Abstract
The transport receptor Mex67–Mtr2 functions in mRNA export, and also by a loop-confined surface on the heterodimer binds to and exports pre-60S particles. We show that Mex67–Mtr2 through the same surface that recruits pre-60S particles interacts with the Nup84 complex, a structural module of the nuclear pore complex devoid of Phe-Gly domains. In vitro, pre-60S particles and the Nup84 complex compete for an overlapping binding site on the loop-extended Mex67–Mtr2 surface. Chemical crosslinking identified Nup85 as the subunit in the Nup84 complex that directly binds to the Mex67 loop. Genetic studies revealed that this interaction is crucial for mRNA export. Notably, pre-60S subunit export impaired by mutating Mtr2 or the 60S adaptor Nmd3 could be partially restored by second-site mutation in Nup85 that caused dissociation of Mex67–Mtr2 from the Nup84 complex. Thus, the Mex67–Mtr2 export receptor employs a versatile binding platform on its surface that could create a crosstalk between mRNA and ribosome export pathways.
Keywords: Mex67–Mtr2, mRNA export, NPC, Nup84 complex, ribosome export
Introduction
Nuclear pore complexes (NPCs) are the gates that allow passage of molecules and macromolecules across the nuclear envelope. NPCs are composed of about 30 nucleoporins (Nups) that form a conserved supramolecular assembly with eight-fold symmetry of ∼60 MDa in yeast and ∼120 MDa in vertebrates (Hetzer et al, 2005; Tran and Wente, 2006). Electron microscopy studies revealed the overall architecture of the NPC, which consists of a central spoke–ring assembly to which the cytoplasmic filaments and the nuclear basket are attached (Akey and Radermacher, 1993; Yang et al, 1998; Allen et al, 2000; Fahrenkrog and Aebi, 2003). A central transport channel is formed by the spoke–ring complex, which is filled up by a meshwork of natively unfolded Phe-Gly (FG) repeats that are part of a class of nucleoporins (Patel et al, 2007). Current models emphasize how the FG repeats are organized within the central transport channel to generate the permeability barrier for passive diffusion of molecules through the NPC (Shulga et al, 2000; Ribbeck and Gorlich, 2001; Rout et al, 2003; Patel et al, 2007). However, a large cargo (>40 kDa) can overcome this barrier by employing nuclear transport receptors (karyopherins), which can pass through the FG meshwork by facilitated diffusion. The role of FG nucleoporins in receptor-mediated transport of large proteins and RNAs is well established by the finding that the shuttling transport receptors can interact by low-affinity contact with the phenylalanine residues of the FG repeat meshwork (Rexach and Blobel, 1995; Bayliss et al, 2000). In contrast, it is less clear what role the non-FG nucleoporins play in nucleocytoplasmic transport, except their structural requirement in constituting the NPC scaffold. Thus, deletion of the large non-FG structural Nup188 or Nup170 can increase the permeability barrier for nuclear protein import and export (Shulga et al, 2000). Moreover, structural nucleoporins lacking FG domains can bind to FG repeats and hence could contribute to the permeability barrier (Patel et al, 2007). This observation might explain that the deletion of a structural Nup170 can affect nucleocytoplasmic transport by altering the organization of the FG repeat meshwork. Moreover, a role of the non-FG repeat domain of Nup2 in cargo release and karyopherin recycling required for nuclear protein import has been reported (Matsuura et al, 2003). Finally, the non-FG repeat part of Nup53 binds to karyopherin Kap121, which in turn inhibits the Kap121-dependent import pathway during mitosis (Makhnevych et al, 2003).
Structural nucleoporins implicated in nuclear export of mRNA has also been described. In particular, several subunits of the Nup84 complex (i.e. Nup85, Nup120, Nup145C and Nup133) when mutated exhibited strong mRNA export defects, whereas nuclear protein import or ribosomal export was not or only partially affected (Doye et al, 1994; Fabre et al, 1994; Heath et al, 1995; Goldstein et al, 1996; Siniossoglou et al, 1996; Segref et al, 1997; Hurt et al, 1999). The Nup84 complex is a conserved essential structural module of the NPC spoke–ring assembly, which consists of seven non-FG nucleoporins (Nup133, Nup84, Nup120, Nup85, Nup145C, Seh1 and Sec13) and exists in 16 copies per NPC (Siniossoglou et al, 1996; Rout et al, 2000). This module like its metazoan counterpart, the Nup107–Nup160 complex, plays a key role in NPC biogenesis (Siniossoglou et al, 1996; Harel et al, 2003; Walther et al, 2003). Electron microscopy revealed that the Nup84 complex exhibits a Y-shaped structure (Siniossoglou et al, 2000; Lutzmann et al, 2002) and structural predictions suggested that the subunits of the Nup84 complex fold into α-solenoid and β-propellers (Rout et al, 2000; Devos et al, 2004). Moreover, the Nup84 complex was reconstituted in vitro from its pre-assembled modules produced in Escherichia coli (Lutzmann et al, 2002). However, till date it remained unclear whether the Nup84 complex is directly involved in mRNA export or merely by causing structural distortions indirectly affects this transport pathway.
Notably, genetic and biochemical interactions have been observed between the general mRNA export receptor Mex67–Mtr2 and the Nup84 complex (Segref et al, 1997; Santos-Rosa et al, 1998; Lutzmann et al, 2005). Mex67–Mtr2 is a heterodimeric complex, which is structurally related to the family of Ran-independent transport receptors that exhibit an NTF2-like fold (Segref et al, 1997; Santos-Rosa et al, 1998; Herold et al, 2000; Fribourg et al, 2001; Fribourg and Conti, 2003). Recent findings indicated that the Mex67–Mtr2 heterodimer also functions as an export receptor for the pre-60S ribosomal subunit. In this role, yeast-specific loops inserted into the NTF2-like domains of Mex67 and Mtr2 were shown to be involved in the binding to 5S rRNA and pre-60S particles (Yao et al, 2007).
