Abstract
Asn-glycosylation is widespread not only in eukaryotes but also in archaea and some eubacteria. Oligosaccharyltransferase (OST) catalyzes the co-translational transfer of an oligosaccharide from a lipid donor to an asparagine residue in nascent polypeptide chains. Here, we report that a thermophilic archaeon, Pyrococcus furiosus OST is composed of the STT3 protein alone, and catalyzes the transfer of a heptasaccharide, containing one hexouronate and two pentose residues, onto peptides in an Asn-X-Thr/Ser-motif-dependent manner. We also determined the 2.7-Å resolution crystal structure of the C-terminal soluble domain of Pyrococcus STT3. The structure-based multiple sequence alignment revealed a new motif, DxxK, which is adjacent to the well-conserved WWDYG motif in the tertiary structure. The mutagenesis of the DK motif residues in yeast STT3 revealed the essential role of the motif in the catalytic activity. The function of this motif may be related to the binding of the pyrophosphate group of lipid-linked oligosaccharide donors through a transiently bound cation. Our structure provides the first structural insights into the formation of the oligosaccharide–asparagine bond.
Keywords: crystal structure, N-glycosylation, oligosaccharide–asparagine bond, oligosaccharyltransferase, STT3
Introduction
Asparagine-linked glycosylation (N-glycosylation), the most ubiquitous protein modification, is essential for protein folding, oligomerization, quality control, sorting, and the transport of secretory and membrane proteins (Kukuruzinska and Lennon, 1998; Burda and Aebi, 1999; Hebert et al, 2005). In accordance, it is not surprising that N-glycosylation is widespread not only in eukaryotic organisms but also in archaeal and some eubacterial organisms (Szymanski et al, 2003; Upreti et al, 2003; Szymanski and Wren, 2005; Kelleher and Gilmore, 2006). The consensus motif of N-glycosylation (referred to as the sequon) is represented by Asn-X-Thr/Ser, where X can be any residue except Pro (Gavel and von Heijne, 1990), throughout all three domains of life. Statistical analyses revealed that just 60–65% of the potential sequons are occupied (Petrescu et al, 2004).
In eukaryotes, N-glycosylation is a highly coordinated and complex process that involves many glycotransferases and glycosidases in the lumen of the endoplasmic reticulum (ER) and in the lumina of the cis-, medial- and trans-Golgi vesicles (Kukuruzinska and Lennon, 1998; Burda and Aebi, 1999). The defining event in N-glycosylation is clearly the formation of the oligosaccharide–amino-acid bond, which is catalyzed by oligosaccharyltransferase (OST) (Knauer and Lehle, 1999; Yan and Lennarz, 2005; Kelleher and Gilmore, 2006). The OST transfers oligosaccharides onto asparagine residues in the consensus sequon in polypeptide chains. The oligosaccharide chain is preassembled on a lipid carrier and consists of 14 sugar residues in higher eukaryotes, and is transferred en bloc to a polypeptide by OST. In contrast, threonine/serine-linked (O-linked) sugars are added one at time, and each sugar transfer is catalyzed by a different enzyme (Spiro, 2002; Wopereis et al, 2006). N-Oligosaccharyl transfer occurs co-translationally on the lumenal side of ER, as the nascent polypeptide chain emerges from the translocon in the ER membrane in eukaryotic cells (Chavan and Lennarz, 2006). Exceptionally, in the eubacterium Campylobacter jejuni, N-oligosaccharyl transfer can occur independently of the protein translocation across the membranes (Kowarik et al, 2006).
The OST-catalyzed reaction is quite unique with respect to the substrate recognition. Since the reaction is co-translational, the active site of OST must recognize glycosylatable sequons in rapidly growing polypeptide chains efficiently, and ensure the rapid product discharge from the active site. It is obvious that the secondary and tertiary structures around a glycosylation site in the final, folded structure will have no direct effect on whether that site is occupied, but the local conformation of the unfolded polypeptide chain in the active site has a decisive role in determining whether a sequon will be modified (Knauer and Lehle, 1999). In this sense, although genetic and biochemical studies have provided considerable information about the OST enzyme, structural information about the active site of OST is essential for understanding the mechanism of Asn-glycosylation.
The OST enzyme is a multi-subunit protein complex residing in the ER membrane in eukaryotic cells. In Saccharomyces cerevisiae (yeast), the OST complex consists of eight subunits, STT3, OST1, OST2, OST3 or OST6, OST4, OST5, WBP1, and SWP1, which all contain one or more predicted transmembrane segments (Knauer and Lehle, 1999). OST3 and OST6 are homologous, interchangeable subunits. Recently, the yeast OST was reported to be a dimer of the octamer when solubilized in digitonin (Chavan et al, 2006), but other previous studies showed that it is a monomer of the octamer (Kelleher and Gilmore, 1994; Karaoglu et al, 2001). Among them, STT3 is the only conserved subunit in the three domains of life (Zufferey et al, 1995). In accordance, STT3 has been shown to be the catalytic subunit of the OST complex from yeast (Yan and Lennarz, 2002; Nilsson et al, 2003; Karamyshev et al, 2005) and C. jejuni (Wacker et al, 2002; Glover et al, 2005). The Campylobacter OST consists of the STT3 protein alone, and can function within Escherichia coli cells (Feldman et al, 2005).
The oligosaccharide donor is a dolichol pyrophosphate-linked oligosaccharide (OS-PP-Dol) in eukaryotes: Glc3Man9GlcNAc2-PP-Dol for most eukaryotes, but similar sugar lipids with smaller oligosaccharides are used by lower eukaryotes (Samuelson et al, 2005). The oligosaccharide donor in C. jejuni is GalNAc2GlcGalNAc3Bac-PP-Und, where Bac and Und represent bacillosamine (2,4-diacetoamido-2,4,6-trideoxyglucopyranose) and undecaprenol, respectively (Wacker et al, 2002; Young et al, 2002).
Although the prokaryal type of N-glycosylation was first described in archaea (Mescher and Strominger, 1976; Lechner and Sumper, 1987), little is known about N-glycosylation in this form of life. Recently, the inactivation of the stt3 gene (alias, aglB) in an archaeon, Methanococcus voltae, resulted in the underglycosylation of flagellin and S-layer proteins (Chaban et al, 2006). This is the first experimental evidence for the involvement of the STT3 protein in the archaeal glycosylation process. Both dolichol-phosphate- and dolichol-pyrophosphate-linked oligosaccharides were detected in extracts of archaeal cells (Mescher et al, 1976; Lechner et al, 1985; Lechner and Wieland, 1989; Kuntz et al, 1997). The glycan structure of the oligosaccharide donors is highly diverse, considering the huge variety of the N-glycan structures on asparagine residues (Schaffer and Messner, 2004; Eichler and Adams, 2005). Thus, the chemical structure of the oligosaccharide donor is quite variable in archaea.
