Abstract
In most eukaryotes, cyclin-dependent kinases (Cdks) play a central role in control of cell-cycle progression. Cdks are inactivated from the end of mitosis to the start of the next cell cycle as well as during sexual differentiation. The forkhead-type transcription factor Fkh2p is required for the periodic expression of many genes and for efficient mating in the fission yeast Schizosaccharomyces pombe. However, the mechanism responsible for coordination of cell-cycle progression with sexual differentiation is still unknown. We now show that Fkh2p is phosphorylated by Cdc2p (Cdk1) and that phosphorylation of Fkh2p on T314 or S462 by this Cdk blocks mating in S. pombe by preventing the induction of ste11+ transcription, which is required for the onset of sexual development. We propose that functional interaction between Cdks and forkhead transcription factors may link the mitotic cell cycle and sexual differentiation.
Keywords: Cdk1, development, phosphorylation, transcription factor, cell cycle
Introduction
The mechanism responsible for the switch from growth to sexual development has been studied in many organisms including the fission yeast Schizosaccharomyces pombe. In fission yeast, the onset of sexual development requires both a pheromone signal and the depletion of nutrients, especially that of nitrogen (Yamamoto, 1996; Yamamoto et al, 1997). If cells of the opposite mating type are available, those that have committed to sexual development conjugate to form diploids. These diploid cells then undergo meiosis and complete sexual development.
The transcription factor Ste11p plays a central role in commitment to sexual development in fission yeast (Sugimoto et al, 1991; Yamamoto, 1996; Yamamoto et al, 1997). It regulates the transcription of many genes required for the initiation and progression of conjugation and meiosis (Mata and Bahler, 2006; Xue-Franzen et al, 2006). Expression of ste11+ itself is regulated by several pathways (Yamamoto, 1996; Yamamoto et al, 1997), including the cyclic AMP (cAMP) pathway. Nutrient exhaustion results in a decrease in the intracellular concentration of cAMP and a consequent inactivation of cAMP-dependent protein kinase (PKA). The transcriptional activator Rst2p, which is negatively regulated by PKA, then binds to the upstream region of ste11+ and induces the production of ste11+ mRNA (Kunitomo et al, 2000; Higuchi et al, 2002). In addition, a stress signal mediated by the mitogen-activated protein kinase pathway is required for the induction of ste11+ mRNA in response to nutrient deprivation (Takeda et al, 1995; Kato et al, 1996; Shiozaki and Russell, 1996). In addition to the regulation of ste11+ transcription, the activity of the encoded protein (Ste11p) is regulated at a post-translational level (Li and McLeod, 1996; Kitamura et al, 2001; Qin et al, 2003; Kjaerulff et al, 2005).
In fission yeast, the single cyclin-dependent kinase (Cdk) Cdc2p controls cell-cycle progression in a manner dependent on various internal and external conditions including nutrient availability (MacNeill and Nurse, 1997). Both nitrogen starvation and pheromone induce arrest in G1 phase of the cell cycle by inhibiting the activity of Cdc2p, an effect in turn mediated by cyclin degradation and upregulation of Cdk inhibitors. Pheromone induces degradation of the B-type cyclins Cig2p and Cdc13p as well as upregulation of the Cdk inhibitor Rum1p (Stern and Nurse, 1997, 1998). Nitrogen exhaustion also promotes the degradation of both Cig2p and Cdc13p by activating the anaphase-promoting complex (Yamaguchi et al, 1997; Kitamura et al, 1998; Yamano et al, 2004). Linkage between cell-cycle control and sexual development is likely provided by Cig2p. Loss of Cig2p function promotes mating, whereas overproduction of this cyclin negatively regulates sexual differentiation (Obara-Ishihara and Okayama, 1994).
Members of the forkhead-box family of transcription factors are present in almost all eukaryotes (Costa, 2005; Costa et al, 2005; Wang et al, 2005). More than 50 such proteins that share homology in the winged-helix DNA-binding domain have been identified in higher eukaryotes. This family of transcription factors is implicated in the regulation of a variety of cellular processes, including the cell cycle, apoptosis, DNA repair, stress resistance, and metabolism. The Sanger Center database (http://www.sanger.ac.uk/) indicates the existence of four forkhead proteins in fission yeast: Fkh2p, Fhl1p, Sep1p, and Mei4p (Bahler, 2005). Fkh2p is required for efficient G2–M transition, normal septation, and periodic gene expression (Buck et al, 2004; Bulmer et al, 2004; Rustici et al, 2004; Szilagyi et al, 2005). It is also required for efficient mating (Szilagyi et al, 2005). However, the mechanism responsible for the partial sterile phenotype of fkh2 mutant cells has remained unknown.
