Abstract
The genomes of birds are much smaller than mammalian genomes, and transposable elements (TEs) make up only 10% of the chicken genome, compared with the 45% of the human genome. To study the mechanisms that constrain the copy numbers of TEs, and as a consequence the genome size of birds, we analyzed the distributions of LINEs (CR1's) and SINEs (MIRs) on the chicken autosomes and Z chromosome. We show that (1) CR1 repeats are longest on the Z chromosome and their length is negatively correlated with the local GC content; (2) the decay of CR1 elements is highly biased, and the 5′-ends of the insertions are lost much faster than their 3′-ends; (3) the GC distribution of CR1 repeats shows a bimodal pattern with repeats enriched in both AT-rich and GC-rich regions of the genome, but the CR1 families show large differences in their GC distribution; and (4) the few MIRs in the chicken are most abundant in regions with intermediate GC content. Our results indicate that the primary mechanism that removes repeats from the chicken genome is ectopic exchange and that the low abundance of repeats in avian genomes is likely to be the consequence of their high recombination rates.
LONG interspersed nuclear elements (LINEs), and their parasites short interspersed nuclear elements (SINEs), are the most successful transposable elements (TEs) in warm-blooded vertebrates. The abundance of LINEs and SINEs seems to be high in most mammals, including monotremes (platypus) and marsupials (Margulies et al. 2005); the ∼550,000 insertions of the L1 and the 1,100,000 Alu elements make up almost 30% of the human genome (Lander et al. 2001). SINEs use the enzymatic machinery of LINEs for replication and insertion (Smit et al. 1995; Jurka 1997; Dewannieux et al. 2003; Dewannieux and Heidmann 2005), and therefore the two classes of TEs might be expected to have similar distributions in the genome. However, their distributions are very different; in primates and rodents, SINEs insert into AT-rich regions of the genome and accumulate in gene-rich regions with high GC content, while LINEs reside in AT-rich regions (Soriano et al. 1983; Lander et al. 2001; Pavlicek et al. 2001; Yang et al. 2004; Hackenberg et al. 2005) and show only modest GC enrichment over time. This pattern has received considerable attention in recent years, but there is still no consensus on the mechanism causing it. It has been proposed that the accumulation of Alu's in gene-rich regions may reflect a so far unidentified genomic function and therefore that Alu's are beneficial for the host (Lander et al. 2001). However, the accumulation of Alu's in gene-rich regions is still slower than the time necessary for the fixation of neutral alleles (Brookfield 2001), which seems to question this possibility. An alternative hypothesis is that deletions (most likely by ectopic exchange between repeats) drive the accumulation of repeats in gene-rich regions (Lobachev et al. 2000; Brookfield 2001; Lander et al. 2001; Stenger et al. 2001; Batzer and Deininger 2002; Hackenberg et al. 2005; Abrusan and Krambeck 2006). According to this theory, deletions are more deleterious in gene- and GC-rich regions of the genome than in the gene-poor, AT-rich regions, because they may result in loss of selectively important sequences. In consequence, repeats are lost at a higher rate from AT-rich regions, which shift the distribution of repeats toward GC-rich regions over time. A third hypothesis—that repeats are removed more efficiently from AT-rich regions due to short deletions—was rejected recently by Belle et al. (2005).
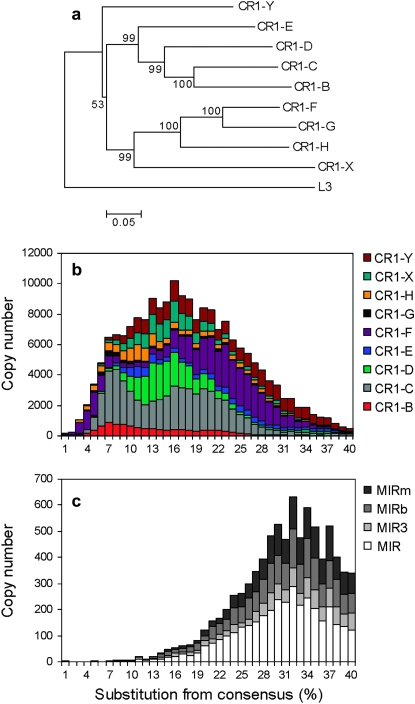
The chicken genome, the only avian genome sequenced so far, is approximately one-third the size of the human genome (International Chicken Genome Sequencing Consortium 2004), and repetitive elements make up only 10% of it, compared with the 40–50% in most mammals (International Chicken Genome Sequencing Consortium 2004; Hughes and Piontkivska 2005; Wicker et al. 2005). The majority of TEs in the chicken genome (80%, or 200,000 copies) belong to the CR1 families of LINEs. Unlike in primates and rodents, where the phylogeny of LINEs forms a single lineage (Smit et al. 1995; Furano 2000), chicken CR1 elements form several distinct lineages that are considerably more diverged from each other than mammalian L1's (International Chicken Genome Sequencing Consortium 2004; Figure 1), and some of them have coexisted (it is unclear whether any of the chicken CR1 families are active at present) since the bird–reptile split (Vandergon and Reitman 1994; Kajikawa et al. 1997; International Chicken Genome Sequencing Consortium 2004). The abundance of CR1 elements peaked ∼45 MYA (Figure 1b, substitution level ∼16%, assuming a substitution rate of 3.6 × 10−9 year−1; Axelsson et al. 2004) and since then gradually declined. A difference compared to mammalian genomes is that all detectable SINEs (MIRs) are ancient, present in low copy numbers, and inactive (International Chicken Genome Sequencing Consortium 2004; Figure 1).