In the present study, we selected the thermophilic archaeon, Pyrococcus furiosus, as a source of OST. First, we showed that the STT3 protein is the catalytic subunit of P. furiosus OST, as in eukaryotes and bacteria. This experimental proof is important, because of the low sequence homology. We also studied the sequon sequences for N-glycosylation and the glycan structure using the purified OST/STT3 enzyme. We further determined the crystal structure of the C-terminal soluble domain of P. furiosus STT3. So far, only the NMR structure of a 36-residue OST4, a non-essential subunit of yeast OST, has been reported (Zubkov et al, 2004). Therefore, this is the first structure of the catalytic subunit of OST. With reference to the structure, we have identified a new conserved motif (DK motif, Asp-X-X-Lys), which is located close to the known WWDYG (Trp-Trp-Asp-Tyr-Gly) motif in the tertiary structure. Unfortunately, a mutagenesis study of the Pyrococcus OST/STT3 enzyme was not possible, because the soluble domain expressed in E. coli cells for the structure determination lacked activity. Instead, a mutagenesis study of yeast STT3 confirmed the important role of the new motif in the OST reaction. Our results provide the first structural basis for the occurrence of Asn-glycosylation.
Results
Immunoaffinity purification of OST from P. furiosus cells
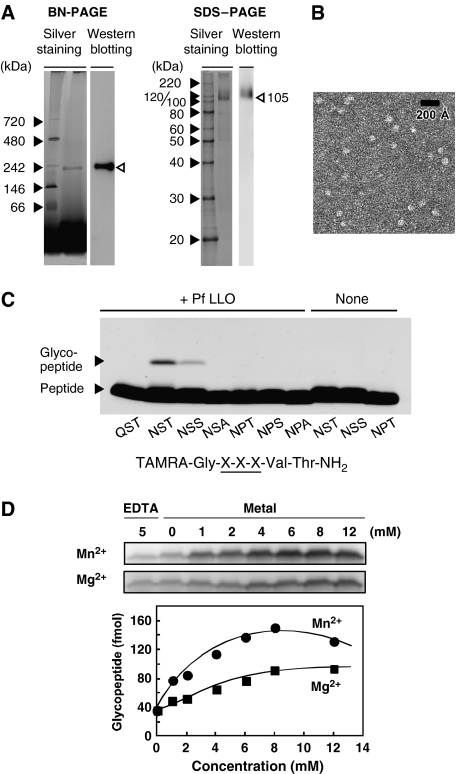
In a previous study, we detected OST activity in a Triton X-100-solubilized membrane fraction from P. furiosus cells (Kohda et al, 2007). Here, we performed one-step immunoaffinity purification of the OST enzyme from the membrane fraction, using an anti-P. furiosus STT3 polyclonal antibody. The purified protein appears as a single band both on a Blue Native (BN)–polyacrylamide gel electrophoresis (PAGE) gel and on an SDS–PAGE gel, by silver staining and immunoblotting (Figure 1A). The comparison of the apparent molecular weight, 230 kDa on BN-PAGE and 105 kDa on SDS–PAGE using soluble protein standards, might suggest the dimer formation, but the uncertain contribution of bound detergents may account for the slow migration in BN-PAGE despite the monomeric structure. A further study is necessary with respect to the quaternary structure of P. furiosus STT3. The band on the BN-PAGE gel was excised from the gel, and the protein was extracted. The detection of OST activity (data not shown) indicated that P. furiosus OST was composed of STT3 alone in the presence of Triton X-100 micelles, and that the other protein subunits were not necessary for the OST activity. It was possible that the other subunits were lost under the acidic elution conditions employed during the immunoaffinity purification, but the apparent molecular weight on the BN-PAGE gel did not change before and after the immunopurification (Supplementary Figure 1), suggesting no tightly bound subunit other than STT3 in the OST. The purified OST/STT3 appeared to be homogenous in an electron micrograph after uranyl acetate staining, with well-separated, individual particles of similar size (Figure 1B). The estimated diameter of the roughly spherical particles is ca. 80 Å.
Figure 1.
Oligosaccharyltransferase purified from Pyrococcus furiosus cells. (A) BN-PAGE and SDS–PAGE analyses of OST purified from P. furiosus cells by immunoaffinity, using an anti-P. furiosus sSTT3 antibody. Proteins on the gels were visualized by silver staining and western blotting, using an anti-P. furiosus sSTT3 antibody as the primary antibody. (B) Electron micrograph of the OST enzyme embedded in uranyl acetate stain. The purified STT3 protein in Triton X-100 micelles appeared as individual particles. Scale bar, 200 Å. (C) Effects of the amino-acid substitutions of the N-glycosylation sequon on the N-oligosaccharyltransferase reaction. The reaction mixtures were incubated for 16 h at 65°C in the presence and absence of P. furiosus LLO. (D) Metal ion dependence of the reaction and plot of the glycopeptide formation as a function of the final concentration of ions. The quantity of the glycopeptide was estimated using a TAMRA-labeled 22-residue peptide as an external standard, as described in Kohda et al (2007). The reaction mixtures were incubated for 1 h at 65°C.
We also examined the requirement of the canonical Asn-X-Thr/Ser motif for the OST reaction. Figure 1C shows the results of the OST assay using our new assay method, which uses fluorescent dye-labeled peptides as a substrate, and SDS–PAGE as the separation mechanism (Kohda et al, 2007). Previously, we showed the necessity of the N-glycosylation sequon, using a crude membrane fraction (Kohda et al, 2007). Here, we assayed the OST activity using the purified STT3 in the presence and absence of lipid-linked oligosaccharide (LLO), prepared from P. furiosus cells, as an oligosaccharide donor. A single product band was observed when the peptide substrate contained the Asn-X-Thr or Asn-X-Ser sequence, in the presence of LLO. The Asn-X-Thr sequence is a better substrate than the Asn-X-Ser sequence. The replacement of the first residue, Asn, with Gln, and that of the third residue, Thr/Ser, with Ala resulted in the disappearance of the product bands. The replacement of the second residue, Ser, with Pro also completely blocked the product formation. Thus, the amino-acid requirement is fully consistent with the N-glycosylation consensus. The addition of exogenous Mn2+ ions stimulated the formation of the glycopeptide product (Figure 1D). Mg2+ ions also had the same effect, but were less efficient. The addition of 5 mM EDTA in the reaction mixture slightly reduced the formation of the product, but did not completely inhibit the reaction, in contrast to yeast OST (Kohda et al, 2007).