We have now investigated the role of Cdc2p in coordination of cell-cycle control and sexual differentiation in fission yeast. We show here that the forkhead transcription factors, including Fkh2p, are responsible for mediating a signal from the kinase Cdc2p to the transcription factor Ste11p.
Results
Forkhead transcription factors are required for mating
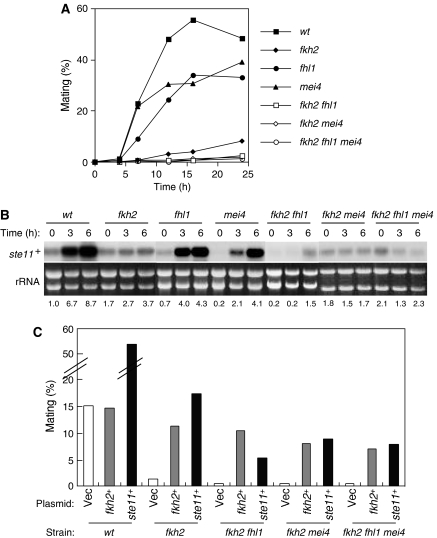
Fkh2p, Fhl1p, Mei4p, and Sep1p are the forkhead transcription factors in fission yeast. Given that fkh2-deleted cells show a partial sterile phenotype (Szilagyi et al, 2005), we investigated the role of forkhead transcription factors in mating in fission yeast. We constructed homothallic strains in which each gene for the forkhead transcription factors was individually deleted. Consistent with previous observations (Ribar et al, 1999), sep1-deleted cells manifested a pronounced septation defect and slow growth. We, therefore, did not further characterize the role of sep1+ in mating. In addition to fkh2-deleted cells, we found that fhl1-deleted and unexpectedly mei4-deleted cells exhibited a partial sterile phenotype, whereas fkh2 fhl1, fkh2 mei4 and fhk2 fhl1 mei4 mutant cells showed a more pronounced sterile phenotype than did cells lacking either gene alone (Figure 1A). These results suggested that, among the forkhead transcription factors, Fkh2p plays the predominant role in mating, with Fhl1p and Mei4p having minor roles that partially overlap with that of Fkh2p.
Figure 1.
Forkhead transcription factors are required for the induction of ste11+ mRNA and efficient mating. (A) wt (HM6), fkh2 (HM5657), fhl1 (HM4837), mei4 (HM50), fkh2 fhl1 (HM4887), fkh2 mei4 (HM5515), or fkh2 fhl1 mei4 (HM5544) cells were grown in EMM2 medium to a density of 1 × 107 cells/ml, washed, and resuspended at a density of 2 × 107 cells/ml in EMM2 medium lacking nitrogen. They were then cultured at 30°C and samples were collected at the indicated times for determination of mating frequency. Data are from representative experiments. (B) Total RNA was extracted from cells treated as in (A), and the abundance of ste11+ mRNA was examined by northern blot analysis. Ethidium bromide staining of rRNA is shown as a loading control. The ratios of intensities of ste11+ to rRNA signals were used to calculate the relative fold enrichment, shown below the rRNA. The samples from wt to fkh2, from fhl1 to mei4, from fkh2 mei4 to fkh2 fhl1 mei4 were from the same gel. All the samples were treated equally and the exposure time was the same. (C) Cells were transformed with pcL-ste11+ (ste11+), pAL-fkh2+ (fkh2+), or the empty vector pcL-X (Vec) and were cultured as in (A) for the determination of mating efficiency at 24 h after transfer to EMM2 medium without nitrogen. Data are from representative experiments.
Given that the induction of ste11+ mRNA plays a central role in mating, we monitored the abundance of this mRNA in the various mutant strains (Figure 1B). In contrast to wt cells, the induction of ste11+ mRNA was greatly delayed or virtually abolished in fkh2, fkh2 fhl1, fkh2 mei4, or fkh2 fhl1 mei4 mutant cells. In fhl1-deleted cells, the increase in ste11+ mRNA was apparent, but slightly reduced. In mei4-deleted cells, the increase in ste11+ mRNA was apparent but delayed. Ectopic expression of ste11+ indeed restored fertility not only to the fkh2-deleted cells but also to fkh2 fhl1, fkh2 mei4, and fkh2 fhl1 mei4 mutant cells to an extent similar to that observed with ectopic expression of fkh2+ (Figure 1C). We thus concluded that the sterility of the forkhead mutant cells was caused largely by poor induction of ste11+, not by slow growth. In addition, the sterility was not attributable to a defect in the induction of G1 arrest by nitrogen starvation, given that the forkhead mutant cells arrested in G1 phase in a manner similar to that of wt cells (Supplementary Figure 1).
