 | 5.9 | 7.6 | ||||||
| 8 | Qmyco1 | Emb48*** | ef × eg | ee | ef | eg | fg | ||
| (sr) (ss) | (ss) | (sr) | (ss) | (rs) | |||||
| 6.7 | 2.7 | 8.7 | 3.9 | ||||||
| 2 | 1 | Qmyco2 | Emb12** | ab × cd | ac | ad | bc | bd | 2.5 |
| (ss) (rs) | (sr) | (ss) | (sr) | (ss) | |||||
| 1.9 | 5.9 | 2.6 | 5.4 | ||||||
| 8 | Qmyco1 | Emb48* | ab × cd | ac | ad | bc | bd | ||
| (sr) (ss) | (ss) | (ss) | (rs) | (rs) | |||||
| 5.3 | 3.9 | 2.3 | 1.8 | ||||||
| 3 | 1 | Qmyco2 | Emb180** | ef × eg | ee | ef | eg | fg | 1.9 |
| (rs) (rs) | (rr) | (rs) | (rs) | (ss) | |||||
| 2.3 | 1.9 | 1.7 | 3.8 | ||||||
Linkage group.
Kruskal–Wallis significance levels *P < 0.05, **P < 0.01, and ***P < 0.001.
Hypothesized genotypes are in parentheses, where r denotes a resistance allele and s a susceptibility allele.
The use of transferable microsatellite markers allowed validation of the two major QTL for myco in two independent families. Both of the major QTL for the severity of M. cryptica damage (myco) were validated in family 2 [Qmyco2 (Emb12, P < 0.01) and Qmyco1 (Emb48, P < 0.05)] by the Kruskal–Wallis test (Table 2). The QTL on linkage group 1 (Qmyco2) was also validated in family 3 (Emb180, P < 0.005; Table 2). Assuming resistance is dominant and a single Mendelian gene controls each QTL, both families 1 and 2 appeared to inherit the alleles for resistance at each QTL from only one heterozygous parent, resulting in segregation of two resistant genotypes to two susceptible ones. In contrast, both parents in family 3 appeared to be heterozygous for resistance at Qmyco2 (Table 2), resulting in segregation of three resistant genotypes to one susceptible.
DISCUSSION
This study is the first report of QTL affecting disease resistance in E. globulus and is the second such report for a eucalypt species. Two major QTL regions have been detected for M. cryptica severity (myco), both of which explain a large proportion of the phenotypic variance. Contrary to predictions from an infinitesimal (or polygenic) model, QTL studies of numerous traits (excluding growth) in forest trees have often suggested oligogenic or major gene control, with a few major genes responsible for much of the total phenotypic variability (reviewed by Sewell and Neale 2000). However, the common finding of oligogenic control must be interpreted with caution in light of the fact that the power of QTL detection is influenced by numerous factors including the sample size, the magnitude of the QTL effect, and the heritability of the trait (Beavis 1998; Kearsey and Farquhar 1998; Erickson et al. 2004). Furthermore, only loci that are heterozygous with a detectable effect in the crosses and environments studied will be discovered (Marques et al. 2002). In combination, these factors can result in an oversimplified view of the genetic control of quantitative traits, by underestimating the number of QTL affecting a trait and overestimating their phenotypic effects.
Nonetheless, oligogenic control is the most likely hypothesis to explain the discovery of just two major QTL for resistance to myco. These explained a large proportion of the phenotypic variance in family 1 and were also significant in at least one other controlled-cross family. This hypothesis is consistent with the moderate to high heritability previously reported for damage from MLD within E. globulus (narrow-sense heritability = 0.12–0.60; Dungey et al. 1997; Milgate et al. 2005b). However, the possibility that the regions with QTL of large effect represent clusters of closely linked genes cannot be dismissed since resistance genes are commonly clustered in plants (Wang et al. 2001; Chu et al. 2004; Mondragon-Palomino and Gaut 2005). Due to the lack of QTL studies for disease resistance in Eucalyptus, comparative mapping is not possible. However, Junghans et al. (2003) reported that one major gene controlled a large proportion of the phenotypic variation for rust resistance in E. grandis, which appeared to display Mendelian inheritance. In other forest trees, QTL studies have commonly found that few genomic regions explain a large proportion of the phenotypic variation in disease resistance. For example, one or two major QTL explained most of the variance in resistance to rust in poplar (Newcombe et al. 1996; Lefèvre et al. 1998; Dowkiw and Bastien 2004; Jorge et al. 2005) and pines (Devey et al. 1995). Similar results have also been found in QTL studies investigating quantitative disease resistance in crop plants (reviewed by Young 1996). Hence, the control of quantitative disease resistance by one or two major loci, in combination with a few minor ancillary loci, appears representative of many pathosystems.