Glycan structure of the OST reaction product
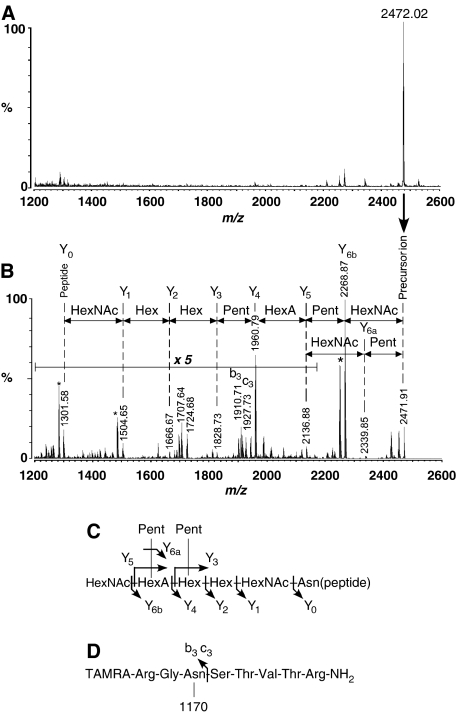
For the analysis of the N-glycan structure, the product of an overnight reaction was purified by reverse-phase HPLC chromatography. The product peptide eluted from the reverse-phase column as a single peak, faster than the substrate peptide. The MALDI-quadruple ion trap (QIT)-TOF MS spectrum showed a single peak of monoisotopic mass, 2472.0 Da (Figure 2A). MS/MS analysis of this ion provided a fragmentation series composed of sequential losses of N-acetylhexosamine (HexNAc, 203 Da), pentose (Pent, 132 Da), hexouronic acid (HexA, 176 Da), and hexose (Hex, 162 Da) (Figure 2B). The glycan structure is a single oligosaccharide chain composed of two HexNAc, two Hex, one HexA, and two Pent. The fragmentation pattern suggests a branched structure at the HexA residue (Figure 2C). The observation of the strong ion at m/z 1960.8 confirmed this branching form, because one fragmentation event at the HexA–Hex linkage generated this fragment ion. The second Pent attaches to the Hex residue next to the HexA residue, based on the MS/MS analysis of partially degraded glycopeptides formed by a crude membrane fraction, as the source of the OST enzyme (data not shown).
Figure 2.
MS/MS analysis of P. furiosus N-glycan structure. (A) MALDI-QIT-TOF MS spectrum of the OST reaction product. (B) MALDI-QIT-TOF MS/MS spectrum of the precursor ion at m/z 2472. The fragment ions originating from the sequential loss of oligosaccharide residues are indicated in the spectrum. Asterisks indicate other prominent peaks generated by dehydration (m/z 2250.9 and 1486.6) and the loss of ammonium (m/z 1284.6). (C) Estimated P. furiosus N-glycan structure, and its fragmentation pattern. (D) The observation of b3 and c3 fragment ions confirmed that the glycopeptide product is Asn-linked. TAMRA denotes carboxytetramethylrhodamine. ‘1170' represents the molecular mass of the attached oligosaccharide moiety.
The mass of the peptide portion of this glycopeptide is m/z 1301.6, which corresponds exactly to the mass of the unglycosylated TAMRA-labeled peptide. The observation of the b3 and c3 fragment ions at m/z 1910.7 and 1927.7, respectively, confirmed that the oligosaccharide is N-linked (Figure 2D). The linkage between the reducing terminal sugar residue and the asparagine residue was inferred not to be the eukaryotic type, GlcNAc-β-Asn, from the following reasons. (1) An ion resulting from the cross-ring cleavage of the innermost GlcNAc residue (Kuster et al, 1996), for which the m/z value would be the peptide ion (1301.6 Da) plus 83 Da, was not observed. (2) The glycopeptide bond was not cleaved by PNGase F, which cleaves between the innermost GlcNAc and asparagine residues of the eukaryotic type N-linked glycopeptides. (3) P. furiosus OST was unable to utilize LLO prepared from yeast, in place of LLO from P. furiosus. The P. furiosus glycopeptide linkage is probably GalNAc-Asn, as in Halobacterium salinarum (Lechner and Wieland, 1989; Messner, 1997).
Overall structure of the C-terminal soluble domain of P. furiosus STT3
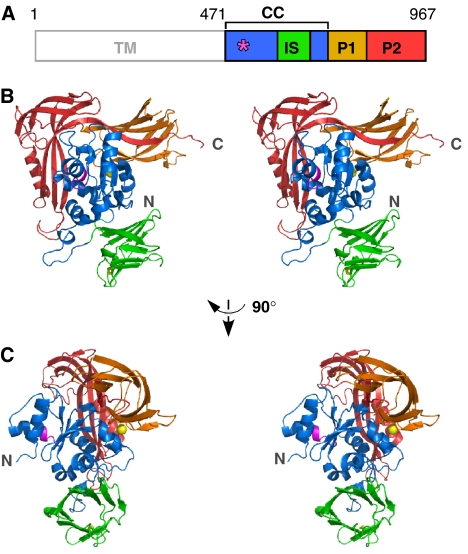
P. furiosus STT3 protein consists of 13 deduced transmembrane helices in the N-terminal half (470 residues) of the primary sequence, and a soluble domain in the C-terminal half (497 residues) (Figure 3A). We expressed, purified, and crystallized the C-terminal soluble domain of the STT3 protein (sSTT3) (Igura et al, 2007). The 2.7-Å resolution crystal structure reveals a compact, globular structure. The primary structure of the sSTT3 can be divided into four regions with reference to the tertiary structure (Figure 3A). The well-conserved WWDYG motif is contained in the central core (CC) domain. The CC domain mainly consists of α-helices (Figure 3B and C). Interestingly, a 10-stranded antiparallel β-barrel structure is inserted into the CC domain (Supplementary Figure 2), and thus we designated it as the IS (insert) domain. This 80-residue amino-acid sequence exists in the STT3s of the genus Pyrococcus and its closely related genus Thermococcus (Supplementary Figure 3). A PSI-BLAST search failed to find similar sequences in other organisms. The IS domain contains a disulfide bond, C638-C658, but the pair of cysteines is not conserved, even in Thermococcus (Supplementary Figure 3). The remaining amino-acid sequence forms two peripheral domains, P1 and P2. These two domains mainly consist of β-strands, and surround the CC domain.
Figure 3.
Crystal structure of the C-terminal soluble domain of STT3. (A) Domain structure of P. furiosus STT3. TM, transmembrane domain; CC, central core domain, residues 471–600+683–725; IS, insertion domain, residues 601–682; P1, peripheral domain 1, residues 726–821; P2, peripheral domain 2, residues 822–967. The position of the WWDYG motif is indicated by an asterisk. (B) Stereoview of the overall structure of the C-terminal soluble domain of STT3 (residues 471–967). The WWDYG motif is shown in magenta. The disulfide bond between C638 and C658 is shown as yellow sticks. A bound metal cation is shown as a yellow sphere. (C) Different view from (B).
A cation is coordinated to the backbone carbonyl groups of A759 and Y793, and to the side-chain carboxyl groups of E554 and E796, and it seems to stabilize the domain–domain interaction between CC and P1 (Supplementary Figure 4). We estimated that the bound cation was Ca2+ by an inductively coupled plasma (ICP)-MS measurement (See Supplementary Results). Despite stimulatory effects by divalent metal cations (Figure 1D), the bound ion does not appear to be directly involved in the enzymatic activity, because the position of the bound ion is distant from the putative catalytic site.