Fkh2p binds to a FLEX element upstream of ste11+ both in vivo and in vitro
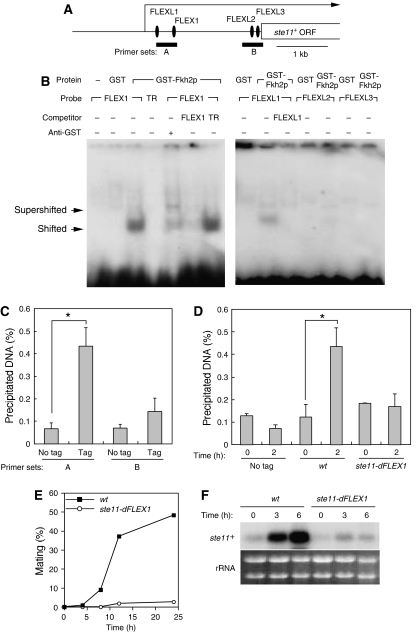
Given that the forkhead family of transcription factors recognizes the core sequence GTAAAYA (Pierrou et al, 1994), we searched for this sequence in the vicinity of the genomic locus of ste11+. One such sequence, designated FLEX1, was detected in the putative 5′ regulatory region of ste11+ (Figure 2A). If one mismatched base is allowed, three FLEX-like sequences—designated FLEXL1, FLEXL2, and FLEXL3—were also apparent in this region.
Figure 2.
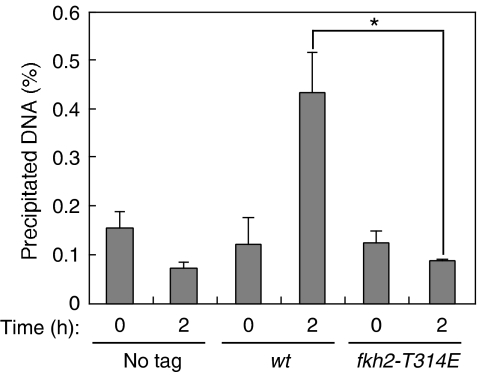
Fkh2 binds to FLEX and FLEXL sequences in the putative promoter region of ste11+ and thereby induces ste11+ mRNA. (A) Schematic representation of the region upstream of the open reading frame (ORF) of ste11+ showing FLEX and FLEXL sequences. The major transcription initiation site of ste11+ is indicated by the arrow, and the regions targeted by primer sets in ChIP analysis are also shown. (B) An EMSA was performed with recombinant GST–Fkh2p(216–330) (or GST alone) and with FLEX1, FLEXL1, FLEXL2, FLEXL3, or TR (negative control) probes labeled with 32P. Competition was evaluated with excess amounts of unlabeled FLEX1, TR, or FLEXL1 oligonucleotides, and supershift analysis was performed with antibodies to GST, as indicated. The positions of shifted and supershifted bands are shown. (C) No tagged cells (no tag, HM6) and cells expressing GFP-tagged Fkh2p (Tag, HM5719) were grown to late log-phase, washed, and resuspended in medium without nitrogen. After incubation for 2 h at 30°C, cells were collected and analyzed by ChIP with antibodies to GFP and with the primer sets indicated in (A). Data are means±s.e. *P<0.006 (Student's t-test). (D) No tagged cells (no tag, HM6) and cells expressing GFP-tagged Fkh2p (wt; HM5719 and ste11-dFLEX1: HM6124) were treated and analyzed as in (C) with primer set A. Samples were collected at 0 and 2 h after nitrogen withdrawal. Data are means±s.e. of values from three separate experiments. *P<0.007 (Student's t-test). (E) wt (HM6) or ste11-dFLEX1 (HM5832) cells were treated and analyzed for mating efficiency as in Figure 1A. (F) Total RNA was extracted from cells treated as in (E) and was subjected to northern blot analysis of ste11+ mRNA.
To examine whether Fkh2p binds to these FLEX or FLEX-like elements, we prepared a fusion protein consisting of glutathione-S-transferase (GST) and the forkhead DNA-binding domain of Fkh2p (amino acids 216–330) and performed an electrophoretic mobility-shift assay (EMSA) with this protein and radioactive oligonucleotides containing the FLEX1 or FLEXL sequences as probes (Figure 2B). Shifted bands were observed with FLEX1 and, to a lesser extent, with FLEXL1, but they were not detected with FLEXL2, FLEXL3, or an unrelated (TR) probe. The shifted bands were specific for Fkh2p and for FLEX1 or FLEXL1, given that they were not observed with GST in place of the fusion protein and that the corresponding unlabeled oligonucleotides, but not an unrelated oligonucleotide (TR), inhibited the binding of the GST–Fkh2 fusion protein to the labeled probes. The shifted band observed with the FLEX1 probe was also supershifted in the presence of antibodies to GST. These results thus indicated that the GST–Fkh2 fusion protein directly binds to FLEX1 and, to a lesser extent, to FLEXL1 in vitro.