Here, we demonstrate that the loop-confined surface on the Mex67–Mtr2 heterodimer not only recruits the pre-60S particles but also binds to the Nup84 complex. The latter interaction depends on the positively charged amino acids that are clustering in a long loop, which protrudes from the NTF2-like domain of Mex67. In vitro crosslinking revealed that the Nup85 subunit of the Nup84 complex directly interacts with the loop region of Mex67. In vivo, deletion of the Mex67 loop when combined with Nup85 N-terminal truncations generated a synergistically enhanced growth retardation and inhibition of mRNA export. Concomitantly, a synergistically decreased binding of Mex67–Mtr2 to the Nup84 complex was observed. Thus, a versatile interaction surface on the heterodimeric Mex67–Mtr2 receptor is involved in mRNA and ribosome export.
Results
The Mex67–Mtr2 complex binds to the Nup84 complex
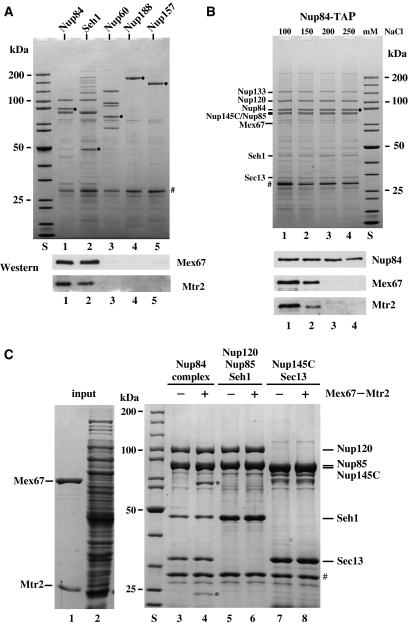
Previous studies indicated an interaction of Mex67–Mtr2 with the Nup84 complex (Santos-Rosa et al, 1998; Lutzmann et al, 2005). To test for the specificity of this interaction, members of the Nup84 complex (Nup84-TAP and Seh1-TAP) and nucleoporins that are not part of this module (i.e. Nup60-TAP, Nup188-TAP and Nup157-TAP) were affinity purified and examined for enrichment of Mex67–Mtr2. This analysis showed that Mex67–Mtr2 was specifically co-enriched in the Nup84 complex (Figure 1A). Notably, none of the subunits of the Nup84 complex contains FG repeats, suggesting a new type of interaction of Mex67–Mtr2 with the NPC. This interaction is insensitive to RNAse (data not shown), but can be dissociated by increasing amounts of salt (Figure 1B).
Figure 1.
The Mex67–Mtr2 heterodimer binds to the Nup84 complex in a salt-dependent way. (A) TAP of nucleoporins via the indicated genomic TAP-tagged bait proteins. Eluates were analyzed by SDS–PAGE and Coomassie staining (upper part) or western blotting (lower part) using antibodies against Mex67 and Mtr2. S, protein standard. Lanes 1–5, bait proteins (•). (B) Salt-sensitive binding of Mex67–Mtr2 to the Nup84 complex. TAP of the Nup84 complex was performed in buffer with increasing concentrations of NaCl (lanes 1–4) before it was eluted with the TEV protease. Eluates were analyzed by SDS–PAGE and Coomassie staining (upper part; indicated are the positions of Nup84, Nup133, Nup120, Seh1, Sec13 and Mex67) or western blotting (lower part) using antibodies against Mex67, Mtr2 and CBP (epitope at the C-terminal of Nup84 via TAP tag). (C) Only the assembled Nup84 complex binds to the Mex67–Mtr2 heterodimer. Recombinant pentameric Nup120–Nup85–Seh1–Nup145C–Sec13 complex, designated Nup84 complex (lanes 3 and 4), recombinant Nup120–Nup85–Seh1 complex (lanes 5 and 6) and recombinant Nup145C–Sec13 complex (lanes 7 and 8) were immobilized on GSH beads (via GST-Nup145C or GST-Nup85, respectively) and incubated with an E. coli lysate (lane 2) that contained (+) or did not contain (−) recombinant Mex67–Mtr2 heterodimer (lane 1). Proteins bound to the beads were eluted with TEV protease and analyzed by SDS–PAGE and Coomassie staining. The position of Mex67 and Mtr2 bound to the assembled Nup84 complex is indicated by *. TEV protease is indicated by #.
To gain insight into how the Nup84 complex interacts with Mex67–Mtr2, we sought to reconstitute these interactions in vitro. Previously, we showed that the Nup84 complex can self-assemble into a Y-shaped structure from the recombinant proteins (Lutzmann et al, 2002). When the reconstituted pentameric Nup84 complex (Nup145C-GST, Nup120, Nup85, Seh1 and Sec13) was incubated with recombinant Mex67–Mtr2, a significant interaction could be reconstituted (Figure 1C). Notably, only the assembled Nup84 complex (Nup120–Nup85–Seh1–Nup145C–Sec13), but not the individual modules that constitute this complex (heterotrimeric Nup120–Nup85–Seh1 and heterodimeric Nup145C–Sec13), could bind in vitro to the Mex67–Mtr2 heterodimer (Figure 1C). Moreover, a minimal Mex67–Mtr2 complex consisting of the NTF2-like Mex67 middle domain and Mtr2 was still bound to the Nup84 complex (data not shown). In contrast, the recombinant human TAP–p15 complex or Mtr2 alone did not associate with the Nup84 complex (data not shown). These data suggest that the Mex67–Mtr2 heterodimer can only bind to the assembled Nup84 complex, but not its individual subunits (see Discussion).