Structure-based identification of a new motif
The eukaryotic STT3 proteins share high sequence homologies over their entire primary sequences. For descriptive purposes, the STT3 sequence is divided into three segments. The M-region (95 residues) is defined as extending 20 residues toward the N terminus from the well-conserved WWDYG motif and 70 residues toward the C terminus (Supplementary Figure 5). The N- and C-regions are defined as the N- and C-terminal segments that flank the M-region. When yeast STT3 is compared with human STT3-A, the M-region has the highest similarity (amino-acid identity 74%). The N-region is 53% identical, and the C-region is 41% identical. The two paralogs of the human STT3 proteins, STT3-A and STT3-B, share similar high identity levels. In contrast, the archaeal and bacterial STT3s show very limited similarities to the eukaryotic STT3s. The M-region has the highest similarity again, but the identity is just 24–25% between yeast and P. furiosus, and between yeast and C. jejuni. The identities of the N-region are less than 20%, and no similarity is detected in the C-region. There are two STT3 paralogs in P. furiosus, STT3 (used in this study) and STT3-2 (STT3-related protein), which share similar low levels of identities.
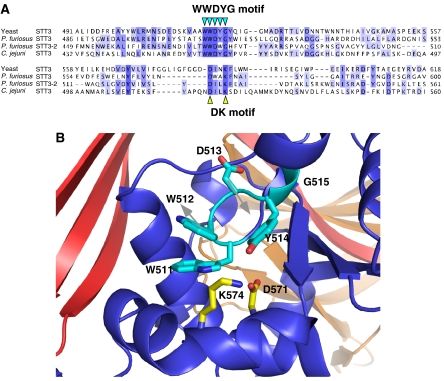
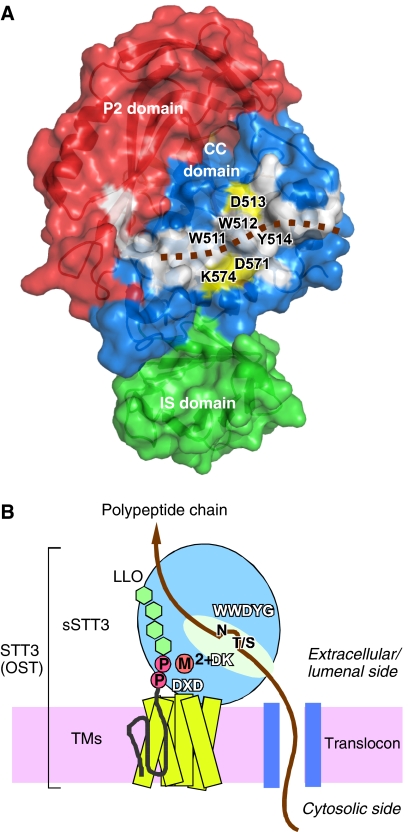
Despite the low sequence homologies, all the STT3 proteins from the three kingdoms conserve the WWDYG motif, in which the aspartate residue is thought to function as a catalytic residue. Thus, they should share a common catalytic mechanism, which prompted us to reexamine the multiple sequence alignment of the M-region, with reference to the tertiary structure of P. furiosus STT3 (Figure 4A). We expected that the low sequence homologies among the three domains of life would highlight the functionally essential residues. A short motif, DxxK, was identified, where x can be any residue. Both the aspartate and lysine residues are close to the WWDYG motif in the tertiary structure, suggesting that the DK motif comprises the active site of OST/STT3 with the WWDYG motif (Figure 4B).
Figure 4.
Putative active site of oligosaccharyltransferase comprising the two conserved motifs. (A) Sequence alignment of the region containing the known WWDYG motif and the newly found DK motif. An initial alignment was obtained with the Mafft algorithm (Katoh et al, 2005) in the program Jalview, and then was edited manually. (B) Close-up view of the putative active site, with the WWDYG motif in cyan (W511, W512, D513, Y514, and G515), and the DK motif in yellow (D571 and K574). Alternative possible side-chain directions of W512 and D513 in the absence of crystal packing effects are indicated by gray arrows.
The DK motif is functionally important in yeast STT3
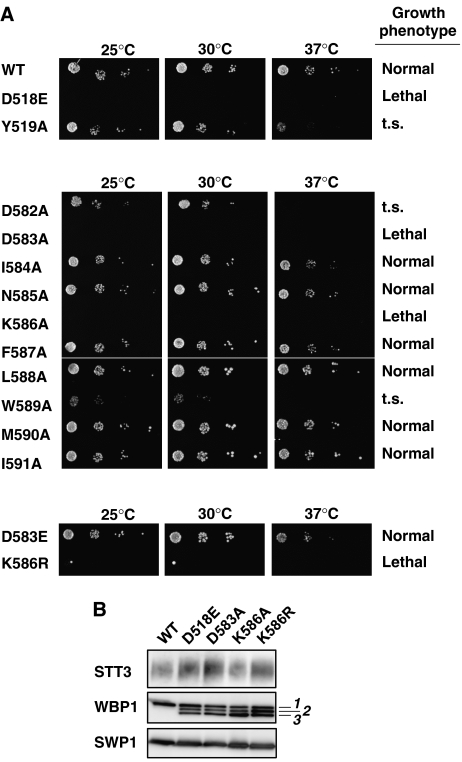
We did not detect the OST activity with the soluble domain of P. furiosus STT3. It is probable that the N-terminal TM region is necessary for the OST activity. Therefore, a mutagenesis study of the DK motif of yeast STT3 was carried out, using yeast cells. Mutagenesis of the essential STT3 gene is well established (Yan and Lennarz, 2002). We mutated a total of 10 consecutive residues in and near the DK motif, and some residues in the WWDYG motif as a control. All the mutant plasmids were generated using a 3 × HA-tagged STT3 construct, pRS313(His)-STT3HA. These mutants were transformed into a Δstt3 null strain containing wild-type STT3 on a URA3 plasmid, pRS316(Ura)-STT3. Yeast cells carrying both plasmids were patched on −His+FOA plates to delete the wild-type gene, and were incubated at 25, 30, and 37°C for 2 days (Figure 5A). The two mutants, D518E and Y519A, in the WWDYG motif displayed a lethal phenotype and a less significant defect, a temperature-sensitive (t.s.) growth phenotype, respectively. This result is consistent with the previous report (Yan and Lennarz, 2002). Cells carrying the DK-motif mutants, D583A and K586A, did not grow at any temperature tested, and thus were lethal. The mutations of D582A, a preceding residue of D583, and W589A each led to a t.s. phenotype. Yan and Lennarz showed that the I593A mutation also led to a t.s. phenotype (data summary in Supplementary Table 1). All the remaining alanine mutations showed a normal phenotype. The residues that resulted in lethal and t.s. phenotypes are spaced three or four residues apart in the primary sequence: DDINKFLWMIRI. Considering the fact that corresponding segment adopts consecutive 310 and α-helical conformations in P. furiosus STT3 (Supplementary Figure 3), these residues are positioned on the same side of a long helix and form a functional surface of yeast STT3. In the DK motif, a glutamate residue was tolerated at the aspartate position, but an arginine residue was unable to substitute for the lysine residue and resulted in a lethal phenotype (Figure 5A, lower panel).