To examine whether Fkh2p binds to the FLEX or FLEX-like sequences upstream of ste11+ in vivo, we performed a chromatin immunoprecipitation (ChIP) assay with cells expressing green fluorescent protein (GFP)-tagged Fkh2p by nmt41 promoter (Figure 2C). In cells expressing GFP-tagged Fkh2p, the mating efficiency and the induction of ste11+ mRNA were comparable to those in wt cells, suggesting that GFP-tagged Fkh2p functions like wt protein (Supplementary Figure 2). Immunoprecipitation with antibodies to GFP revealed that GFP-Fkh2p associates with genomic DNA containing both FLEX1 and FLEXL1 (primer set A), whereas association with genomic DNA containing both FLEXL2 and FLEXL3 (primer set B) is little. The amount of either region of genomic DNA immunoprecipitated with the antibodies to GFP was greatly reduced for cells not expressing GFP-Fkh2p. These results showed that Fkh2p binds in vivo to the genomic locus containing the FLEX1 and FLEXL1 elements upstream of ste11+. To test whether such binding depends on nutrient conditions, we measured the binding activity of Fkh2p in cells subjected to nitrogen deprivation. The ChIP assay revealed that nitrogen withdrawal resulted in an increase in the binding of GFP-Fkh2p to genomic DNA containing FLEX1 and FLEXL1 (Figure 2D), but not to the region upstream of cdc15+(Supplementary Figure 3). The amount of genomic DNA containing FLEX1 and FLEXL1 immunoprecipitated with the antibodies to GFP was low for cells not expressing GFP-Fkh2p upon nitrogen starvation. These results suggest that Fkh2p associates with the upstream region of ste11+ in vivo when cells are able to mate.
To examine the role of FLEX1 in mating, we deleted the 7-bp core sequence of this site from its chromosome locus. The mating efficiency of the resulting mutant strain (ste11-dFLEX1) was greatly reduced compared with that of wt cells (Figure 2E; Supplementary Figure 2). This sterility was not attributable to a defect in induction of G1 arrest (Supplementary Figure 4). These observations suggested that the core sequence of FLEX1 is required for efficient mating. In addition, induction of ste11+ mRNA by nitrogen withdrawal was largely abolished in ste11-dFLEX1 cells (Figure 2F; Supplementary Figure 2), suggesting that the core sequence of FLEX1 is also required for activation of ste11+ expression. The ChIP assay revealed that the core FLEX deletion resulted in a decrease in the binding of GFP-Fkh2p to genomic DNA around FLEX1 (Figure 2D), suggesting that the core FLEX1 is required for Fkh2p to associate with the upstream region of ste11+ in vivo.
Effects of phosphorylation of Fkh2p by Cdc2p
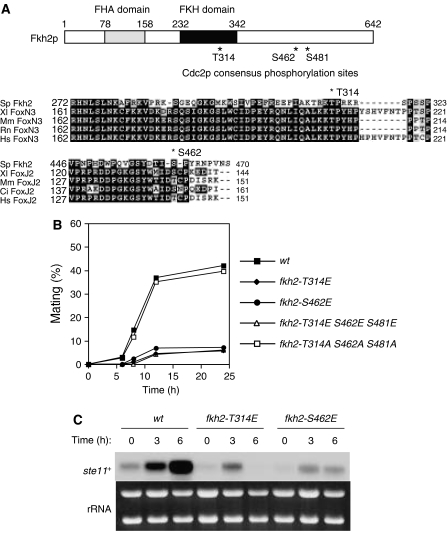
Cdks regulate forkhead transcription factors in various organisms, and Fkh2p has been shown to be a phosphoprotein in fission yeast (Buck et al, 2004; Bulmer et al, 2004). A search of the Fkh2p sequence for consensus phosphorylation sites for Cdc2p, (pS/pT)-P-X-(R/K) (Nigg, 1993), revealed three such sites at residues T314, S462, and S481 (Figure 3A). The sequence surrounding T314 in the DNA-binding domain of Fkh2p is conserved among members of the forkhead-box family of other species, especially those of the FoxN subfamily (Mazet et al, 2003), although the consensus phosphorylation site sequence is not fully conserved. S462 is conserved, but not as a Cdc2p phosphorylation site, among members of the FoxJ subfamily of transcription factors (Mazet et al, 2003). To assess the potential function of these putative Cdc2p phosphorylation sites in Fkh2p, we changed the serine or threonine residues to glutamic acid by site-directed mutagenesis of the chromosome to mimic the effect of Cdc2p phosphorylation in vivo. Cells in which T314 (fkh2-T314E) or S462 (fkh2-S462E) of Fkh2p was replaced