The suggestion of oligogenic control is consistent with specialized associations that may exist between hemibiotrophic pathogens such as Mycosphaerella and their host. Biotrophic and hemibiotrophic pathogens invade living cells and subvert metabolism to favor their growth and reproduction; hence minor differences in either organism can upset the balance (Hammond-Kosack and Jones 1997). The genetics of host resistance and pathogen virulence in biotrophic pathogens often fit the classic gene-for-gene model (Flor 1971), whereby, due to coevolution of host and parasite, complementary pairs of dominant genes in both the pathogen and the host govern resistance. This model may apply to the control of resistance to M. cryptica in E. globulus. In support of this hypothesis, the specialization of different biotypes of M. cryptica on resistant vs. susceptible trees in the same trial and the same disease epidemic as this study (Milgate et al. 2005a) suggests a degree of specificity in the resistance of E. globulus to M. cryptica, similar to loci for rust resistance in poplar (Tabor et al. 2000; Dowkiw and Bastien 2004) and pine (Wilcox et al. 1996).
The genotype means for both of our major QTL fit a model of Mendelian inheritance in each family with resistance dominant over susceptibility (Table 2). This fits the gene-for-gene model and observations in other forest trees (e.g., Devey et al. 1995; Junghans et al. 2003). The differences in mean level of resistance of families in the present study (Table 2) are consistent with one QTL segregating in family 3 (assuming it is fixed for the resistant allele at Qmyco1), two in family 2, and two, plus minor QTL, segregating in family 1. This model also accords with the increasing variability within these families (Figure 2). The higher resistance of family 3 is likely to reflect its pedigree of solely King Island grandparents, since trees from King Island are generally more resistant than those from Taranna (Dungey et al. 1997).
The specificity of QTL or genes for disease resistance in their interaction with different races of a pathogen has implications for their durability. The literature abounds with examples from crop plants (e.g., Cowger et al. 2000; Talukder et al. 2004) and forest trees such as poplar (e.g., Pinon 1995; Frey et al. 2005) of breeding programs concentrating on selection for major genes controlling qualitative race-specific resistance, only for the pathogen to evolve new virulent types, resulting in the resistance being overcome. However, although the genetic basis of durable resistance is not fully understood, it is believed that durable host resistance is often conferred by minor genes (Newcombe 1998). Hence durable resistance may be achieved from a combination of major “pivotal genes” and ancillary genes or QTL of lesser effect (Newcombe 1998), as is evident in this study. The low amount of genetic variability and prevalence of asexual reproduction evident in a preliminary study of the pathogen M. cryptica (Milgate et al. 2005a) also bode well for the durability of resistance. A greater understanding of the potential durability of the QTL for resistance to M. cryptica identified herein will require testing against multiple races and/or species of MLD and more knowledge regarding the genetic variability and adaptive potential of the pathogen.