Figure 1.—
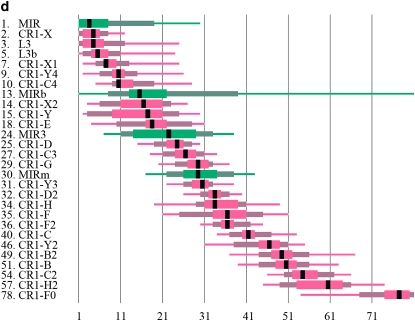
The evolution and age of CR1's and MIRs. (a) Neighbor-joining tree of the ORF2's of the chicken CR1 families, with the human L3 element as outgroup (International Chicken Genome Sequencing Consortium 2004). Bootstrap values (1000 replicates) are indicated on the nodes. (b) The age distribution of CR1 families in the chicken genome in bins corresponding to 1% increments in substitution levels. (c) The age distribution of MIRs. MIRs were most active ∼90 MYA (substitution level ∼32%) and apparently went extinct at the time when CR1's were most active. (d) The rank order of the age of CR1, L3, and MIR families in the chicken. Note that the ages of MIRs, L3's, and three CR1 families (CR1-Y2, CR1-H2, CR1-F0) are probably underestimated compared to other repeats due to their shortness and low connectedness with other repeats (supplemental Table 1 at http://www.genetics.org/supplemental/).
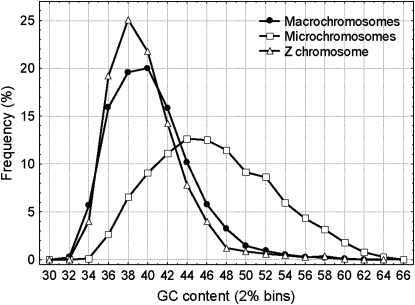
The 30 sequenced chicken chromosomes are considerably more diverse than the human chromosomes in several properties (International Chicken Genome Sequencing Consortium 2004). Their size spans almost two orders of magnitude, from the 188 Mb of chromosome 1 to 1 Mb of chromosome 32. Autosomes are classified into macrochromosomes (1–5), intermediate chromosomes (6–10), and microchromosomes (11–32) (International Chicken Genome Sequencing Consortium 2004). Several biologically important traits covary with chromosome size (International Chicken Genome Sequencing Consortium 2004): GC content (Figure 2), gene density, substitution rate, and recombination rate correlate negatively (making sequence divergence a less accurate tool for TE age determination than in mammalian genomes), while the amount of noncoding material (the abundance of repetitive elements and intron length) correlates positively (Axelsson et al. 2004; International Chicken Genome Sequencing Consortium 2004).
Figure 2.—
The GC distributions of macro-, micro-, and sex chromosomes.
Female birds are heterogametic (Z and W chromosomes), but unlike in mammals, males are the homogametic sex (ZZ) and females are the ZW. Like the mammalian Y, the W chromosome is genetically degenerate (although it is larger than some of the microchromosomes) and repeat rich (International Chicken Genome Sequencing Consortium 2004). Similarly to the mammalian X and Y chromosomes (Lahn and Page 1999), the cessation of recombination between the Z and W chromosomes was gradual (Ellegren and Carmichael 2001), which has led to the formation of evolutionary strata in Z–W divergence (Handley et al. 2004).
In this article we characterize the evolution of LINE and SINE families of the chicken genome, and their chromosomal distributions on macro-, micro-, and Z chromosomes in relation to GC content. We determine the chronological order of all repeats in the chicken genome, using a novel method of age determination (Giordano et al. 2007). The method does not rely on sequence divergence from the consensus; therefore it is not biased by the large differences in the recombination rates in the chicken genome. We show that CR1's decay faster in GC-rich regions than in AT-rich regions, but the decay is highly asymmetric: 5′-ends of the repeats (in relation to their consensus sequence) are lost much faster than 3′-ends, and the CR1 repeats are most abundant in AT-rich and GC-rich regions. We argue that ectopic exchange between repeats is the main force that removes repeats from the chicken genome.