The loops in the Mex67–Mtr2 heterodimer facilitate recruitment to the Nup84 complex
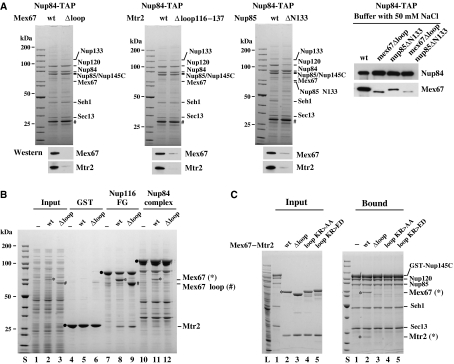
Previous work indicated that the positively charged Mex67 loop participates in binding of Mex67–Mtr2 to pre-60S particles (Yao et al, 2007). To test whether the same loop in Mex67 is also involved in binding to the Nup84 complex, the Mex67Δloop mutation was expressed in yeast and the Nup84 complex was affinity purified. This Mex67Δloop–Mtr2 heterodimer, which was normally expressed and assembled in vivo, did not co-enrich with the Nup84 complex under standard tandem affinity purification (TAP) conditions (i.e. 100 mM NaCl; Figure 2A). However, under lower ionic strength (e.g. 50 mM NaCl), a reduced but significant interaction could be observed (Figure 2A). When the Nup84 complex was affinity purified from the mtr2Δloop116–137 mutant, which is also impaired in the interaction with pre-60S particles (Yao et al, 2007), coprecipitation of Mex67–Mtr2 was reduced albeit less strongly (Figure 2A). Consistent with the in vivo data, in vitro reconstitution showed that the wild-type Mex67–Mtr2 complex, but not a Mex67–Mtr2 complex that has the Mex67 loop deleted (Figure 2B) or harbors point mutations within the loop (Figure 2C), bound to the Nup84 complex. However, the deletion of the Mex67 loop did not impair binding of Mex67–Mtr2 to FG repeats (Figure 2B). These data suggest that electrostatic interactions involving the positively charged Mex67 loop significantly contribute to the binding of Mex67–Mtr2 to the Nup84 complex.
Figure 2.
The loop-confined surface on the Mex67–Mtr2 heterodimer is involved to bind to the Nup84 complex. (A) Deletion of the loop region of Mex67 or Mtr2, or the N-terminus of Nup85 impaired the interaction of Mex67–Mtr2 and the Nup84 complex. Nup84 complex was affinity purified under standard TAP conditions (100 mM NaCl) or low-salt conditions (50 mM NaCl) by genomic TAP-tagged Nup84 from the indicated wild-type and mutant cells. Mutant cells include the loop deletion of Mex67 (408–435 aa), the partial loop deletion of Mtr2 (116–137 aa) and the truncation of the first 133 amino acids of Nup85. Eluates by TEV protease were analyzed by SDS–PAGE and Coomassie staining and western blotting using antibodies against Mex67 and Mtr2. Also a protein standard is shown. Prominent bands are indicated on the right. TEV protease is indicated by #. (B) The Mex67 loop deletion decreases binding to the Nup84 complex but not to FG repeats in vitro. GST alone (lanes 4–6), recombinant GST-Nup116 FG repeats (lanes 7–9) and the reconstituted Nup84 complex (lanes 10–12) were immobilized on GSH beads and incubated with an E. coli lysate devoid of Mex67–Mtr2 (input in lane 1; binding in lanes 4, 7 and 10) or containing recombinant Mex67–Mtr2 (input in lane 2; binding in lanes 5, 8 and 11) or mutant Mex67Δloop-Mtr2 (input in lane 3; binding in lanes 6, 9 and 12). Proteins bound to the beads were eluted in SDS sample buffer and analyzed by SDS–PAGE and Coomassie staining. Indicated is the position of the GST and GST-tagged proteins (filled circles), of wild-type Mex67 (*), Mex67Δloop (#) and Mtr2. (C) Mutation of positively charged amino acids in the Mex67 loop inhibits binding of Mex67–Mtr2 to the Nup84 complex. Recombinant Nup84 complex immobilized on GSH beads (bound; lanes 1–5) was incubated with an E. coli lysate in the absence (bound; lanes 1) or the presence recombinant wild-type Mex67–Mtr2 (input; lane 2), Mex67Δloop-Mtr2 (input; lane 3), Mex67 loop KR>AA-Mtr2 (input; lane 4) and Mex67 loop KR>ED-Mtr2 (input; lane 5). Proteins bound to the beads were eluted in SDS sample buffer and analyzed by SDS–PAGE and Coomassie staining. Indicated are Mex67 and Mtr2.
Pre-60S particles and the Nup84 complex compete for an overlapping binding site on the Mex67–Mtr2 heterodimer
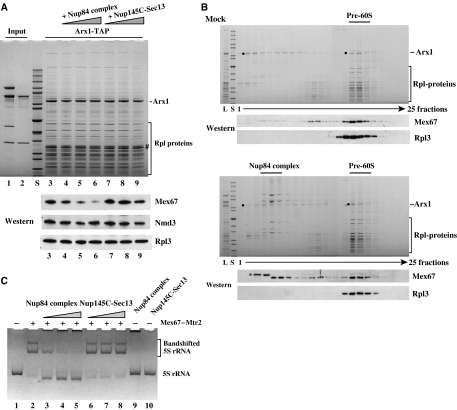
To find out whether pre-60S particles and the Nup84 complex compete for an overlapping binding site on the Mex67–Mtr2 heterodimer, we performed in vitro displacement studies. Purified pre-60S particles with co-enriched Mex67–Mtr2 heterodimer were immobilized on beads (via the Arx1-TAP bait) and incubated with increasing amounts of the reconstituted Nup84 complex. As shown in Figure 3A, the pentameric Nup84 complex could displace the Mex67–Mtr2 heterodimer, but not the Nmd3 export adaptor or the ribosomal protein Rpl3 from the pre-60S particle. In contrast, the Nup145C–Sec13 module that was unable to bind to Mex67–Mtr2 (see above) did not displace Mex67–Mtr2 from the pre-60S subunit (Figure 3A).
Figure 3.