A limitation of QTL analysis is that the QTL are often specific to the pedigree, the environment, and the developmental stage in which they are found. This is particularly true in forest trees as opposed to crop plants, due to their longevity, genetic heterogeneity, and the variety of sites in which they are grown (Grattapaglia 2000; Sewell and Neale 2000). However, in this study, the validation of the major QTL for M. cryptica resistance is promising, suggesting the QTL may affect disease severity in trees with a range of genetic backgrounds. QTL regions influencing disease resistance in several different pedigrees have also been demonstrated in forest trees such as E. grandis (Junghans et al. 2003) and hybrid Populus (Lefèvre et al. 1998). The results of this study are also promising in terms of the environmental stability of MLD resistance. Strong intercorrelations were observed across three widely separated locations (Figure 1). Furthermore, these correlations were between predicted breeding values (from Ridgley) and observed disease severity in different but related genetic material (grown at Woolnorth and Weilangta). Hence, these findings suggest that the genetic control of disease resistance may be stable in different environments and also provide further evidence for stability of the genetic control of resistance in different genetic backgrounds. Environmental stability of MLD resistance has also been previously suggested between families from a single provenance (Reinoso 1992). While the direct application of QTL for marker-assisted selection in forest trees is limited (Strauss et al. 1992; Henery et al. 2007), in part due to the limitations mentioned above, QTL studies can still provide substantial indirect benefits for tree breeding. In the present case, the suggestion that resistance to Mycosphaerella in E. globulus is controlled by few genes reinforces the findings of quantitative genetic studies that there is ample scope to breed for increased disease resistance. Importantly, the fact that resistance appears to be dominant at both major QTL suggests the level of resistance could easily be improved in breeding programs by conventional selection of elite genotypes following disease outbreaks. Furthermore, the apparent stability of resistance in different environments and genetic backgrounds increases the value of resistant genotypes.
In summary, this is the first study to report QTL influencing the severity of damage by M. cryptica in Eucalyptus and the second report of loci influencing disease resistance in the genus. The discovery, and independent validation, of two major QTL explaining a large proportion of the phenotypic variance in M. cryptica severity may be indicative of oligogenic control for the severity of damage by M. cryptica. The severity of damage in different genotype classes at each major QTL is consistent with Mendelian control, with resistance dominant. The validation of both of the major QTL for the severity of damage by M. cryptica in at least one independent pedigree and the demonstration that disease expression is stable across different environments add substantially to the findings of this study by suggesting the QTL may be influential beyond the mapping population. Furthermore, the presence of microsatellites close to the major QTL will allow them to be tested more widely, in different pedigrees of E. globulus and other eucalypt species.
Acknowledgments
We thank Kelsey Joyce and Gunns Ltd. for access to the field trials and assistance with their establishment; Andrew Milgate and Paul Tilyard for data collection; and Adam Smolenski, Rebecca Jones, Dorothy Steane, Merv Shepherd, and Haobing Li for their help in the laboratory or with analysis. We also acknowledge financial support from the Australian Research Council (project DP0773686).
References
- Asins, M. J., 2002. Present and future of quantitative trait locus analysis in plant breeding. Plant Breed. 121: 281–291. [Google Scholar]
- Beavis, W. D., 1998. QTL analysis: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by H. A. Patterson. CRC Press, Boca Raton, FL.
- Brooker, M. I. H., 2000. A new classification of the genus Eucalyptus L'Her. (Myrtaceae). Aust. Syst. Bot. 13: 79–148. [Google Scholar]
- Burdon, R. D., 2001. Genetic diversity and disease resistance: some considerations for research, breeding, and deployment. Can. J. For. Res. 31: 596–606. [Google Scholar]
- Burgess, T., and M. J. Wingfield, 2002. Impact of fungal pathogens in natural forest ecosystems: a focus on Eucalyptus, pp. 285–306 in Microorganisms in Plant Conservation and Biodiversity, edited by K. Sivasithamparam, K. W. Dixon and R. L. Barrett. Kluwer Academic Publishers, The Netherlands.