MATERIALS AND METHODS
Transposon (RepeatMasker) and gene (RefSeq) annotation files and the sequence of the chicken genome (release galGal2, February 2004, and release galGal3, May 2006) were downloaded from the University of California Santa Cruz Genome Browser at http://genome.ucsc.edu (Karolchik et al. 2003). Release galGal2 was used in the evolutionary analysis of chicken repeat families (Figure 1; supplemental Figure 1 at http://www.genetics.org/supplemental/) and the GC distribution of MIRs (Figure 5b, Figure 6, c, f, and i), while release galGal3, which contains no MIRs, was used in the analysis of CR1's (with the exception of Figure 1). Preliminary analyses (G. Abrusán, unpublished results) showed that the chromosomes of intermediate size (6–10) show a qualitatively similar (intermediate) pattern to macro- and microchromosomes, and therefore we did not include their detailed analysis in this article. The age of CR1 families was determined in two independent ways: using their divergence from the consensus and using an interruptional analysis (Giordano et al. 2007). Divergence levels provided in the RepeatMasker annotation (D) were corrected for the CpG content of each insertion by DCpG = D/(1 + 9FCpG) (Mouse Genome Secquencing Consortium 2002), where FCpG is the frequency of CpG dinucleotides in the consensus, and DCpG was corrected with the Jukes–Cantor formula for multiple substitutions (Mouse Genome Secquencing Consortium 2002). No further corrections for regional or chromosomal differences in substitution rates (Axelsson et al. 2005; Webster et al. 2006) were made. The detailed methodology of the transposon-interruption analysis and the software used is described elsewhere (Giordano et al. 2007). In short, the method uses the information from transposon clusters—TEs that insert into other TEs—to determine the age of families. A TE that interrupts another TE by necessity is younger than the interrupted one. Using the interruptions from the entire genome, we determined the rank order of the age of TE families of the chicken genome. The family with rank 1 is the oldest one, and the family with the highest rank is the youngest; the error bars are generated by an iterative process and represent 100, 90, and 50% confidence intervals of the position of the repeat families in the rank order (see Giordano et al. 2007 for details). The bootstrap neighbor-joining tree (1000 replicates, Figure 1) of CR1 families was constructed with MEGA3 (Kumar et al. 2004) and is based on all the ORF2's of the CR1 consensus; these were aligned with ClustalX.
Figure 5.—
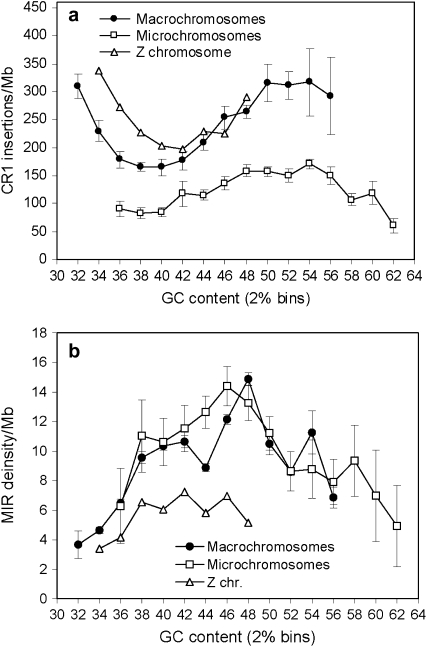