Pre-60S particles and the Nup84 complex compete for an overlapping site on the Mex67–Mtr2 heterodimer. (A) The Nup84 complex displaces Mex67–Mtr2 from the pre-60S particle. Reconstituted pentameric Nup84 complex (Nup120–Nup85–Seh1–Nup145C–Sec13) (lane 1; input) or Nup145C–Sec13 heterodimer (lane 2; input) were incubated with the immobilized pre-60S particle (Arx1-TAP bound to IgG-Sepharose; lanes 3–9). After incubation and washing, the Arx1 particle was eluted by the TEV protease, and eluates were analyzed by SDS–PAGE and Coomassie staining (upper part) or western blotting (lower part) using antibodies against Mex67, Nmd3 and Rpl3. Lanes 3–9, Arx1-TAP incubated with mock buffer (lane 3), increasing amounts of Nup84 complex (lanes 4–6) or Nup145C–Sec13 heterodimer (lanes 7–9). S, protein standard (10 kDa ladder). (B) Mex67–Mtr2 can be exchanged from the pre-60S particle to the Nup84 complex. Sedimentation of the Arx1-containing pre-60S particle on sucrose gradients pre-incubated with mock-buffer (upper part) or with the reconstituted Nup84 complex (lower part). The load (L), a protein standard (S) and the fractions from the sucrose gradient (1–25) were analyzed by SDS–PAGE and Coomassie staining or western blotting (anti-Mex67 and anti-Rpl3 antibodies). The positions of Arx1 (•), the Nup84 complex, the pre-60S particle and Rpl-proteins are indicated. * Indicates an E. coli contaminant (derived from the addition of the recombinant Nup84 complex), which is crossreactive with the anti-Mex67 antibodies. (C) Binding of 5S rRNA to Mex67–Mtr2 can be displaced by the Nup84 complex. Recombinant and purified wild-type Mex67–Mtr2 (∼250 ng per lane) was incubated with in vitro-transcribed yeast 5S rRNA (∼150 ng) to form the 5S rRNA–Mex67–Mtr2 complex (lane 2). Addition of increasing amounts (665, 1330 and 2660 ng) of Nup84 complex (lanes 3–5) or Nup145C–Sec13 heterodimer (408, 816 and 1632 ng) (lanes 6–8) to the preformed 5S rRNA–Mex67–Mtr2 complex. 5S rRNA was analyzed on a 6% polyacrylamide gel stained with ethidium bromide. As control, 5S rRNA was incubated with the highest amount of the Nup84 complex (lane 9) or Nup145C–Sec13 heterodimer (lane 10) in the absence of Mex67–Mtr2. No protein (lane 1). Non-shifted and bandshifted 5S rRNA are indicated.
To show directly that association of Mex67–Mtr2 can be exchanged from the pre-60S particle to the Nup84 complex, we incubated a purified pre-60S particle containing Mex67–Mtr2 with the reconstituted Nup84 complex or with buffer and analyzed the potential transfer by sucrose gradient sedimentation. In the mock-treated sample, Mex67 co-sediment predominantly with the Arx1-containing pre-60S subunits indicated by ribosomal marker Rpl3 (Figure 3B). However, upon pre-incubation with the reconstituted Nup84 complex, a pool of Mex67 was released from the Arx1 particle and co-sedimented with the NPC module in the upper part of the sucrose gradient (Figure 3B). Altogether, the data revealed that the Nup84 complex and pre-60S particles compete for an overlapping binding site on the Mex67–Mtr2 heterodimer.
To find out whether the loop-confined surface on the Mex67–Mtr2 heterodimer that binds in vitro to 5S rRNA (Yao et al, 2007) overlaps with the protein-binding surface that recruits the Nup84 complex, we performed competition bandshift experiments. For these studies, we first generated a complex between 5S rRNA and Mex67–Mtr2, which shows slower migrating behavior on a native polyacrylamide gel (Figure 3C, lane 2). Upon addition of increasing amounts of Nup84 complex, but not of Nup145C–Sec13, Mex67–Mtr2 was efficiently dissociated from the 5S rRNA (Figure 3, lanes 3–8). However, neither the Nup84 complex nor Nup145C–Sec13 alone could bind to 5S rRNA (Figure 3, lanes 9 and 10). Thus, RNA bound to the loop-confined molecular surface on the Mex67–Mtr2 heterodimer can be displaced by the Nup84 complex.
The Nup85 subunit of the Nup84 complex is crosslinked to the Mex67 loop
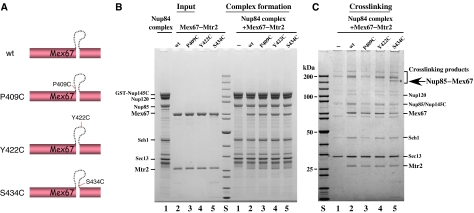
To identify the subunit(s) of the Nup84 complex that bind(s) to the loop-confined surface on the Mex67–Mtr2 heterodimer, we performed chemical crosslinking. By site-specific mutagenesis single cysteine substitutions were generated at discrete positions in the Mex67 loop (Figure 4A, P409>C, Y422>C, S434>C; note that Mex67 has only one cysteine in the LRR domain), which may allow crosslinking to the nearby cysteine residues of the interacting subunit of the Nup84 complex (note that 12, 12 and 18 cysteines, respectively, are present in Nup85, Nup145C and Nup120). None of the generated cysteine substitutions in Mex67 impaired cell growth (data not shown), and all allowed the Mex67–Mtr2 heterodimer to bind to the Nup84 complex (Figure 4B; data not shown). Upon addition of the homobifunctional sulfhydryl-reactive crosslinker BMB (1,4-bis-maleimidobutane) to the in vitro reconstituted assembly of the Nup84 complex and Mex67–Mtr2, a specific crosslink band of approximately 150 kDa was observed after treatment, using the Mex67 S434>C loop mutation (Figure 4C). This band was identified by mass spectrometry as a crosslinked product between Mex67 and Nup85. Thus, the Mex67 loop can directly interact with the Nup85 subunit of the Nup84 complex.
Figure 4.
The Nup85 subunit of the Nup84 complex can be crosslinked to the Mex67 loop. (A) Schematic drawing of the cysteine substitutions P409>C, Y422>C and S434>C in the Mex67 loop. (B) In vitro binding of Mex67–Mtr2 with indicated cysteine substitutions in the Mex67 loop to the Nup84 complex. Recombinant Nup84 complex immobilized on GSH beads was incubated with buffer (lane 1) or recombinant wild-type Mex67–Mtr2 (lane 2), Mex67 P409>C-Mtr2 (lane 3), Mex67 Y422>C-Mtr2 (lane 4) and Mex67 S434>C-Mtr2 (lane 5). After washing, proteins bound to the beads were eluted in SDS sample buffer and analyzed by SDS–PAGE and Coomassie staining. Indicated are the position of the subunits of the Nup84 complex, as well as Mex67 and Mtr2. (C) In vitro crosslinking of S434C residue in the Mex67 loop to Nup85. The reconstituted Nup84 complex was incubated with mock buffer (lane 1) or wild-type Mex67–Mtr2 (lane 2) and the indicated cysteine substitution mutants (lanes 3–5) in the same buffer as used for binding assay. The bivalent cysteine-specific crosslinker BMB was added to a final concentration of 50 μM to the preformed complexes. After crosslinking, the TCA-precipitated proteins were dissolved in SDS sample buffer and analyzed by SDS–PAGE and Coomassie staining. In the range of the crosslinked products, a band indicated by * was shown by mass spectrometry to be a product between Nup85 and Mex67.