- Carnegie, A. J., and P. K. Ades, 2002. Mycosphaerella leaf disease reduces growth of plantation-grown Eucalyptus globulus. Aust. For. 66: 113–119. [Google Scholar]
- Carnegie, A. J., P. J. Keane, P. K. Ades and I. W. Smith, 1994. Variation in susceptibility of Eucalyptus globulus provenances to Mycosphaerella leaf disease. Can. J. For. Res. 24: 1751–1757. [Google Scholar]
- Carnegie, A. J., P. K. Ades, P. J. Keane and I. W. Smith, 1998. Mycosphaerella diseases of juvenile foliage in a eucalypt species and provenance trial in Victoria, Australia. Aust. For. 61: 190–194. [Google Scholar]
- Carnegie, A. J., P. K. Ades and R. Ford, 2001. The use of RAPD-PCR analysis for the differentiation of Mycosphaerella species from Eucalyptus in Australia. Mycol. Res. 105: 1313–1320. [Google Scholar]
- Cervera, M. T., J. Gusmao, M. Steenackers, J. Peleman, V. Storme et al., 1996. Identification of AFLP markers for resistance against Melampsora larici-populina in Populus. Theor. Appl. Genet. 93: 733–737. [DOI] [PubMed] [Google Scholar]
- Chu, Z., Y. Ouyang, J. Zhang, H. Yang and S. Wang, 2004. Genome-wide analysis of defense-responsive genes in bacterial blight resistance of rice mediated by the recessive R gene xa13. Mol. Genet. Genomics 271: 111–120. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossalter, C., and C. Pye-Smith, 2003. Fast Wood Forestry: Myths and Realities. Center for International Forestry Research, Jakarta, Indonesia.
- Cowger, C., M. E. Hoffer and C. C. Mundt, 2000. Specific adaptation by Mycosphaerella graminicola to a resistant wheat cultivar. Plant Pathol. 49: 445–451. [Google Scholar]
- Devey, M. E., A. Delfino-Mix, B. B. Kinloch and D. B. Neale, 1995. Random amplified polymorphic DNA markers tightly linked to a gene for resistance to white pine blister rust in sugar pine. Proc. Natl. Acad. Sci. USA 92: 2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowkiw, A., and C. Bastien, 2004. Characterization of two major genetic factors controlling quantitative resistance to Melampsora larici-populina leaf rust in hybrid poplars: strain specificity, field expression, combined effects, and relationship with a defeated qualitative resistance gene. Phytopathology 94: 1358–1367. [DOI] [PubMed] [Google Scholar]
- Dungey, H. S., B. M. Potts, A. J. Carnegie and P. K. Ades, 1997. Mycosphaerella leaf disease: genetic variation in damage to Eucalyptus nitens, Eucalyptus globulus, and their F1 hybrid. Can. J. For. Res. 27: 750–759. [Google Scholar]
- Erickson, D. L., C. B. Fenster, H. K. Stenøien and D. Price, 2004. Quantitative trait locus analyses and the study of evolutionary processes. Mol. Ecol. 13: 2505–2522. [DOI] [PubMed] [Google Scholar]
- Flor, H. H., 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9: 275–296. [Google Scholar]
- Freeman, J. S., B. M. Potts, M. Shepherd and R. E. Vaillancourt, 2006. Parental and consensus linkage maps of Eucalyptus globulus using AFLP and microsatellite markers. Silvae Genet. 55: 4–5. [Google Scholar]
- Frey, P., P. Gérard, N. Feau, C. Husson and J. Pinon, 2005. Variability and population biology of Melampsora rusts on poplars, pp. 63–72 in Rust Diseases of Willow and Poplar, edited by M. H. Pei and A. R. McCracken. CAB International, Wallingford, UK.
- Grattapaglia, D., 2000. Molecular breeding of Eucalyptus, pp. 451–474 in Molecular Biology of Woody Plants, Vol. 1, edited by S. M. Jain and S. C. Minocha. Kluwer Academic Publishers, The Netherlands.