Figure 5.
Spotting growth assay of yeast STT3 point mutations. (A) Growth phenotype of yeast strains carrying point mutations in yeast STT3. Cells carrying wild type (WT) and mutations were spotted on −His+FOA plates. The growth of the colonies at three different temperatures was compared after 2 days. ‘t.s.' stands for temperature sensitive. Upper panel, WT and two mutations in the WWDYG motif; middle panel, alanine-scanning mutations in and near the DK motif; lower panel, two non-alanine mutations in the DK motif. Note that there was a single large colony at both 25 and 30°C for the K586R mutant. We confirmed that they were revertants, and thus K586R is regarded as a lethal phenotype. (B) The incorporation of mutated STT3 into the yeast OST complex. HA-tagged STT3 mutants in a yeast cell lysate containing 0.15% digitonin were immunoprecipitated under non-denaturing conditions, using an anti-HA antibody. The absorbed proteins were resolved by SDS–PAGE, followed by western blotting using anti-yeast STT3, anti-yeast WBP1, and anti-yeast SWP1 antibodies. Note that the underglycosylation of WBP1 resulted in three bands (labeled as 1, 2, and 3), whereas SWP1 lacks N-glycosylation sequons. STT3 migrated diffusely probably due to its many TM segments, irrespective of glycosylation.
Next, we examined the incorporation of the mutant STT3 proteins that resulted in a lethal phenotype into the OST complex. We used yeast cells carrying both pRS313-STT3*HA (asterisk denotes a mutation) and pRS316-STT3 (this supports the cell viability). After the digitonin solubilization of the cell lysate, the HA-tagged STT3* mutant protein was immunoprecipitated using an anti-HA antibody, and the co-immunoprecipitation of essential subunits of yeast OST, WBP1 and SWP1, was examined by western blotting (Figure 5B). The OST complexes from all of the lethal mutants, D518E, D583A, K586A, and K586R, contained WBP1 and SWP1. This indicates that these four mutant STT3s form a stable OST complex that has seriously impaired OST activity, thus suggesting the functional importance of these residues.
Discussion
There are many potential Asn-glycosylation sites in an extracellular protein, but only about two-third of them are utilized. OST determines the occurrence of Asn-glycosylation. We purified the OST enzyme from a hyperthermophilic archaeon, P. furiosus, to homogeneity. P. furiosus OST in the presence of Triton X-100 is composed of the STT3 protein alone, and appears as a uniform particle in EM-negative staining, with a diameter of 80 Å (Figure 1A and B). In contrast, yeast OST solubilized in digitonin contains nine different polypeptides, including STT3 (Chavan et al, 2006). The larger size (120 Å diameter) of the particles in EM is consistent with the multi-subunit structure. The bacterial OST from C. jejuni also consists of the STT3 protein alone (Feldman et al, 2005). With respect to the subunit composition, the archaeal OST resembles the bacterial OST.
The purified P. furiosus STT3 protein, embedded in detergent micelles, can catalyze the OST reaction in vitro in the N-glycosylation sequon-dependent manner, in the presence of LLO and divalent metal cations (Figure 1C and D). This clearly demonstrates that the STT3 protein functions as the catalytic subunit of the OST enzyme in archaeal organisms, as in eukaryotes and bacteria. This confirmation is necessary because the sequence identities between the eukaryotic and archaeal STT3s are marginally meaningful (Supplementary Figure 5). The STT3 proteins are characterized by 11–13 transmembrane helices in the N-terminal half of the amino-acid sequence, and a globular domain in the C-terminal half (Kim et al, 2005). We prepared the C-terminal globular domain of P. furiosus STT3 as a recombinant protein in E. coli cells. The outstanding stability of the P. furiosus proteins permits heat treatment of the E. coli cell lysate, and further purification without any affinity tag.
The STT3 crystal structure, determined for the first time, is shown in Figure 3. The overall structure of the sSTT3 consists of four structural domains. The mainly α-helical CC domain is located at the center, and three β-sheet-rich domains, IS, P1, and P2, encircle the CC domain. The overall architecture of sSTT3 appears to be novel, as no equivalent fold was found in a DALI search (Holm and Sander, 1998). At the domain level, the P1 domain has an immunoglobulin (IG)-like β-sandwich fold, and the IS domain is weakly related to β-barrel folds. The IS domain intimately contacts the CC domain of another STT3 molecule in the crystal (Supplementary Figure 6). However, this dimer formation in the crystal does not seem to be biologically relevant, because sSTT3 behaves as a monomeric protein in gel filtration chromatography (Igura et al, 2007).
The mutations in the WWDYG motif in the CC domain abolished or substantially reduced the OST activity in yeast and C. jejuni STT3 (Yan and Lennarz, 2002; Glover et al, 2005). Thus, the WWDYG sequence is thought to be directly involved in the catalysis. We searched for the amino-acid residues that are spatially close to the WWDYG motif in the tertiary structure, and are conserved among the STT3 proteins in the three domains of life (Figure 4A). We found a pair of aspartate and lysine residues spaced three amino acids apart, which we refer to as the ‘DK motif'. The two residues reside on a kinked helix and may form a salt bridge (Figure 4B). We propose that the WWDYG and DK motifs constitute the active site of the OST/STT3 enzyme. Indeed, we showed that the mutations of the aspartate and lysine residues in the DK motif resulted in a lethal phenotype in yeast STT3 (Figure 5). This effect was very specific to the two residues, because other mutations in and near the DK motif did not cause lethality.
It should be noted that the conformations of the WWDYG and DK motifs are both unusual. An analysis with the program DSSP indicated that the N-terminal part of the WWDYG motif, W511WD513, adopts a rare left-handed helical conformation (Supplementary Figure 3 and the Ramachandran plot in Supplementary Figure 7). This unusual conformation is apparently stabilized by the crystal packing interaction in the crystallographic dimer (Supplementary Figure 8). Without the crystal packing effects, the side chain of D513 would face inwardly, as the backbone conformation took a usual right-handed helix (Figure 4B, gray arrows). The DK motif, L569NDWAK574, forms an unusually long, six-residue 310 helix. We postulate that these intrinsically unstable conformations in the crystal reflect some flexibility in the active site, and hence facilitate the large conformational changes that are functionally coupled with the OST reaction cycle.
There are two proposed catalytic mechanisms for the OST reaction (Bause and Legler, 1981; Imperiali et al, 1992). Both models assume the existence of a specific turn conformation of the Asn-X-Thr/Ser sequon. The complete inactivation of the OST enzyme by subtle amino-acid substitutions, such as aspartate to glutamate in the WWDYG motif (Yan and Lennarz, 2002; Glover et al, 2005), and threonine to allothreonine in the sequon (Bause et al, 1995), demonstrates the strict geometric requirement of the turn structure intermediately formed in the active site. The side-chain carboxylate group of the central aspartate in the WWDYG motif probably functions as a catalytic base during the glycosylation reaction. We suggest that the DK motif is the binding site for the pyrophosphate group of LLO, through a transiently bound Mn2+ or Mg2+.