Functional interaction of Nup85 with the Mex67 loop is required for mRNA export
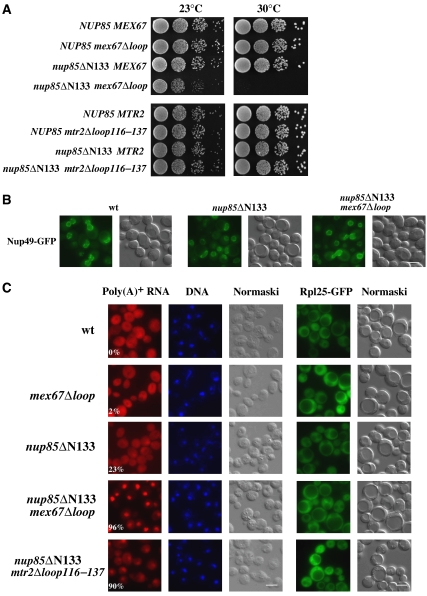
Next, we sought to test whether the interaction between Nup85 and the Mex67 loop is required for 60S subunit or mRNA export or both. Nup85 is an elongated protein with a predicted α-solenoid fold that binds by its N-terminal domain to Seh1 and by its C-terminal domain assembles into the Nup84 complex (Siniossoglou et al, 2000; Lutzmann et al, 2002; Devos et al, 2006). Hence, we sought to generate N-terminal truncation mutants of Nup85 that are not severely impaired in Nup84 core complex assembly except for loss of Seh1 (Siniossoglou et al, 2000) and analyze them for genetic interaction with the mex67Δloop allele. The deletion of the first 133 residues from the Nup85 N-terminus yielded viable cells that were not impaired in cell growth at 30°C (Figure 5A). Moreover, nup85ΔN133 cells did not exhibit an NPC-clustering phenotype (Figure 5B), in contrast to C-terminal deletion mutants, which are known to affect NPC formation (Goldstein et al, 1996; Siniossoglou et al, 2000). Also, the nup85ΔN133 mutant, in contrast to the nup170Δ mutant, did not show an altered permeability barrier of the NPC as shown by an NLS-GFP reporter leakage assay (Shulga et al, 2000; Supplementary Figure S1).
Figure 5.
Functional interaction of the Nup85 N-terminus with the Mex67 loop is essential for nuclear mRNA but not pre-60S subunit export. (A) Growth analysis of the indicated wild type, single- and double-mutant strains. Serial 10−1 dilutions of cells were spotted onto YPD plates and were grown for 2 days at 23 or 30°C. (B) NPC distribution in wild type, nup85ΔN133 single-mutant and nup85ΔN133 mex67Δloop double-mutant cells. Nup49-GFP, which was expressed as NPC marker, was analyzed by fluorescence microscopy. Wild-type and nup85ΔN133 cells were analyzed at 30°C, the double mutant was grown at 23°C and shifted for 2 h to 30°C. Scale bar, 5 μm. (C) Analysis of nuclear mRNA export (left panel) and 60S subunit export (right panel) in the indicated wild type, single- and double-mutant strains. Poly(A)+ RNA was detected by in situ hybridization with a Cy3-labeled oligo(dT) probe and DNA was stained with DAPI. Subcellular location of the Rpl25-GFP (60S reporter) was analyzed by fluorescence microscopy. Cells were grown at 30°C except for the double-mutant nup85ΔN133 mex67Δloop, which was grown at 23°C and shifted for 2 h to 30°C. The numbers indicate the percentage of cells (∼500 were counted in each case) that exhibit an apparent mRNA export defect. Scale bar, 5 μm.
However, binding of Mex67–Mtr2 to the Nup84 complex was significantly reduced in cells expressing the Nup85ΔN133 construct (Figure 2A). Moreover, combining the nup85ΔN133 mutation with the mex67Δloop allele induced a strongly enhanced growth defect with no NPC-clustering phenotype (Figure 5A and B). Concomitantly, a synergistically decreased binding of Mex67–Mtr2 to the Nup84 complex was observed under conditions of reduced ionic strength (e.g. 50 mM NaCl) (Figure 2A). Additionally, mutant alleles of the Nup84 complex (e.g. nup145-11, nup120-1) and also the nup116Δ, nup82-27 or nup159-1 mutants, which are known to impair mRNA export, exhibited a synthetically enhanced phenotype when combined with the mex67Δloop mutation (data not shown). In contrast, mex67Δloop was not found to be sl/se with the nup84Δ, nup188Δ and seh1Δ disruption alleles.
To find out whether the synergistically enhanced growth inhibition between the nup85ΔN133 and mex67Δloop alleles resulted in inhibition of nuclear export pathways, we analyzed nuclear exit of poly(A)+ RNA, pre-60S subunits and tRNA. Whereas 60S subunit and tRNA export were not impaired, export of mRNA was significantly inhibited in the nup85ΔN133 mex67Δloop double mutant in comparison with the single mutants (Figure 5C; data not shown). In contrast, when the mtr2Δloop116–137 deletion, which was previously shown to be synthetically lethal when combined with the nmd3ΔNES1 mutation (Yao et al, 2007), was combined with the nup85ΔN133 allele (Figure 5A), the strength of nuclear mRNA accumulation was less enhanced (Figure 5C) and no synergistic growth defect was observed (Figure 5A). The specific link of the Nup85 N-domain to the Mex67-dependent mRNA export route but not to other transport pathways was further substantiated by the observation that nup85ΔN133 did not enhance the growth defect of the ribosome export mutant nmd3ΔNES1 (Figure 6A) or the tRNA export receptor mutant los1Δ (data not shown). Altogether, these data suggest that the Mex67 loop in conjunction with the Nup85 N-terminal domain is specifically involved in nuclear mRNA export through the NPCs.