Chromosomal densities of CR1 and MIR insertions in regions of different GC content. Error bars show standard errors. CR1 densities in macro- and microchromosomes are significantly different in every GC bin (P < 0.05, two-sample t-tests). The Z chromosome differs significantly (P < 0.05, one-sample t-tests) from macrochromosomes in regions with GC content <40%. MIR densities of macro- and microchromosomes do not differ significantly, but the Z chromosome has significantly lower MIR densities than the autosomes (P < 0.05, one-sample t-tests).
Figure 6.—
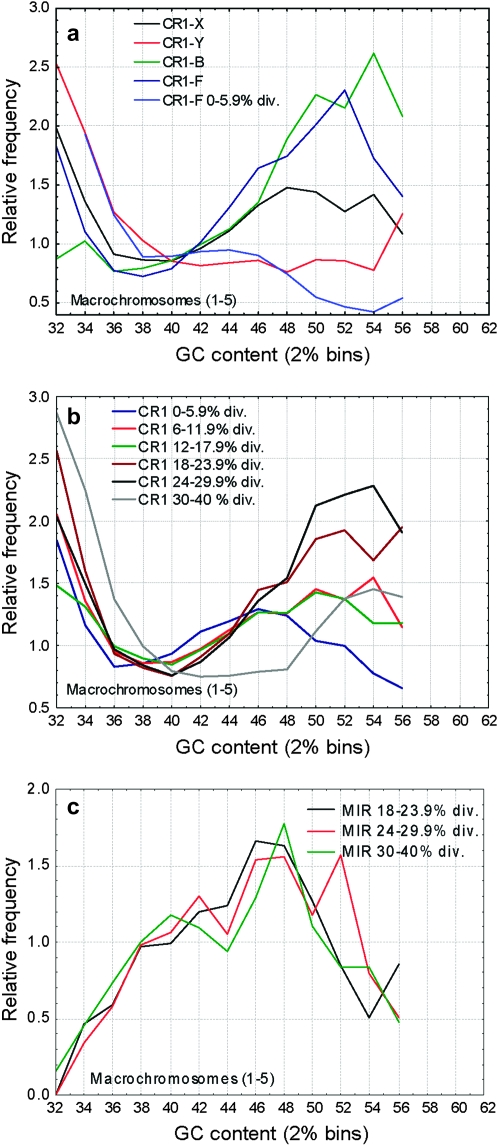
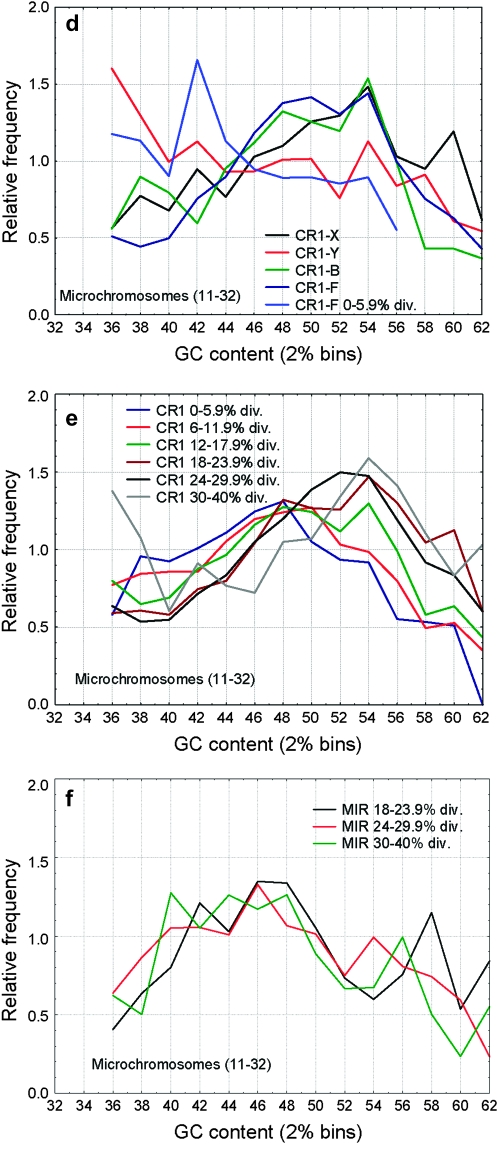
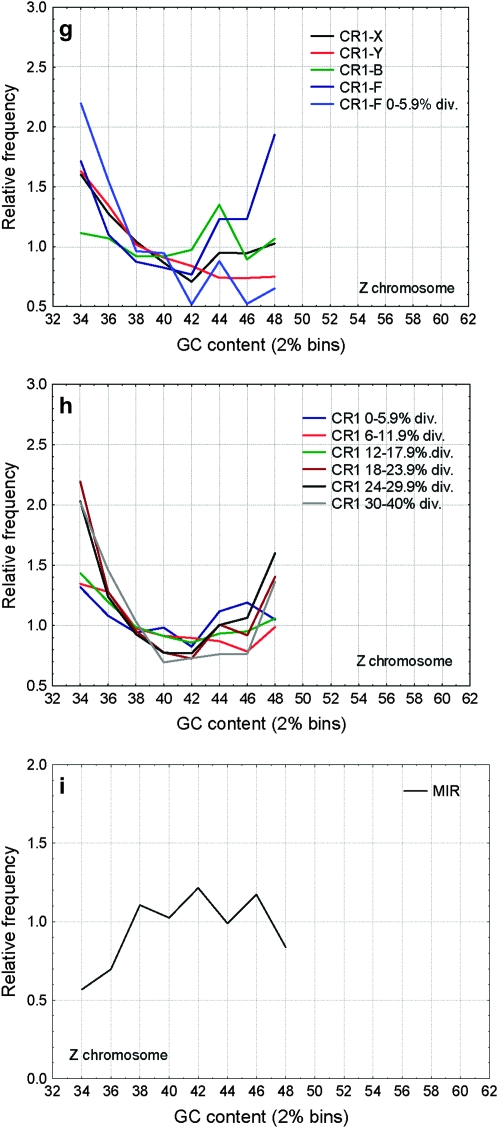
Frequencies of CR1 and MIR families in regions of different GC content. The different CR1 families show different GC distributions (a, d, and g), the oldest families (CR1-Y, CR1-X) being more frequent in AT-rich regions than the younger families (CR1-B, CR1-F, P < 0.001 for all possible comparisons with CR1-Y and CR1-X families). Similarly to human Alu's, the youngest insertions of the recently active CR1-F family show a rapid shift toward AT-rich regions: CR1-F insertions diverged <6% show a clear bias toward AT-rich regions (a, d, and g), but not older insertions (P < 0.001 for the difference between their GC distributions on all chromosome classes). However, in addition to this initial change in their distribution, CR1's show no consistent shift toward regions of high GC content with increasing divergence in any of the chromosome classes (P > 0.05 for most comparisons; b, e, and h). Unlike most mammalian SINEs, MIRs in the chicken genome are most frequent in regions with intermediate GC content (c and f) and show no significant change in their distribution with time (P > 0.05 among the age classes).
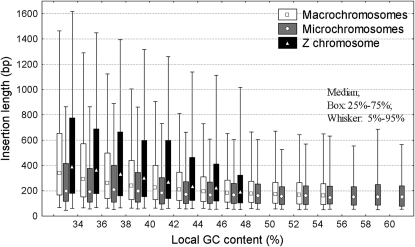
The GC distributions of the chromosomes (GCchr, Figure 2) were calculated by dividing the entire genome into 30-kb nonoverlapping windows, excluding repetitive elements (in consequence, the total nucleotide counts in the windows were typically 27–28 kb). The local GC content of repeats (GCrep) was calculated in 2- × 15-kb windows adjoining every TE insertion, and fragmented repeats were treated as one insertion. The length of TE copies was determined using their chromosomal coordinates; for fragmented repeats, the sum of their fragments was used. To test for interactions between the length of CR1's and their local GC distribution on different chromosome classes, we used general linear models (Figure 3). We tested whether CR1's decay symmetrically (i.e., both sides of the insertion shorten at a similar rate). The frequency distributions of the positions of 5′-ends and 3′-ends of CR1's were calculated by grouping them into bins every 50 bases (Figure 4). Differences between the medians of the distributions were determined with Mann–Whitney tests.