with glutamic acid exhibited a reduced mating efficiency (Figure 3B). In contrast, similar mutation of S481 of Fkh2p (fkh2-S481E) did not substantially affect mating efficiency (data not shown). In addition, the mating efficiency of fkh2-T314E S462E S481E mutant cells was similar to that of fkh2-T314E or fkh2-S462E cells (Figure 3B). These results suggested that dephosphorylation of Fkh2p at T314 and S462 is required for efficient mating. However, unphosphorylated form of Fkh2p (fkh2-T314A S462A S481A) failed to enhance mating efficiency, suggesting that an additional mechanism is required for ectopic mating (Figure 3B). The induction of ste11+ mRNA in response to nitrogen deprivation was greatly reduced in fkh2-T314E or fkh2-S462E cells compared with that apparent in wt cells (Figure 3C). These observations thus suggested that dephosphorylation of Fkh2p on T314 and S462 is required for efficient induction of ste11+ mRNA.
Figure 3.
Phosphorylation of Fkh2p by Cdc2p negatively regulates mating. (A) A schematic representation of Fkh2p indicating consensus phosphorylation sites (T314, S462, S481) for Cdc2p as well as the FHA and FKH domains is shown in the upper panel. Multiple alignment of Fkh2, FoxN3, and FoxJ2 proteins of S. pombe (Sp), Xenopus laevis (Xl), Mus musculus (Mm), Rattus norvegicus (Rn), Homo sapiens (Hs), and Ciona intestinalis (Ci) is shown in the lower panels. Identical (shaded black) and similar (shaded gray) amino acids as well as two Cdc2p consensus phosphorylation sites (T314 and S462) of Fkh2p are indicated. Dashes represent gaps introduced to optimize alignment. (B) Cells expressing wild type (wt, HM5145) or T314E (fkh2-T314E, HM5910), S462E (fkh2-S462E, HM5911), T314E S462E S481E (fkh2-T314E S462E S481E, HM5827) or T314A S462A S481A (fkh2-T314A S462A S481A, HM5722) mutant forms of Fkh2p were treated and analyzed for mating efficiency as in Figure 1A. (C) Total RNA was extracted from cells treated as in (B) and was subjected to northern blot analysis of ste11+ mRNA.
Other defects of fkh2-deleted cells, such as abnormal morphology or septation defect, were less than 1% in wt, fkh2-T314E, or fkh2-S462E cells. In addition, cell length is similar in wt, fkh2-T314E, or fkh2-S462E cells (Supplementary Figure 5). These facts suggest that these point mutations specifically affect mating. To confirm that these point mutations do not affect cell-cycle progression and transcriptional activity during the normal mitotic cell cycle, we measured the timing of mitotic entry and the mRNA levels of cdc15+, spo12+, and slp1+, as Fkh2p is required for periodic expression of these mRNAs (Buck et al, 2004; Bulmer et al, 2004). Cells were transiently arrested in late G2 by the inactivation of cdc25+ and released to the permissive temperature to enter a synchronous cell cycle. In contrast to fkh2-deleted cells that showed the severe delay in entry into mitosis (Buck et al, 2004), wt, fkh2-T314E, or fkh2-S462E cells entered mitosis almost with the same timing as indicated by the coincidence of the peak of septa (Supplementary Figure 6). Additionally, the periodic expressions of cdc15+, spo12+, and slp1+ mRNAs were observed in wt, fkh2-T314E, or fkh2-S462E cells (Supplementary Figure 6). These results suggest that these point mutations specifically affect ste11+ mRNA expression but not other mitotic genes expression. The poor mating efficiency of the fkh2-T314E or fkh2-S462E mutants was not due to a defect in induction of cell-cycle arrest in G1 phase (Supplementary Figure 7). In addition, the abundance of the mutant proteins was similar to that of the wild-type protein (Supplementary Figure 8), suggesting that the poor mating efficiency of the mutant cells was not attributable to a reduced protein level.