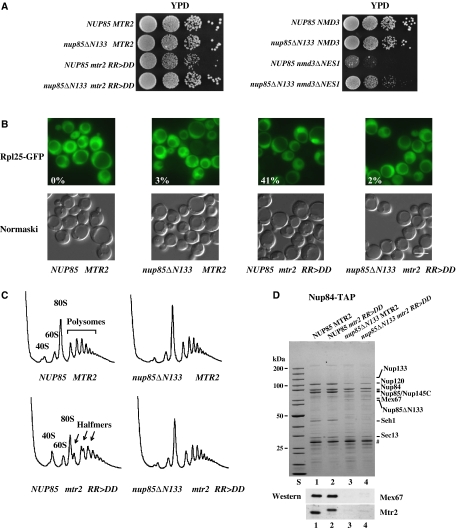
Figure 6.
Suppression of the ribosomal export defect induced by the mtr2 RR>DD mutation upon deletion of the N-terminus from Nup85. (A) Suppression of the growth defect of mtr2 RR>DD or nmd3ΔNES1 mutant cells by the nup85ΔN133 truncation. Growth analysis of the indicated wild type, single- and double-mutant strains. Serial 10−1 dilutions of cells were spotted onto YPD plates. It was grown for 2 days at 30°C. (B) Analysis of 60S subunit export in the nup85ΔN133 and mtr2 RR>DD single and nup85ΔN133 mtr2 RR>DD double mutants. Cells were grown at 30°C before the subcellular location of the Rpl25-GFP (60S subunit) reporter was determined by fluorescence microscopy. The numbers indicate the percentage of cells (∼200 were counted in each case) that exhibit nuclear accumulation of Rpl25-GFP. Scale bar, 5 μm. (C) Analysis of ribosome and polysome profile (OD254 nm) of the indicated wild type and mutants by sucrose gradient centrifugation. 40S subunits, 60S subunits, 80S ribosomes, polysomes and halfmer polysomes are indicated. (D) Nup85 N-terminal deletion reduced the binding of Mex67–Mtr2 RR>DD to the Nup84 complex. Nup84 complex was affinity purified under standard TAP conditions (100 mM NaCl) from the indicated wild-type or mutant cells. TEV eluates were analyzed by SDS–PAGE and Coomassie staining or western blotting using antibodies against Mex67 and Mtr2. Also a protein standard is shown. Prominent bands are indicated on the right. TEV protease is indicated by #.
Pre-60S subunit export impaired by mutating the loop in Mtr2 or the NES in Nmd3 can be rescued by the nup85ΔN133 allele
Previously, we have shown that the mtr2 RR>DD mutation mapping in the loop region of Mtr2 caused a reduced Mex67–Mtr2 binding to pre-60S particles and a pre-60S subunit export defect, but mRNA export was not impaired (Yao et al, 2007). Thus, we sought to test whether the pre-60S export defect generated by the mtr2 RR>DD mutation can be rescued by inducing a release of a pool of Mex67–Mtr2 from the Nup84 complex. The mtr2 RR>DD mutant shows a reduced growth at 30°C. However, this growth defect can be significantly rescued by combining the mtr2 RR>DD allele with the nup85ΔN133 mutation (Figure 6A). Concomitant to a restored cell growth, nuclear export of pre-60S subunits was improved, which can be seen as reduced nuclear accumulation of the Rpl25-GFP reporter (Figure 6B) and disappearance of ‘halfmer' polysomes (‘halfmers' are 40S subunits bound to mRNA, but lacking 60S subunits) when compared with the mtr2 RR>DD single mutant (Figure 6C). Additionally, the gain-of-function in ribosome export was partly counterbalanced by a slightly impaired mRNA export in the double mutant (data not shown). These data suggest that a pool of Mex67–Mtr2 when released from the Nup84 complex by deleting the N-terminal domain from the Nup85 subunit can be used to improve hindered pre-60S subunit export. Consistent with these data, the nup85ΔN133/nmd3ΔNES1 double mutant grows significantly better than the single nmd3ΔNES1 mutant (Figure 6A), suggesting again that dissociation of Mex67–Mtr2 from the mutant Nup84 complex could rescue the pre-60S subunit export defect.
Discussion
Our study has revealed that the loop-confined surface on the Mex67–Mtr2 export receptor not only binds to pre-60S particles (Yao et al, 2007) but also to the Nup84 complex. Thus, a versatile molecular surface exists on the Mex67–Mtr2 heterodimer that can accommodate either protein or RNA (Supplementary Figure S2). We speculate that the elongated Nup85 molecule (Lutzmann et al, 2005) with its predicted α-solenoid fold (Devos et al, 2006) interacts along its longitudinal axis with the extended surface on the Mex67–Mtr2 heterodimer (Supplementary Figure S2).
It was unexpected to find that Mex67–Mtr2 binds to Nup85 only when assembled into the Nup84 complex. This suggests that Nup85 changes its conformation after incorporation into the Nup84 complex. Consistent with this model, limited proteolysis revealed a different conformation of another subunit, Nup145C, in the assembled Nup84 complex in comparison with the Nup145C–Sec13 heterodimer (ML, unpublished data).
It is not known to what extent Mex67–Mtr2 has direct contact with the Nup84 complex in vivo, as this module is thought to be tightly embedded into the structural core of the NPC. Nevertheless, the Nup84 complex, which lacks FG repeats, is crucially involved in nuclear mRNA export via its core subunits Nup85, Nup120 and Nup145C (Doye et al, 1994; Heath et al, 1995; Dockendorff et al, 1997; Segref et al, 1997). On the other hand, nup85, nup120 and nup145C mutants are also impaired in NPC biogenesis and induce clustering of NPCs (Heath et al, 1995; Goldstein et al, 1996; Siniossoglou et al, 1996, 2000). Notably, the nup85ΔN133 mutant described in this study generates a synergistically enhanced mRNA export defect without NPC clustering when combined with the mex67Δloop mutation. Thus, we conclude that the recruitment of Mex67–Mtr2 to the Nup84 complex is crucial for nuclear mRNA export.