Figure 3.—
Relationships between the length of CR1 insertions and their local GC content on the different chromosome classes. The data were analyzed with general linear models, but are shown as box plots for clarity. Log-transformed insertion length was used as the dependent variable, chromosome class as the categorical predictor, and local GC content as continuous predictor. The analyses were done separately for GC ranges of 32–46% (n = 124,862) and 48–56% (n = 11,204). Macro- and microchromosomes differ in both the intercept (P < 0.001) and the slopes (P < 0.001). The Z chromosome differs significantly from the autosomes in both slope and intercept (P ≪ 0.001). The slope of the regression between local GC content and repeat length is significant for every chromosome class (P ≪ 0.001). Within the GC range of 48–56% there is no significant difference between the slopes (P = 0.15), but the intercepts of macro- and microchromosomes are significantly different (P < 0.001).
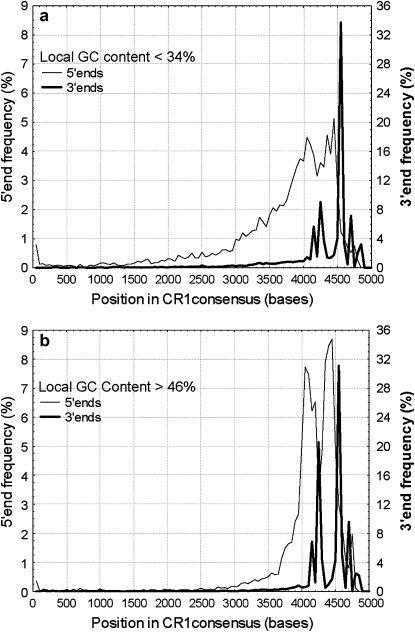
Figure 4.—
Biased shortening of CR1's on macrochromosomes. (a) Distribution of 5′-ends and 3′-ends (in relation to the CR1 consensus) of all CR1's with local GC content <34%. The multiple peaks, particularly easily visible in the distributions of 3′-ends, correspond to different CR1 families, which have slightly different consensus lengths. (b) Distribution of 5′-ends and 3′-ends of all CR1's with local GC content >48%. Both the distributions of 5′-ends and 3′-ends differ highly significantly from each other (Mann–Whitney tests, P < 0.001, n = 14,282).
Absolute repeat densities of CR1's and MIRs (Figure 5) were standardized with the GC content of the chromosome by dividing the number of repeats with local GC content falling into a GC range (e.g., 38–40%) by the total amount of sequence having similar GC content. In addition, using the RefSeq gene annotations, we determined the “location” of every insertion, i.e., whether it is between genes or is present in introns. Repeat densities were calculated separately for each chromosome, with the exception of some microchromosomes, which were pooled due to their small size: chromosomes 15–16, chromosomes 21–22, chromosomes 23–25, and chromosomes 26–32. Differences in repeat densities were tested with two-sample t-tests (macro vs. micro) and one-sample t-tests (Z vs. macrochromosomes).
To compare the distributions of CR1 elements of different age or from different families, we used the method of Yang et al. (2004): the frequency of GCrep falling into a bin of its frequency distribution was divided by the frequency of GCchr falling into the same bin of the GCchr distribution (Figure 6). In addition to standardizing for GC content, this method corrects for the differences in absolute repeat densities as well. The statistical significance of the changes in the GC distributions within a chromosome class was tested with Kruskall–Wallis tests.
RESULTS
The analysis of the divergence and the interruptional analysis of CR1's and MIRs confirms that most of the CR1 lineages differentiated early and have coexisted for a long period in the chicken and that MIRs are among the oldest detectable TEs (Figure 1; supplemental Figure 1 and supplemental Table 1 at http://www.genetics.org/supplemental/). In contrast to their phylogeny (note the low 53% bootstrap support for the first node in Figure 1), the interruption analysis suggests that the oldest CR1 family is CR1-X (Figure 1). The rank order of all repeats in the chicken is presented in supplemental Figure 1 and supplemental Table 1.
CR1's of different lengths are distributed unevenly on the chromosomes, according to their local GC content (Figure 3). Unlike in humans and the mouse, where L1's are longest in regions with intermediate GC content (38–40%) (Mouse Genome Secquencing Consortium 2002), in the chicken genome, CR1 length decreases monotonically with decreasing AT content (Figure 3). In the GC range of 32–46%, there is a significant negative correlation between the local GC content of CR1's and their length on all chromosomes (Figure 3). In the GC range of 48–54%, CR1's are slightly but significantly longer on macrochromosomes than on microchromosomes (P = 0.002 for the intercepts), but there is no difference in the slopes (Figure 3). Due to the inefficiency of reverse transcription that results in insertion of incomplete, “dead on arrival” CR1's, the vast majority of CR1 copies are 5′ truncated (Wicker et al. 2005). However, in addition to this initial loss of 5′-ends, we observed a surprising pattern in the erosion of the repeats: the shortening of CR1 repeats after insertion in the GC-rich regions is also highly biased; the 5′-ends of the insertions are being further lost, but not the 3′-ends (the reference being the consensus sequence: the first base of the 5′-UTR of the consensus is position 1 and the last base of the 3′-UTR is 4200–4500, depending on the CR1 family; in Figure 4, the medians of the distributions differ significantly by Mann–Whitney tests, P < 0.001). This is not specific for chicken CR1's; in the human genome, primate-specific L1's show a similar, although less pronounced, bias in their shortening (G. Abrusán, unpublished results). The distributions of 3′-end positions have multiple peaks (Figure 4) due to the different lengths of the consensus sequences of the various CR1 families, and the distributions of CR1 3′-end positions are not significantly different when CR1 families are analyzed independently (G. Abrusán, unpublished results).