- Hammond-Kosack, K. E., and J. D. G. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Henery, M. L., G. F. Moran, I. R. Wallis and W. J. Foley, 2007. Identification of quantitative trait loci influencing foliar concentrations of terpenes and formylated phloroglucinol compounds in Eucalyptus nitens. New Phytol. 176: 82–95. [DOI] [PubMed] [Google Scholar]
- Jansen, R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., 1994. Controlling the type I and type II errors in mapping quantitative trait loci. Genetics 138: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., and P. Stam, 1994. High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge, V., A. Dowkiw, P. Faivre-Rampant and C. Bastien, 2005. Genetic architecture of qualitative and quantitative Melampsora larici-populina leaf rust resistance in hybrid poplar: genetic mapping and QTL detection. New Phytol. 167: 113–127. [DOI] [PubMed] [Google Scholar]
- Junghans, D. T., A. C. Alfenas, S. H. Brommonschenkel, S. Oda, E. J. Mello et al., 2003. Resistance to rust (Puccinia psidii Winter) in Eucalyptus: mode of inheritance and mapping of a major gene with RAPD markers. Theor. Appl. Genet. 108: 175–180. [DOI] [PubMed] [Google Scholar]
- Keane, P. J., G. A. Kile, F. D. Podger and B. N. Brown, 2000. Diseases and Pathogens of Eucalypts. CSIRO, Melbourne.
- Kearsey, M. J., and A. G. L. Farquhar, 1998. QTL analysis in plants; Where are we now? Heredity 80: 137–142. [DOI] [PubMed] [Google Scholar]
- Kubisiak, T. L., F. V. Hebard, C. D. Nelson, J. Zhang, R. Bernatzky et al., 1997. Molecular mapping of resistance to blight in an interspecific cross in the genus Castanea. Phytopathology 87: 751–759. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre, F., M. C. Goué-Mourier, P. Faivre-Rampant and M. Villar, 1998. A single resistance gene cluster controls incompatibility and partial resistance to various Melampsora larici-populina races in hybrid poplar. Phytopathology 88: 156–163. [DOI] [PubMed] [Google Scholar]
- Li, H., H. Amerson and B. Li, 2006. Genetic models of host-pathogen gene interactions based on inoculation of loblolly pine seedlings with the fusiform rust fungus. New For. 31: 245–252. [Google Scholar]
- Marques, C. M., R. P. V. Brondani, D. Grattapaglia and R. Sederoff, 2002. Conservation and synteny of SSR loci and QTLs for vegetative propagation in four Eucalyptus species. Theor. Appl. Genet. 105: 474–478. [DOI] [PubMed] [Google Scholar]
- Milgate, A. W., R. E. Vaillancourt, C. Mohammed, M. Powell and B. M. Potts, 2005. a Genetic structure of a Mycosphaerella cryptica population. Aust. Plant Pathol. 34: 345–354. [Google Scholar]
- Milgate, A. W., B. M. Potts, K. Joyce, C. Mohammed and R. E. Vaillancourt, 2005. b Genetic variation in Eucalyptus globulus for susceptibility to Mycosphaerella nubilosa and its association with tree growth. Aust. Plant Pathol. 34: 11–18. [Google Scholar]
- Milgroom, M. G., and T. L Peever, 2003. Population biology of plant pathogens. Plant Dis. 87: 608–617. [DOI] [PubMed] [Google Scholar]
- Mohammed, C., T. Wardlaw, A. Smith, E. Pinkard, M. Battaglia et al., 2003. Mycosphaerella leaf diseases of temperate eucalypts around the Southern Pacific Rim. NZ J. For. Sci. 33: 362–372. [Google Scholar]
- Mondragon-Palomino, M., and B. S. Gaut, 2005. Gene conversion and the evolution of three leucine-rich repeat gene families in Aribopsis thaliana. Mol. Biol. Evol. 22: 2444–2456. [DOI] [PubMed] [Google Scholar]
- Newcombe, G., 1998. Association of Mmd1, a major gene for resistance to Melampsora medusae f.sp. deltoidae, with quantitative traits in poplar rust. Phytopathology 88: 114–121. [DOI] [PubMed] [Google Scholar]
- Newcombe, G., H. D. Bradshaw, G. A. Chastagner and R. F. Stettler, 1996. A major gene for resistance to Melampsora medusae f.sp. deltoidae in a hybrid poplar pedigree. Phytopathology 86: 87–94. [Google Scholar]
- Park, R. F., 1988. Effect of certain host, inoculum, and environmental factors on infection by two Mycosphaerella species. Trans. Br. Mycol. Soc. 90: 221–228. [Google Scholar]
- Park, R. F., P. J. Keane, M. J. Wingfield and P. W. Crous, 2000. Fungal diseases of Eucalyptus foliage, pp. 153–240 in Diseases and Pathogens of Eucalypts, edited by P. J. Keane, G. K. Kile, F. D. Podger and B. N. Brown. CSIRO, Melbourne.