We have not detected the OST activity with the C-terminal soluble domain of P. furiosus STT3, which was used for the structure determination, despite many trials. For example, we used high concentrations of sSTT3 to compensate the possible low activity, and added various detergents to mix the soluble protein and hydrophobic LLO. The inactive sSTT3 implies the direct involvement of the TM region in the OST reaction. Recently, STT3 was shown to be distantly related to a mannosyltransferase, which catalyzes the O-mannosylation of proteins using D-mannose-P-dolichol as a sugar donor, and was classified to the glycosyltransferase GT-C superfamily (A Bateman, http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PMT). The GT-C superfamily contains diverse glycosyltransferases that possess 8–13 predicted TM segments, and displays a DXD motif in the first extracellular/lumenal loop (Liu and Mushegian, 2003). In fact, the Pyrococcus STT3 proteins possess a DPD sequence just after the first TM helix (Supplementary Figure 3). In eukaryotic STT3s, an EXD sequence was found instead of DXD. The role of this motif is unclear, but it may be related to the binding of dolichyl-(pyro)phosphate, in a coordinated or sequential manner with the DK motif.
The visual inspection of the STT3 structure revealed a large, elongated hydrophobic patch mainly on the surface of the CC domain (Figure 6A). The direct contact of STT3 with a nascent polypeptide chain was revealed by a photo-crosslinking experiment, using dog pancreas STT3 (Nilsson et al, 2003). We postulate that the hydrophobic patch interacts with polypeptides. The putative active site residues are located at the center of the hydrophobic patch. The overall structure and the location of the conserved motifs in the full-length STT3, and the putative substrate-binding modes are schematically summarized in Figure 6B.
Figure 6.
Large hydrophobic surface and schematic of the overall structure of STT3. (A) Surface representation of the C-terminal soluble domain of P. furiosus STT3, colored according to the domain structure in Figure 3. The P1 domain is behind the molecule, and is not visible. A continuous, hydrophobic surface (white) consists of residues W491, W511, W512, Y514, G515, Y516, W517, I518, L522, L523, Y568, A577, I578, Y580, L581, and Y590 in the CC domain, and L872, F887, and I939 in the P2 domain. The brown dotted line indicates a hypothetical binding mode of a substrate polypeptide chain. Note that the putative catalytic amino-acid residues, D513 in the WWDYG motif, and D571 and K574 in the DK motif, shown in yellow, line both sides of the hydrophobic patch. (B) The overall structure of the full-length STT3, and the location of the three conserved motifs, WWDYG, DK, and DXD. A possible binding mode of a nascent polypeptide chain emerged from a translocon, and that of an oligosaccharide-PP-dolichol/undecaprenol are shown. The aspartate in the WWDYG motif functions as a catalytic base. The DK and DXD motifs are the binding sites for the pyrophosphate group of lipid-linked oligosaccharide donor, through a transiently bound Mn2+ or Mg2+, in a coordinated or sequential manner.
To elucidate the precise catalytic mechanism of the OST reaction, high-resolution crystal structures of the STT3 protein in complexes with substrates or their analogs are needed. A simple co-crystallization or soaking of OST/STT3 with polypeptide substrates will not work, because the sequons are transiently recognized during the OST reaction. The expression and crystallization of the full-length STT3 protein are also desired for the preparation of complexes with LLOs. The preliminary characterization of the glycan structure (Figure 2) is the first step for the appropriate design of inhibitors, suicide substrates, or transient-state analogs for future work.
Materials and methods
Polyclonal antibodies
The C-terminal soluble domain (residues 471–967) of P. furiosus STT3 (sSTT3) was produced in E. coli cells at 37°C as inclusion bodies (Igura et al, 2007). The rabbit polyclonal antibody against sSTT3 was raised using the inclusion bodies dissolved in 8 M urea (1 mg protein/ml). The Ig fraction was isolated from the antiserum by chromatography on protein G-Sepharose (GE Healthcare), and was stored at 10 mg/ml in PBS containing 0.1% NaN3. The soluble domains of yeast STT3 (residues 485–718), WBP1 (residues 21–392 as an N-terminally His-tagged protein), and SWP1 (residues 20–193 as an N-terminally His-tagged protein) were all expressed in inclusion bodies, and were used to raise rabbit polyclonal antibodies.
Affinity purification from the membrane fraction of P. furiosus cells
A Triton X-100-solubilized membrane fraction from 1 g of P. furiosus wet cells was obtained, as described in Kohda et al (2007). The rabbit polyclonal antibody against sSTT3 was immobilized to the Protein A gel, using the crosslinker DSS (PIERCE). A 2-ml aliquot of the membrane fraction was mixed with 500 μl of 50% antibody-protein A-gel slurry in 20 mM Tris–HCl, pH 7.5, containing 0.5 M NaCl and 0.05% Triton X-100. After the resin was washed five times in the same buffer, the bound materials were eluted by 0.3 ml of ImmunoPure IgG elution buffer (pH 2.8, PIERCE) plus 0.05% Triton X-100, and were neutralized immediately by the addition of 30 μl of 1 M Tris–HCl, pH 8.0. The elution was repeated two times, and the eluted fractions were combined. After centrifugation, the supernatant was kept at 4°C. The OST assay was performed as described in Kohda et al (2007). LLOs were isolated from P. furiosus cells, as described for yeast LLO (Kelleher et al, 2001). See Supplementary Materials and methods for details.
MS/MS analysis of the product of the OST assay
The OST reaction was carried out on a large scale. After an incubation at 65°C for 18 h, the reaction was stopped by mixing with an equal volume of 0.1% TFA, followed by centrifugation to remove insoluble materials. The supernatant was immediately subjected to reverse-phase HPLC for the separation of the product from the substrate peptide (TAMRA-Arg-Gly-Asn-Ser-Thr-Val-Thr-Arg-NH2, monoisotopic mass: 1301.6 Da) with a linear gradient of acetonitrile in 0.1% TFA. The magenta color of the TAMRA fluorescent dye (558 nm) facilitated the identification of the product. The glycopeptide product eluted first, and was collected, dried in a SpeedVac concentrator, and dissolved in water at 0.5 pmol/μl. The matrix used was DHB (2,5-dihydroxybenzoic acid, Shimadzu GLC) dissolved at 10 mg/ml in 0.1% TFA:acetonitrile (6:4, v/v). Equal volumes (0.5 μl each) of the sample and the matrix solution were mixed on a target plate and dried. MS and MS/MS spectra were acquired in the positive ion reflection mode, using a MALDI-QIT-TOF mass spectrometer (AXIMA QIT, Shimadzu). We detected no degradation of the peptide by contaminant proteases, or deamination of the asparagine/C-terminal amide groups during the incubation.