The binding of Mex67–Mtr2 to Nup85 at the NPC could serve many functions, perhaps this interaction helps to target Mex67–Mtr2 to the NPCs at an early stage or release Mex67–Mtr2 from FG repeats at a late step during mRNA export. It is also conceivable that Mex67–Mtr2 could play a role in nuclear translocation of the assembled Nup84 complex to the inner site of the NPC during NPC assembly. Last but not least the interaction of Mex67–Mtr2 with the Nup84 complex might be needed during transcription-coupled mRNA export. It was recently found that the Nup84 complex recruits the transcriptional factors Rap1/Gcr1/Gcr2 to the NPC (Menon et al, 2005). Moreover, Mex67 was found to associate with transcribing genes, perhaps at the nuclear periphery, to assist in transcription-coupled mRNA export (Gwizdek et al, 2006; Hobeika et al, 2007). Thus, an interaction of Mex67–Mtr2 with the Nup84 complex might link transcription with mRNA export at the nuclear face of the NPC.
An extensive network of coupling exists among the gene expression machines that synthesize, modify and process mRNAs before they are exported to the cytoplasm for translation. Intriguingly, our study has uncovered an overlapping binding site on the Mex67–Mtr2 receptor that could create a crosstalk between mRNA and ribosome export. It waits to be determined whether this additional interplay of factors involved in mRNA formation (mRNA export) and ribosome production (60S subunit export) helps to further coordinate gene expression and thus cell growth.
Materials and methods
Yeast strains and plasmids
Genomic C-terminal TAP tag integrations were performed using PCR-based DNA constructs for homologous recombination as described previously (Puig et al, 1998). The yeast strains used for TAP tagging were derived from wild-type yeast strain DS1-2b (Lutzmann et al, 2005). Other strains used in this study are listed in Supplementary Table S1, and used plasmids are listed in Supplementary Table S2. Deletion or point mutations were created by fusion PCR with primers containing the planned nucleotide exchange, and the mutations were verified by DNA sequencing. Primer sequences used in this study are available upon request. Standard methods for yeast growth, transformation and test for synthetic lethality were used (Santos-Rosa et al, 1998).
Purification and analysis of TAP-tagged nucleoporins
Affinity purification of TAP-tagged nucleoporins was carried out as described previously (Lutzman et al, 2005). TAP purifications were performed in LB buffer (50 mM Tris-Cl pH 7.4, 100 mM NaCl, 1.5 mM MgCl2 and 0.15% NP-40) or where indicated in LB buffer with higher NaCl. The TEV-eluted proteins were analyzed on an SDS 4–12% polyacrylamide gradient gel (Invitrogen) stained with Coomassie R250 (Sigma). For western blotting, the following primary antibodies were used in the indicated dilution: anti-Mex67 (1:5000), anti-Mtr2 (1:1000), anti-Nmd3 (1:3000), anti-CBP (1:4000) and anti-Rpl3 (1:3000). The secondary anti-rabbit HRP-conjugated antibody was used in a 1:3000 dilution.
Protein expression in E. coli
The subunits of the Nup84 complex and the Mex67–Mtr2 heterodimer were expressed in minimal medium in E. coli BL21 codon plus RIL cells (Stratagene) as described previously (Strasser et al, 2000; Lutzmann et al, 2002). Expression was induced by the addition of 0.8 mM IPTG at 23°C for 3 h, except for the expression of the pentameric Nup84 complex, which was induced at 16°C with a growth time of 14 h. Cells expressing GST-tagged nucleoporins were resuspended in lysis buffer (150 mM NaCl, 50 mM KOAc, 20 mM Tris-Cl pH 7.5, 2 mM Mg(OAc)2 0.1% NP-40 and 1 mM dithiothreitol (DTT)) containing a protease inhibitor cocktail (SERVA). Cells were lysed by sonication and centrifuged at 20 000 g for 20 min. The supernatant was incubated with Glutathione-Sepharose 4B (Amersham) at 4°C for 1–2 h. After extensive washing with lysis buffer, the purified Nup84 complex was eluted by TEV protease at 16°C for 1–2 h, and the complex was further purified by FPLC gel filtration (Superdex 200 HR 30/10 column, Amersham). Cells expressing untagged Mex67 or its mutants and His6-tagged Mtr2 were purified by Ni-NTA agarose (Qiagen), affinity purification and ion-exchange chromatography with MonoS column (Amersham) as described previously (Strasser et al, 2000).
In vitro binding and competition assays
The reconstituted Nup84 complex, which was bound by GST-Nup145C to 50 μl GSH beads per binding assay, was incubated with wild-type or mutant Mex67–Mtr2 complex in the presence of 20 μl E. coli lysate (to compete for unspecific binding). After incubation at 4°C for 1 h with beads washed with lysis buffer (150 mM NaCl, 50 mM KOAc, 20 mM Tris-Cl pH 7.5, 2 mM Mg(OAc)2 0.1% NP-40 and 1 mM DTT), the bound proteins were eluted with SDS-sample buffer or TEV protease. For the competition binding assay, pre-60S particles were isolated from yeast via Arx1-TAP affinity purification on IgG-Sepharose. Purified recombinant pentameric Nup84 complex or Nup145C–Sec13 heterodimer was added to the beads and further incubated for 1 h at 4°C. After washing, the pre-60S particles were eluted from the IgG-Sepharose by incubation with TEV protease at 16°C for 1 h.
Sucrose gradient sedimentation analysis was performed using purified pre-60S particles via Arx1 TAP-tagged bait followed by TEV cleavage. A 100 μl volume of TEV eluate was incubated at 4°C for 1 h with 100 μl of LB buffer or purified Nup84 complex dialyzed against the same buffer. The binding mixture was loaded onto a 10–30% (w/w) sucrose density gradient prepared in LB buffer. It was centrifuged at 27 000 r.p.m. for 16 h in an SW40 rotor (Beckman) and the fractions were collected. After trichloroacetic acid (TCA) precipitation, proteins from each fraction were analyzed by SDS–PAGE and western blot.