- Pinkard, E. A., and C. L. Mohammed, 2006. Photosynthesis of Eucalyptus globulus Labill. with Mycosphaerella leaf disease. New Phytol. 170: 119–127. [DOI] [PubMed] [Google Scholar]
- Pinon, J., 1995. Variability in poplar rusts and evolution of their populations. Consequences for control strategies. C. R. Seances Acad. Agric. Fr. 81: 99–109. [Google Scholar]
- Potts, B. M., R. E. Vaillancourt, G. Jordan, G. Dutkowski, J. Costa e Silva et al., 2004. Exploration of the Eucalyptus globulus gene pool, pp. 46–61 in Eucalyptus in a Changing World, edited by N. Borralho, J. S. Periera, C. Marques, J. Coutinho, M. Madeira et al. Proceedings of the IUFRO Conference, RAIZ, Instituto Investigação de Floresta e Papel, Aveiro, Portugal.
- Reinoso, C., 1992. Variation in Eucalyptus globulus in susceptibility to Mycosphaerella leaf diseases. Honours Thesis, University of Melbourne, Melbourne.
- Sewell, M. M., and D. B. Neale, 2000. Mapping quantitative traits in forest trees, pp. 407–423 in Molecular Biology of Woody Plants, Vol. 1, edited by S. M. Jain and S. C. Minocha. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Strauss, S. H., R. Lande and G. Namkoong, 1992. Limitations of molecular marker-aided selection in forest tree breeding. Can. J. For. Res. 22: 1050–1061. [Google Scholar]
- Tabor, G. M., T. L. Kubisiak, N. B. Klopfenstein, R. B. Hall and H. S. J. McNabb, 2000. Bulked segregant analysis identifies molecular markers linked to Melampsora medusae resistance in Populus deltoides. Phytopathology 90: 1039–1042. [DOI] [PubMed] [Google Scholar]
- Talukder, Z. I., D. Tharreau and A. H. Price, 2004. Quantitative trait loci analysis suggests that partial resistance to rice blast is mostly determined by race-specific interactions. New Phytol. 162: 197–209. [Google Scholar]
- Van Ooijen, J. W., 1999. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., M. P. Boer, C. Jansen and C. Maliepaard, 2002. MapQTL 4.0, Software for the Calculation of QTL Position on Genetic Maps. Plant Research International, Wageningen, the Netherlands.
- Van Zyl, L. M., T. A. Coutinho, M. J. Wingfield, K. Pongpanich and B. D. Wingfield, 2002. Morphological and molecular relatedness of geographically diverse isolates of Coniothryium zuluense from South Africa and Thailand. Mycol. Res. 106: 51–59. [Google Scholar]
- Wang, Z., G. Taramino, D. Yang, G. Liu, S. V. Tingey et al., 2001. Rice ESTs with disease-resistance gene- or defence-response gene-like sequences mapped to regions containing major resistance genes or QTLs. Mol. Genet. Genomics 265: 302–310. [DOI] [PubMed] [Google Scholar]
- Wilcox, P. L., H. V. Amerson, E. G. Kuhlman, B. H. Liu, D. M. O'Malley et al., 1996. Detection of a major gene for resistance to fusiform rust disease in loblolly pine by genomic mapping. Proc. Natl. Acad. Sci. USA 93: 3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. E., and M. I. H. Brooker, 1997. Eucalypts: an introduction, pp. 1–15 in Eucalypt Ecology: Individuals to Ecosystems, edited by J. E. Williams and J. C. G. Woinarski. Cambridge University Press, Cambridge, UK.
- Williams, K., and B. M. Potts, 1996. The natural distribution of Eucalyptus species in Tasmania. Tasforests 8: 39–164. [Google Scholar]
- Young, N. D., 1996. QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 34: 479–501. [DOI] [PubMed] [Google Scholar]