Unlike in mice and humans, where L1 repeats are most abundant in AT-rich regions (Mouse Genome Secquencing Consortium 2002), on macrochromosomes and the Z chromosome CR1 repeats show a bimodal pattern; repeats are abundant both in AT-rich and GC-rich regions and have the lowest densities in regions with intermediate GC content (Figure 5a). On microchromosomes, even when standardized with the local GC content, CR1 densities are much lower (Figure 5a) and, due to the high GC content of these chromosomes, the peak in the AT-rich region is missing. The different GC content of chromosomes does not explain the differences in repeat density; CR1 density is significantly lower in every GC bin on microchromosomes (P < 0.05, two-sample t-tests, Figure 5a). CR1 density on the Z chromosome is significantly higher than on macrochromosomes in regions with low GC content (<38%, one-sample t-tests), but not in regions with higher GC content. In contrast to CR1's, MIRs are most abundant in regions with intermediate GC content (46–48%, Figure 5b). There are no significant differences between the densities of MIRs in macro- and microchromosomes (Figure 5b), but their abundance is significantly lower on the Z, independently of the local GC content (Figure 5b).
The distribution of CR1 elements shows considerable differences between families: relatively young families like CR1-F or CR1-B are more enriched in regions of high GC content than the oldest families such as CR1-X and CR1-Y (Figure 6), which is the opposite to the pattern observed in the human and rodent genomes. However, the CR1-F family that had the most recent burst of activity in the chicken shows a similar shift toward regions of high GC content as SINEs in the mammalian genomes. The pattern is similar on the Z chromosome and the macrochromosomes, but less pronounced on the microchromosomes (Figure 6). The distribution of MIRs changes minimally over time; only the oldest insertions (30–40% divergence) are slightly (but statistically significantly) shifted toward AT-rich regions (Figure 6, c and f). On microchromosomes (Figure 6, d and f), repeats show less pronounced differences between regions of different GC content, and above the GC content of 52–54%, the relative frequency of CR1 repeats declines (in the case of MIRs from 48%).
DISCUSSION
Evolution of CR1 families:
The evolutionary analysis of chicken repeats shows that the three methods used supplement each other and that the interruptional analysis provides useful information on CR1 evolution where the other two methods are not decisive. There are inconsistencies between the phylogeny of the repeats and their divergence. For example, the CR1-F family is one of the youngest families in the phylogeny (Figure 1a); nevertheless, the divergence of most CR1-F insertions from their consensus sequence is comparable to the older families (Figure 1b). In addition, the split between the oldest families (CR1-X and CR1-Y) is not resolved well by their phylogeny, and the large spatial variation in the nucleotide substitution rates on the chicken chromosomes (Axelsson et al. 2005; Webster et al. 2006) makes it particularly difficult to make inferences about the real age of old, diverged families. The transposon-interruption analysis provides a picture of the evolutionary history of repeats qualitatively similar to the one obtained by the phylogeny. The method is able to resolve the history of the oldest CR1 families, and unlike the phylogeny that it supports, the CR1-X family is the oldest. Since the interruptional analysis is not influenced by spatial or temporal variability of substitution rates, it is well suited to resolve evolutionary relationships between the repeats where phylogenetic trees or substitution rates do not lead to clear conclusions and can be successfully used in phylogenetic inferences on the species level as well (see Giordano et al. 2007).
Implications of CR1 length for their activity and mechanisms constraining their abundance:
Since different chicken chromosomes have very different GC contents (Figure 2), any differences in the length of CR1 elements could be a simple by-product of chromosomal GC distributions if copies of different length are distributed unevenly according to the local GC content. Indeed, CR1's grow short with increasing GC content on all chromosomes (Figure 3). However, the different GC content is not sufficient to explain the differences of CR1 length, although it accounts for most of the difference between macro- and microchromosomes (75% of the explained variance within the GC range of 32–46%; Figure 3).