Phosphorylation of Fkh2p by Cdc2p in vitro and in vivo
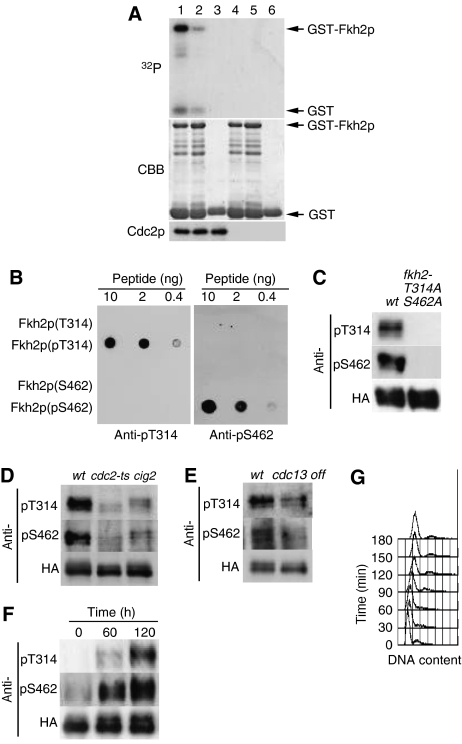
We next tested whether Cdc2p directly phosphorylates GST fusion proteins containing various fragments of Fkh2p (residues 305–492, 216–330, or 317–479) in vitro (Figure 4A; Supplementary Figure 9). Cdc2p precipitated from cell extracts with anti-hemagglutinin epitope (HA) antibody or Suc1p-coated beads phosphorylated each of the GST–Fkh2p fusion proteins but not GST alone. We found that mutation to alanine of the consensus phosphorylation sites for Cdc2p in each of the Fkh2p fragments (T314 in Fkh2p (216–330), S462 in Fkh2p (317–479), or T314, S462, and S481 in Fkh2p (305–492)) reduced the extent of phosphorylation by Cdc2p. These results thus suggested that Cdc2p phosphorylates at least T314 and S462 residues of Fkh2p in vitro. In addition, recombinant human Cdc2p complex, but not the kinase inactive complex, phosphorylated Fkh2p, suggesting that Cdc2p directly phosphorylates Fkh2p (Supplementary Figure 9).
Figure 4.
Phosphorylation of Fkh2p on T314 and S462 by Cdc2p in vitro and in vivo. (A) Kinase assays were performed with Cdc2p precipitates prepared from protein extracts of exponentially growing cells expressing hemagglutinin epitope (HA)-tagged forms of Cdc2p (HM6118; lanes 1–3) or not expressing HA (HM6; lanes 4–6) with anti-HA antibody. Substrates (lanes 1–3, respectively) included GST–Fkh2p(305–492), GST–Fkh2p(305–492) containing T314A, S462A, and S481A mutations, or GST alone. Reaction mixtures were separated by SDS–polyacrylamide gel electrophoresis, and proteins were detected by staining with Coomassie brilliant blue (CBB) and autoradiography (32P). Arrows indicate GST and the GST–Fkh2p fusion proteins. The Cdc2p input into each reaction mixture was also examined separately by Western blotting. (B) Various amounts (10, 2, or 0.4 ng) of Fkh2p peptides containing phosphorylated or nonphosphorylated T314 or S462 were spotted onto a nitrocellulose membrane and subjected to immunodetection with affinity-purified antibodies (anti-pT314 and anti-pS462) generated in response to the corresponding phosphorylated peptides. (C) Cells expressing HA-tagged forms of wild-type Fkh2p (HM5145) or the Fkh2p(T314A,S462A) mutant (HM5722) were grown to mid-log phase at 30°C. Cell lysates were then subjected to immunoprecipitation with antibodies to HA, and the resulting precipitates were subjected to immunoblot analysis with anti-pT314, anti-pS462, and anti-HA, as indicated. (D) Cells expressing HA-tagged Fkh2p were either grown to mid-log phase in EMM2 at 24°C and then incubated at 36.5°C for 7 h (wt, HM5145; cdc2-ts, HM5444) or grown as in (C) (cig2, HM5530). Cell lysates were subjected to immunoprecipitation and immunoblot analysis as in (C). (E) Cells expressing HA-tagged Fkh2p (wt, HM5146; cdc13 off, HM5554) were grown to mid-log phase in EMM2 at 30°C, after which thiamine was added to the culture medium to switch off cdc13+ expression and the cells were incubated for an additional 5 h. Cell lysates were subjected to immunoprecipitation and immunoblot analysis as in (C). (F) Cells expressing HA-tagged Fkh2p (HM6107) were synchronized in G1 by transient temperature arrest and samples taken every 1 h upon release to the permissive temperature. Cell lysates were subjected to immunoprecipitation and immunoblot analysis as in (C). (G) DNA content of the cells in (F) was determined by flow cytometric analysis.