In vitro competing RNA bandshift assay was performed as described (Yao et al, 2007) with minor modifications. 5S rRNA was prepared by in vitro transcription using T7 RNA polymerase (MBI Fermantas) from linearized plasmid DNA (pET9D-5S). Recombinant His6-tagged Mex67–Mtr2 was purified from E. coli strain BL21 by affinity chromatography using Ni-NTA agarose (Qiagen) and followed by ion-exchange chromatography with a MonoS column (Amersham Pharmacia Biotech). All recombinant proteins were dialyzed against RNA-binding buffer (20 mM HEPES pH7.4, 100 mM KCl, 10 mM NaCl, 4 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 1 mM DTT and 0.5% NP-40) overnight at 4°C. The RNA-binding assay was performed at room temperature for 30 min in binding buffer. Samples were analyzed by loading on a 6% polyacrylamide gel (0.5 × Tris-Borate-EDTA) and stained with ethidium bromide to visualize RNA under UV light.
In vitro chemical crosslinking of Nup85 and Mex67
The affinity-purified Nup84 complex was incubated with wild-type or cysteine substitution mutant Mex67–Mtr2 in binding buffer (150 mM NaCl, 50 mM KOAc, 20 mM Tris-Cl pH 7.5 and 2 mM Mg(OAc)2). Chemical crosslinker BMB (10.9 Å; Pierce Chemical Co.) was freshly prepared as a 40 mM stock in DMSO and added to the protein complex at a final concentration of 50 μM. The reaction was incubated at 4°C for 2 h before quenching with 20 mM DTT at room temperature for 10 min. The crosslinked products were TCA-precipitated and loaded on a 4–12% gradient gel (Invitrogen) for further analysis.
Analysis of nucleocytoplasmic transport
The Rpl25-eGFP reporter assays to analyze pre-60S ribosomal subunit export was carried out as described previously (Gadal et al, 2001). Nuclear accumulation of poly(A)+ RNA was determined by in situ hybridization using Cy3-labeled oligo-d(T) probes (Segref et al, 1997). The nuclear leakage assay of an NLS-GFP reporter was performed as described previously (Shulga et al, 1996, 2000). Cells were examined by fluorescence microscopy using an Imager Z1 microscope (Carl Zeiss) with a 63 × NA 1.4 Plan-Apo-Chromat Oil immersion len (Carl Zeiss) and DICIII, HEeGFP, DAPI or HECy3 filter sets, respectively. Pictures were acquired with an AxioCamMRm camera (Carl Zeiss) and software AxioVision 4.3 (Carl Zeiss) at resolution 1388 × 1040 (Binning 1 × 1, gain factor 1).
Sucrose gradient sedimentation
Yeast ribosomal and polysomal profiles were analyzed by 7–50% (w/v) sucrose gradient centrifugation in low salt as described previously (Yao et al, 2007). Cells were grown in 200 ml of YPD at 30°C to OD600 nm 0.6–0.8. Cycloheximide (100 μg/ml) was added to the culture before whole-cell lysate extraction. The lysate with an OD260 nm of 8 was loaded onto the sucrose gradient and centrifuged at 38 000 r.p.m. for 225 min in an SW40 rotor (Beckman) and the fractions were analyzed at 254 nm using a density gradient fractionator.
Supplementary Material
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
The excellent technical assistance of Ruth Kunze and the help of Dr Jochen Baßler in preparing Supplementary Figure S2 is acknowledged. Moreover, we thank Sabine Merker and Petra Ihrig under the supervision of Dr J Lechner (Mass Spectrometry Unit, BZH, Heidelberg) for performing the mass spectrometical analysis. We are grateful to Dr Catherine Dargemont (Universités Paris VI and VII, Paris, France) for anti-Mex67, Dr Arlen W. Johnson (University of Texas, Austin, TX) for anti-Nmd3 and Dr Jonathan R Warmer (Albert Einstein College of Medicine, Bronx, NY) for anti-Rpl3 antibodies. EH is recipient of grants from the Deutsche Forschungsgemeinschaft (SFB 638/B2) and Fonds der Chemischen Industrie.
References
- Akey CW, Radermacher M (1993) Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol 122: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW (2000) The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci 113: 1651–1659 [DOI] [PubMed] [Google Scholar]
- Bayliss R, Littlewood T, Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-b in nuclear trafficking. Cell 102: 99–108 [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP (2004) Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA 103: 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN (1997) C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol 17: 906–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC (1994) A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J 13: 6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC (1994) Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell 78: 275–289 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Aebi U (2003) The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 4: 757–766 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E (2001) Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8: 645–656 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Conti E (2003) Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15. EMBO Rep 4: 699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN (1996) Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell 7: 917–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C (2006) Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci USA 103: 16376–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN (1995) Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol 131: 1677–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E (2000) TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol 20: 8996–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW (2005) Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21: 347–380 [DOI] [PubMed] [Google Scholar]
- Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, Stewart M, Divita G, Dargemont C (2007) Coordination of Hpr1 and Ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell 18: 2561–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G (1999) A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol 144: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E (2002) Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J 21: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Stangl K, Stelter P, Toth KF, Bottcher B, Hurt E (2005) Reconstitution of Nup157 and Nup145N into the Nup84 complex. J Biol Chem 280: 18442–18451 [DOI] [PubMed] [Google Scholar]
- Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW (2003) Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115: 813–823 [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M (2003) Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J 22: 5358–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM (2005) Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF (2007) Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96 [DOI] [PubMed] [Google Scholar]
- Puig O, Rutz B, Luukkonen BG, Kandels-Lewis S, Bragado-Nilsson E, Seraphin B (1998) New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast 14: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83: 683–692 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Magnasco MO, Chait BT (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 13: 622–628 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol 18: 6826–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS (2000) Yeast nucleoporins involved in passive nuclear envelope permeability. J Cell Biol 149: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS (1996) In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol 135: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt EC (2000) Structure and assembly of the Nup84p complex. J Cell Biol 149: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loi odice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW (1998) Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell 1: 223–234 [DOI] [PubMed] [Google Scholar]
- Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2