There are two basic mechanisms that can eliminate long CR1 insertions from the genome: short deletions that erode them gradually and ectopic exchange, which can remove larger fragments or entire repeats. Both short deletions (Petrov et al. 2000; Petrov 2002) and ectopic exchange (Langley et al. 1988; Charlesworth et al. 1994; Bartolome et al. 2002) are likely to occur during meiotic recombination, and both have been proposed to be the main mechanism that controls the expansion of noncoding material in the genome. In theory, both mechanisms can explain the biased erosion of the repeats (Figure 4): short deletions can lead to the 5′-end-biased decay if the coding region of CR1's is deleterious, for example, due to interference with the expression of closely linked genes, while 3′-UTRs are not, or less deleterious. In this case, selection will favor the fixation of deletions in the coding regions of the repeats, particularly in gene-rich, highly recombining regions of the genome. We tested this theory using CR1's of the macrochromosomes and found no significant differences in the distribution of 5′-ends and 3′-ends of intergenic and intronic repeats, indicating similar rates of sequence loss (supplemental Figure 3 at http://www.genetics.org/supplemental/); thus this hypothesis alone is not sufficient for explaining the observed pattern. However, selection against long repeats in combination with ectopic exchange offers a possible explanation. Since LINEs are reverse transcribed, CR1 insertions show small variability in the position of their 3′-ends, but due to 5′ truncation, which most likely occurs due to the dissociation of the reverse transcriptase from the mRNA during reverse transcription, insertions show a large variation in their 5′-end positions. In an ectopic exchange event between two copies of unequal length (supplemental Figure 4), one of the repeats is lost (note that both the shorter and the longer insertion can be lost in this way, depending on the order of the repeats). However, if long repeats are more deleterious than short ones, then the likelihood that the deletion containing the longer repeat will reach fixation is higher, which leads to a gradual loss of long CR1 insertions.
Similarly to the mammalian X chromosome (Baker and Wichman 1990; Mouse Genome Secquencing Consortium 2002), CR1's are more abundant on the Z chromosome than on the autosomes (Figure 5), probably due to its low recombination rates. Lyon (1998) has proposed that the high density of LINEs on mammalian X is connected with a function in X inactivation. In birds, it is unclear whether Z inactivation occurs at all (Ellegren 2002); most authors found no evidence of Z inactivation (Baverstock et al. 1982; Kuroda et al. 2001), with the exception of McQueen et al. (2001).
Implications of GC distributions of CR1's for the mechanisms that control their abundance and genome size:
The distribution of CR1's (Figure 5) is different from the distribution of L1's in mammals (see Yang et al. 2004 for the analysis of L1's); CR1's have peak densities in both AT-rich and GC-rich regions. This pattern is most likely caused by several mechanisms: insertion bias, selection against deleterious insertions, and ectopic exchange between repeats. GC-rich regions are also gene rich, and therefore the likelihood that an insertion will be deleterious due to the disruption of selectively important sequences is higher than in AT-rich (gene-poor) regions, so that selection will remove more insertions from GC-rich regions. In contrast, ectopic exchange is expected to remove repeats more efficiently from AT-rich regions, where deletions are less deleterious.
The 5′-end biased shortening of the repeats supports the ectopic exchange hypothesis. However, the high density of old CR1 families in AT-rich regions is the opposite of the pattern observed in mammals. In addition to possible changes in the insertion preference of CR1 families, an alternative explanation is that deletions that reach fixation in the chicken are not AT biased, possibly due to the less-pronounced isochore structure of the chicken genome. In vertebrates, the GC content of a genomic region is positively correlated with its recombination rate (Eyre-Walker 1993; Myers et al. 2005), and the current consensus is that recombination increases the local GC content by biased gene conversion (Marais 2003; Meunier and Duret 2004; Webster et al. 2005). In addition, a continuous loss of AT-rich sequence due to ectopic exchange is likely to contribute to the discrepancy between the observed and the expected GC content of mammalian genomes. Although in the chicken genome repeats are lost from highly recombining regions, probably the same process, i.e., ectopic exchange, is responsible for the removal of the repeats. Since the recombination rates of avian chromosomes are much higher than those of mammalian ones, and ectopic exchange events occur primarily during meiotic recombination, ectopic exchange is likely to be a key factor responsible for the small genome size of birds.
Acknowledgments
We thank Pilar Junier, Friederike Ettwig, the reviewers, and Deborah Charlesworth for valuable comments, which greatly improved the manuscript. G.A. was supported by a postdoctoral fellowship from the Alexander von Humboldt foundation and the Max Planck Society, and P.E.W. and G.A. were supported in part by National Institutes of Health grant RO1 HG02919.