To test whether Fkh2p is phosphorylated on T314 or S462 in vivo, we prepared antibodies to Fkh2p peptides containing phosphorylated (p) T314 or pS462. The antibodies (anti-pT314, anti-pS462) specifically recognized the respective Fkh2p peptides containing pT314 or pS462 but not the corresponding nonphosphorylated peptides (Figure 4B). They also recognized wild-type Fkh2p but not the Fkh2p(T314A,S462A) mutant expressed in fission yeast cells (Figure 4C). Fkh2p exhibited multiple forms because of phosphorylation (Buck et al, 2004; Bulmer et al, 2004). Similarly, multiple bands appeared in Fkh2p(T314A,S462A) mutant (Figure 4C; Supplementary Figure 8), suggesting that multiple bands come from phosphorylation other than these sites.
We, therefore, next examined whether Cdc2p is required for phosphorylation of Fkh2p on T314 or S462 in vivo. We first examined a temperature-sensitive cdc2 mutant. Inactivation of cdc2+ by a temperature shift resulted in a decrease in the level of Fkh2p phosphorylation on each of these two residues (Figure 4D). We then examined a strain in which the B-type cyclin gene cig2+ is deleted and found that the level of Fkh2p phosphorylation on T314 and S462 was also decreased (Figure 4D). In addition, shut off of expression of the B-type cyclin gene cdc13+ induced a slight decrease in the level of Fkh2p phosphorylation on each of these two residues (Figure 4E). To test whether Fkh2p is phosphorylated depending on the cell-cycle stage, cells were transiently arrested in G1 by the inactivation of cdc10+ to induce cyclin degradation and released to the cell cycle (Figure 4F and G). In G1, Fkh2p was found to be dephosphorylated on T314 and S462. On release from G1, S462 was phosphorylated earlier than T314, although both of these residues were eventually phosphorylated. This may be due to the facts that the major cyclin responsible for phosphorylating these residues may be different and that the expression of the cyclin may vary during the cell cycle. On the basis of these results, we concluded that Cdc2p and the B-type cyclins Cig2p and Cdc13p are required for phosphorylation of Fkh2p on T314 and S462 in vivo.
To examine whether phosphorylation of Fkh2p on T314 affects its ability to bind to the upstream region of ste11+ containing the FLEX1 and FLEXL1 sites, we performed ChIP analysis with cells expressing phosphomimetic mutants of Fkh2p (Figure 5). The mating efficiency and the induction of ste11+ were low in GFP-tagged Fkh2p(S462E) cell-like control cells (Supplementary Figure 2; see Figure 3B and C). At 2 h after nitrogen withdrawal, the amount of Fkh2p(T314E) associated with this genomic region failed to increase compared with that of the wild-type protein. These results thus suggested that the poor mating efficiency of, as well as the impaired induction of ste11+ mRNA in, fkh2-T314E mutant cells is due to the reduced ability of the Fkh2p(T314E) mutant protein to bind to the upstream region of ste11+. Given that we showed that Fkh2p binds directly to the FLEX1 element upstream of ste11+ in vitro, we next examined the binding activity of the Fkh2p(T314E) mutant by EMSA analysis with a FLEX1 probe (Supplementary Figure 10). The binding activity of Fkh2p(T314E) was only slightly reduced compared with that of the wild-type protein. This fact suggests that an additional mechanism may operate to regulate the binding of Fkh2p by Cdc2p phosphorylation in vivo. Together, these findings suggest that, during the mitotic cycle, Cdc2p phosphorylates Fkh2p on T314, leading to the failure of its binding to the upstream region of ste11+ at least in vivo.
Figure 5.
Phosphorylation of Fkh2p on T314 reduces its binding to the FLEX1 sequence upstream of ste11+. No tagged cells (No tag, HM6) and cells expressing GFP-tagged wild-type (wt, HM5719) or T314E (fkh2-T314E, HM5912) mutant form of Fkh2p were treated and subjected to ChIP analysis with antibodies to GFP and the primer set A as in Figure 2D. Data are means±s.e. of values from three independent experiments. *P<0.013 (Student's t-test).