Structure determination
The C-terminal soluble domain (residues 471–967) of the STT3 protein (sSTT3) from P. furiosus (accession no. PF0156, Q8U4D2_PYRFU) was expressed using E. coli cells at 16°C, purified from the supernatant fraction, and crystallized, as described in Igura et al (2007). Briefly, the cell lysate was heated at 80°C. The thermostable sSTT3 remained in the solution, but the majority of the E. coli proteins were precipitated. After centrifugation, the sSTT3 was recovered in the supernatant, and was further purified by anion-exchange and gel-filtration chromatography. The native and selenomethionyl (SeMet) derivative crystals grew from a hanging drop with a 1:1 volume ratio (total volume, 1.4 μl) of the protein stock (10 mg/ml, 10 mM Tris–HCl, pH 8.0) and the reservoir solution (0.1 M Tris–HCl, pH 9.0, 22% PEG4000, 0.3 M sodium acetate) at 303 K over 1–3 weeks. The crystals were cryoprotected by increasing the concentration of PEG4000 up to 24%. Data were collected at beamline BL41XU at SPring8 (Harima, Japan). Data were processed with HKL2000 (Otwinowski and Minor, 1997) up to the resolutions of 2.7 and 3.0 Å for the native and SeMet crystals, respectively, and yielded the space group P21212 for the native and SeMet crystals. Both structures contained four molecules in the asymmetric unit cell (59% solvent, VM=3.0 Å3/Da). A site search of the selenium atoms was carried out using the SAD phasing method with SOLVE (Terwilliger and Berendzen, 1999), resulting in 16 out of 20 Se sites, assuming four SeMet from each molecule. Phases were further improved by density modification methods using autoSHARP (Vonrhein et al, 2006). Refinement of the native and SeMet data sets was performed using the program CNS (Brünger et al, 1998), and manual rebuilding was carried out with the program O (Jones et al, 1991). The final R factors obtained were Rwork/Rfree 22.7/27.3% for the native structure, and 22.0/27.2% for the SeMet structure. The statistics of data collection and refinement are summarized in Table I. The four structures in the asymmetric unit are similar, with Cα rmsd values in the range of 0.5–0.8 Å. The N-terminal 6–7 residues and the C-terminal 0–3 residues were disordered. Residues 523–533 were also disordered in the final native structure, and were not included. The stereochemistry of the final coordinates was checked with PROCHECK (Laskowski et al, 1993) in the CCP4 suite. The figures were generated with the program PyMOL (http://pymol.sourceforge.net/), using the coordinates of the native structure supplemented with those of residues 523–533 of the SeMet structure.
Table 1.
Data collection and refinement statisticsa
| SeMet | Native | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P21212 |
| Cell dimensions, a, b, c (Å) | 137.34, 266.60, 73.95 | 136.93, 265.29, 74.13 |
| Molecules per AU | 4 | 4 |
| Wavelength (Å) | 0.97915 (peak) | 0.96419 |
| Resolution range (Å) | 30–3.0 (3.1–3.0) | 40–2.7 (2.8–2.7) |
| No. of measured reflections | 610 027 | 458 643 |
| No. of unique reflections | 55 387 (5441) | 75 226 (7371) |
| Completeness (%) | 100.0 (100.0) | 99.9 (99.4) |
| Multiplicity | 11.0 (11.1) | 6.1 (5.8) |
| Average I/σ(I) | 7.4 (3.8) | 12.4 (3.5) |
| Rmergeb | 0.136 (0.598) | 0.081 (0.443) |
| Mosaicity (deg) | 0.254 | 0.261 |
| Refinement | ||
| Resolution range (Å) | 20.0–3.0 | 20.0–2.7 |
| No. of reflections (work/test) | 51 210/3916 | 71 091/3805 |
| Rwork/Rfreec | 22.0/27.2 | 22.7/27.3 |
| B-factors (Å2) (protein/solvent) | 47.6/50.1 | 44.0/31.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.0082 | 0.0072 |
| Bond angles (deg) | 1.50 | 1.39 |
| Ramachandran plot | ||
| Most favored (%) | 80.5 | 84.1 |
| Additionally allowed (%) | 17.0 | 13.9 |
| Generously allowed (%) | 1.6 | 1.5 |
| Disallowed (%) | 1.0 | 0.5 |
| PDB entry | 2ZAG | 2ZAI |
| Values in parentheses refer to data in the highest resolution shell in each data set. | ||
| aData collection statistics were reported previously (Igura et al, 2007). | ||
| bRmerge= ∑h∑j∣〈I〉h−Ih,j∣/∑h∑i Ih,j, where 〈I〉h is the mean intensity of symmetry-equivalent reflections. | ||
| cRwork=∑h∣∣Fo∣−∣Fc∣∣/∑h∣Fo∣, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively, for reflection h. | ||
Other analytical methods
BN-PAGE was carried out using NativePAGE 4–16% Bis-Tris Gels (Invitrogen). SDS–PAGE was carried out using homogeneous gels (7.5%) and gradient gels (10–20%) (Daiichi or ATTO). The proteins in the gels were detected by silver staining (Daiichi), or were transferred to Immobilon-P PVDF membranes (Millipore), probed with the indicated antibodies, and visualized by ECL Plus or ECL Advance kits (GE Healthcare). The metal content of the protein samples was analyzed by ICP-MS. See Supplementary Materials and methods for details. For negative staining in electron microscopy, a 5-μl drop of the OST sample, containing 0.2% Triton X-100, was applied to a glow-discharged 300-mesh copper grid covered with a thin layer of carbon film. After removing the excess solution by blotting with filter paper, the grid was washed with a 10-μl drop of water. After removing the water by blotting, the grid was stained with a 5-μl drop of a 2% (w/v) uranyl acetate solution. Excess stain was removed by blotting, and the grids were air-dried. The grids were then coated by vacuum deposition of carbon. Images were recorded in a JEOL JEM 2000EX electron microscope operating at 80 kV, with a magnification of 50 000 and under defocus of 65 nm. Spotting growth assay of yeast STT3 mutants, and non-denaturing immunoprecipitation for testing the integration of the STT3 mutants into the yeast OST complex were performed, as described in Yan and Lennarz (2002). See Supplementary Materials and methods for details.
Accession numbers
The atomic coordinates of the native and SeMet derivative of the C-terminal soluble domain of P. furiosus STT3 have been deposited in the Protein Data Bank, with the accession codes 2ZAI and 2ZAG, respectively.