References
- Abrusan, G., and H. J. Krambeck, 2006. The distribution of L1 and Alu retroelements in relation to GC content on human sex chromosomes is consistent with the ectopic recombination model. J. Mol. Evol. 63: 484–492. [DOI] [PubMed] [Google Scholar]
- Axelsson, E., N. G. C. Smith, H. Sundstrom, S. Berlin and H. Ellegren, 2004. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 21: 1538–1547. [DOI] [PubMed] [Google Scholar]
- Axelsson, E., M. T. Webster, N. G. C. Smith, D. W. Burt and H. Ellegren, 2005. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 15: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. J., and H. A. Wichman, 1990. Retrotransposon Mys is concentrated on the sex-chromosomes: implications for copy number containment. Evolution 44: 2083–2088. [DOI] [PubMed] [Google Scholar]
- Bartolome, C., X. Maside and B. Charlesworth, 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926–937. [DOI] [PubMed] [Google Scholar]
- Batzer, M. A., and P. L. Deininger, 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. [DOI] [PubMed] [Google Scholar]
- Baverstock, P. R., M. Adams, R. W. Polkinghorne and M. Gelder, 1982. A sex-linked enzyme in birds: Z-chromosome conservation but no dosage compensation. Nature 296: 763–766. [DOI] [PubMed] [Google Scholar]
- Belle, E. M. S., M. T. Webster and A. Eyre-Walker, 2005. Why are young and old repetitive elements distributed differently in the human genome? J. Mol. Evol. 60: 290–296. [DOI] [PubMed] [Google Scholar]
- Brookfield, J. F. Y., 2001. Selection on Alu sequences? Curr. Biol. 11: R900–R901. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., and T. Heidmann, 2005. L1-mediated retrotransposition of murine B1 and B2SINEs recapitulated in cultured cells. J. Mol. Biol. 349: 241–247. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., C. Esnault and T. Heidmann, 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., 2002. Dosage compensation: Do birds do it as well? Trends Genet. 18: 25–28. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., and A. Carmichael, 2001. Multiple and independent cessation of recombination between avian sex chromosomes. Genetics 158: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., 1993. Recombination and mammalian genome evolution. Proc. R. Soc. Lond. Ser. B Biol. Sci. 252: 237–243. [DOI] [PubMed] [Google Scholar]
- Furano, A. V., 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons, pp. 255–294 in Progress in Nucleic Acid Research and Molecular Biology, Vol. 64, edited by K. Moldave. Elsevier, Amsterdam/New York. [DOI] [PubMed]
- Giordano, J., Y. Ge, Y. Gelfand, G. Abrusán, G. Benson and P. E. Warburton, 2007. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput. Biol. 3(7): e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg, M., P. Bernaola-Galvan, P. Carpena and J. L. Oliver, 2005. The biased distribution of alus in human isochores might be driven by recombination. J. Mol. Evol. 60: 365–377. [DOI] [PubMed] [Google Scholar]
- Handley, L. L., H. Ceplitis and H. Ellegren, 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. L., and H. Piontkivska, 2005. DNA repeat arrays in chicken and human genomes and the adaptive evolution of avian genome size. BMC Evol. Biol. 5: 12. [DOI] [PMC free article] [PubMed]
- International Chicken Genome Sequencing Consortium, 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- Jurka, J., 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 94: 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, M., K. Ohshima and N. Okada, 1997. Determination of the entire sequence of turtle CR1: the first open reading frame of the turtle CR1 element encodes a protein with a novel zinc finger motif. Mol. Biol. Evol. 14: 1206–1217. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., R. Baertsch, M. Diekhans, T. S. Furey, A. Hinrichs et al., 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Kuroda, Y., N. Arai, M. Arita, M. Teranishi, T. Hori et al., 2001. Absence of Z-chromosome inactivation for five genes in male chickens. Chromosome Res. 9: 457–468. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Langley, C. H., E. Montgomery, R. Hudson, N. Kaplan and B. Charlesworth, 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet. Res. 52: 223–235. [DOI] [PubMed] [Google Scholar]
- Lobachev, K. S., J. E. Stenger, O. G. Kozyreva, J. Jurka, D. A. Gordenin et al., 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19: 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, M. F., 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80: 133–137. [DOI] [PubMed] [Google Scholar]
- Marais, G., 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19: 330–338. [DOI] [PubMed] [Google Scholar]
- Margulies, E. H., V. V. B. Maduro, P. J. Thomas, J. P. Tomkins, C. T. Amemiya et al., 2005. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc. Natl. Acad. Sci. USA 102: 3354–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen, H. A., D. McBride, G. Miele, A. P. Bird and M. Clinton, 2001. Dosage compensation in birds. Curr. Biol. 11: 253–257. [DOI] [PubMed] [Google Scholar]
- Meunier, J., and L. Duret, 2004. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 21: 984–990. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Secquencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Myers, S., L. Bottolo, C. Freeman, G. McVean and P. Donnelly, 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Pavlicek, A., K. Jabbari, J. Paces, V. Paces, J. Hejnar et al., 2001. Similar integration but different stability of Alus and LINEs in the human genome. Gene 276: 39–45. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61: 531–544. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl and K. L. Shaw, 2000. Evidence for DNA loss as a determinant of genome size. Science 287: 1060–1062. [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A., G. Toth, A. D. Riggs and J. Jurka, 1995. Ancestral, mammalian-wide subfamilies of line-1 repetitive sequences. J. Mol. Biol. 246: 401–417. [DOI] [PubMed] [Google Scholar]
- Soriano, P., M. Meunierrotival and G. Bernardi, 1983. The distribution of interspersed repeats is nonuniform and conserved in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 80: 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, J. E., K. S. Lobachev, D. Gordenin, T. A. Darden, J. Jurka et al., 2001. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 11: 12–27. [DOI] [PubMed] [Google Scholar]
- Vandergon, T. L., and M. Reitman, 1994. Evolution of chicken repeat-1 (Cr-1) elements: evidence for ancient subfamilies and multiple progenitors. Mol. Biol. Evol. 11: 886–898. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., N. G. C. Smith, L. Hultin-Rosenberg, P. F. Arndt and H. Ellegren, 2005. Male-driven biased gene conversion governs the evolution of base composition in human Alu repeats. Mol. Biol. Evol. 22: 1468–1474. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., E. Axelsson and H. Ellegren, 2006. Strong regional biases in nucleotide substitution in the chicken genome. Mol. Biol. Evol. 23: 1203–1216. [DOI] [PubMed] [Google Scholar]
- Wicker, T., J. S. Robertson, S. R. Schulze, F. A. Feltus, V. Magrini et al., 2005. The repetitive landscape of the chicken genome. Genome Res. 15: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., A. F. Smit, S. Schwartz, F. Chiaromonte, K. M. Roskin et al., 2004. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res. 14: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]