Supplementary Material
Supplementary Figure 1
Supplementary Information
Acknowledgments
We thank Dr Yoshizumi Ishino (Kyushu University) for providing the P. furiosus cells, Mr Masafumi Sasaki (Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University) and Dr Kota Mayanagi (Nagahama Institute of Bioscience and Technology and JST-BIRD) for the electron microscopy measurements, and Dr Midori Watanabe (the Center of Advanced Instrumental Analysis, Kyushu University) for the ICP-MS measurements. We thank the staff at both the SPring8 beamlines BL38B1 and BL41XU, and the Photon Factory beamlines BL-5A and AR-NW12A. The experiments at SPring8 were approved by the Japan Synchrotron Radiation Research Institute (JASRI), as proposals 2005A0877, 2005B0157, 2006A1739, and 2007A1204. The experiments at the Photon Factory were approved by the High Energy Accelerator Research Organization (KEK), as a proposal 2007G208. KM and DK were supported by Grants-in-Aid for Scientific Research in Priority Areas and the National Project on Targeted Protein Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Bause E, Breuer W, Peters S (1995) Investigation of the active site of oligosaccharyltransferase from pig liver using synthetic tripeptides as tools. Biochem J 312 (Pt 3): 979–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E, Legler G (1981) The role of the hydroxy amino acid in the triplet sequence Asn-Xaa-Thr(Ser) for the N-glycosylation step during glycoprotein biosynthesis. Biochem J 195: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M (1999) The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta 1426: 239–257 [DOI] [PubMed] [Google Scholar]
- Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF (2006) Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol 61: 259–268 [DOI] [PubMed] [Google Scholar]
- Chavan M, Chen Z, Li G, Schindelin H, Lennarz WJ, Li H (2006) Dimeric organization of the yeast oligosaccharyl transferase complex. Proc Natl Acad Sci USA 103: 8947–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan M, Lennarz W (2006) The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci 31: 17–20 [DOI] [PubMed] [Google Scholar]
- Eichler J, Adams MW (2005) Posttranslational protein modification in Archaea. Microbiol Mol Biol Rev 69: 393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M (2005) Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA 102: 3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng 3: 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover KJ, Weerapana E, Numao S, Imperiali B (2005) Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem Biol 12: 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M (2005) The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol 15: 364–370 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res 26: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igura M, Maita N, Obita T, Kamishikiryo J, Maenaka K, Kohda D (2007) Purification, crystallization, and preliminary X-ray diffraction studies of the soluble domain of the oligosaccharyltransferase STT3 subunit from the thermophilic archaeon Pyrococcus furiosus. Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiali B, Shannon KL, Rickert KW (1992) Role of peptide conformation in asparagine-linked glycosylation. J Am Chem Soc 114: 7942–7944 [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Karamyshev AL, Kelleher DJ, Gilmore R, Johnson AE, von Heijne G, Nilsson I (2005) Mapping the interaction of the STT3 subunit of the oligosaccharyl transferase complex with nascent polypeptide chains. J Biol Chem 280: 40489–40493 [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R (2001) Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase. Biochemistry 40: 12193–12206 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R (1994) The Saccharomyces cerevisiae oligosaccharyltransferase is a protein complex composed of Wbp1, Swp1, and four additional polypeptides. J Biol Chem 269: 12908–12917 [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Karaoglu D, Gilmore R (2001) Large-scale isolation of dolichol-linked oligosaccharides with homogeneous oligosaccharide structures: determination of steady-state dolichol-linked oligosaccharide compositions. Glycobiology 11: 321–333 [DOI] [PubMed] [Google Scholar]
- Kim H, von Heijne G, Nilsson I (2005) Membrane topology of the STT3 subunit of the oligosaccharyl transferase complex. J Biol Chem 280: 20261–20267 [DOI] [PubMed] [Google Scholar]
- Knauer R, Lehle L (1999) The oligosaccharyltransferase complex from yeast. Biochim Biophys Acta 1426: 259–273 [DOI] [PubMed] [Google Scholar]
- Kohda D, Yamada M, Igura M, Kamishikiryo J, Maenaka K (2007) New oligosaccharyltransferase assay method. Glycobiology 17: 1175–1182 [DOI] [PubMed] [Google Scholar]
- Kowarik M, Numao S, Feldman MF, Schulz BL, Callewaert N, Kiermaier E, Catrein I, Aebi M (2006) N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314: 1148–1150 [DOI] [PubMed] [Google Scholar]
- Kukuruzinska MA, Lennon K (1998) Protein N-glycosylation: molecular genetics and functional significance. Crit Rev Oral Biol Med 9: 415–448 [DOI] [PubMed] [Google Scholar]
- Kuntz C, Sonnenbichler J, Sonnenbichler I, Sumper M, Zeitler R (1997) Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology 7: 897–904 [DOI] [PubMed] [Google Scholar]
- Kuster B, Naven TJ, Harvey DJ (1996) Effect of the reducing-terminal substituents on the high energy collision-induced dissociation matrix-assisted laser desorption/ionization mass spectra of oligosaccharides. Rapid Commun Mass Spectrom 10: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Lechner J, Sumper M (1987) The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem 262: 9724–9729 [PubMed] [Google Scholar]
- Lechner J, Wieland F (1989) Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem 58: 173–194 [DOI] [PubMed] [Google Scholar]
- Lechner J, Wieland F, Sumper M (1985) Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J Biol Chem 260: 860–866 [PubMed] [Google Scholar]
- Liu J, Mushegian A (2003) Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci 12: 1418–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher MF, Hansen U, Strominger JL (1976) Formation of lipid-linked sugar compounds in Halobacterium salinarium. Presumed intermediates in glycoprotein synthesis. J Biol Chem 251: 7289–7294 [PubMed] [Google Scholar]
- Mescher MF, Strominger JL (1976) Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J Biol Chem 251: 2005–2014 [PubMed] [Google Scholar]
- Messner P (1997) Bacterial glycoproteins. Glycoconj J 14: 3–11 [DOI] [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE (2003) Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol 161: 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14: 103–114 [DOI] [PubMed] [Google Scholar]
- Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW (2005) The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA 102: 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer C, Messner P (2004) Surface-layer glycoproteins: an example for the diversity of bacterial glycosylation with promising impacts on nanobiotechnology. Glycobiology 14: 31R–42R [DOI] [PubMed] [Google Scholar]
- Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12: 43R–56R [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Logan SM, Linton D, Wren BW (2003) Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol 11: 233–238 [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW (2005) Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol 3: 225–237 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upreti RK, Kumar M, Shankar V (2003) Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3: 363–379 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2006) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, Aebi M (2002) N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298: 1790–1793 [DOI] [PubMed] [Google Scholar]
- Wopereis S, Lefeber DJ, Morava E, Wevers RA (2006) Mechanisms in protein O-glycan biosynthesis and clinical and molecular aspects of protein O-glycan biosynthesis defects: a review. Clin Chem 52: 574–600 [DOI] [PubMed] [Google Scholar]
- Yan A, Lennarz WJ (2005) Unraveling the mechanism of protein N-glycosylation. J Biol Chem 280: 3121–3124 [DOI] [PubMed] [Google Scholar]
- Yan Q, Lennarz WJ (2002) Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J Biol Chem 277: 47692–47700 [DOI] [PubMed] [Google Scholar]
- Young NM, Brisson JR, Kelly J, Watson DC, Tessier L, Lanthier PH, Jarrell HC, Cadotte N, St Michael F, Aberg E, Szymanski CM (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem 277: 42530–42539 [DOI] [PubMed] [Google Scholar]
- Zubkov S, Lennarz WJ, Mohanty S (2004) Structural basis for the function of a minimembrane protein subunit of yeast oligosaccharyltransferase. Proc Natl Acad Sci USA 101: 3821–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M (1995) STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J 14: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Information