If Cdc2p phosphorylates Fkh2p, which in turn reduces mating efficiency, then Fkh2p functions downstream of Cdc2p. To confirm this notion, we performed a mating assay with several strains harboring a cig2 deletion (Figure 6A and B). The mating efficiency of cig2 fkh2, cig2 fkh2 fhl1, cig2 fkh2 mei4, cig2 fkh2 fhl1 mei4, cig2 fkh2-T314E, or cig2 fkh2-S462E cells was reduced compared with that of the cig2 single mutant, although the difference of the mating efficiency among cig2 fkh2, cig2 fkh2-T314E, cig2 fkh2-S462E, and cig2 cells was not due to a timing in induction of cell-cycle arrest in G1 phase (Supplementary Figures 4 and 7), suggesting that Fkh2p function overlaps with that of Fhl1p and Mei4p and that Fkh2p acts downstream of Cig2p. We also found that the induction of ste11+ mRNA occurred earlier in cig2-deleted cells than in wt cells (Figure 6C), which likely explains the enhanced mating phenotype of the former cells. However, we cannot exclude the possibility that this phenotype of cig2-deleted cells is caused by earlier arrest in G1 phase in response to nitrogen deprivation (Obara-Ishihara and Okayama, 1994; Supplementary Figures 4 and 7). The induction of ste11+ mRNA of cig2 fkh2-T314E or cig2 fkh2-S462E cells was reduced compared with those of cig2-deleted cells (Figure 6D). These results suggested that Fkh2p(314E) or Fkh2p(S462E) acts downstream of Cig2p in both mating and induction of ste11+ mRNA but not in induction of cell-cycle arrest.
Figure 6.
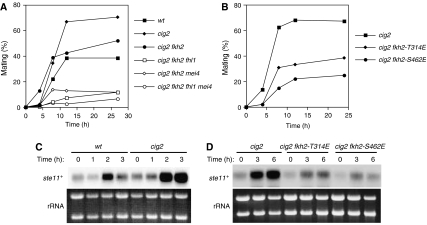
The mating efficiency of cig2 cells is reduced by mutation of forkhead transcription factors. (A) wt (HM6), cig2 (HM5555), cig2 fkh2 (HM5701), cig2 fkh2 fhl1 (HM5702), cig2 fkh2 mei4 (HM5703), or cig2 fkh2 fhl1 mei4 (HM5704) cells were assayed for mating efficiency as in Figure 1. (B) cig2 (HM5530), cig2 fkh2-T314E (HM5924), or cig2 fkh2-S462E (HM5925) cells were assayed for mating efficiency as in Figure 1. (C, D) Total RNA was extracted from cells treated as in (A) or (B) and was subjected to northern blot analysis of ste11+ mRNA.
Discussion
Initiation of sexual development in fission yeast requires the temporal coordination of the induction of many genes with cell-cycle progression, but the molecular events that underlie this coordination are not well understood. We have now provided evidence that Fkh2p is phosphorylated by Cdc2p and that this phosphorylation inhibits sexual development by preventing the induction of ste11+. Although many other processes likely also contribute to control of mating, our data establish a direct connection between initiation of mating and a key regulator of cell-cycle progression.
Our study has revealed the following. (1) The forkhead transcription factors Fkh2p, Fhl1p, and Mei4p have overlapping functions and are required for induction of ste11+ mRNA and efficient mating. Among these factors, Fkh2p plays a major role in mating. (2) Fkh2p binds to the FLEX1 sequence present upstream of ste11+, when the kinase activity of Cdc2p is low, with FLEX1 serving as a cis-acting element for Fkh2p. (3) Cdc2p phosphorylates Fkh2p on T314 and S462 both in vivo and in vitro, and phosphorylation of these residues results in inhibition of both ste11+ induction and mating; phosphorylation on T314 also inhibits the binding of Fkh2p to the FLEX1 sequence in vivo.
On the basis of these results, we propose a model for the roles of Cdc2p and Fkh2p in the control of mating. During the mitotic cycle, Cdc2p is active and phosphorylates Fkh2p on T314 and S462. Phosphorylation of T314 may inhibit the binding of Fkh2p to the FLEX1 site upstream of ste11+ in vivo. The mechanism by which phosphorylation of Fkh2p on S462 inhibits the induction of ste11+ mRNA is unknown, but it is possible that phosphorylation of this residue results in the recruitment of a repressor protein that blocks ste11+ transcription. In support of this notion, human FoxN3, which is homologous to Fkh2p, binds to a histone deacetylase complex (Scott and Plon, 2003). Nutrient exhaustion in fission yeast triggers the degradation of B-type cyclins and the consequent inactivation of Cdc2p. The absence of the kinase activity of Cdc2p allows the dephosphorylation of Fkh2p on T314 and S462 and the consequent activation of this transcription factor. Fkh2p thus binds to the FLEX1 site upstream of ste11+ and induces its transcription, thereby triggering sexual development.
Nutrient exhaustion and Cdk inactivation
We have shown that phosphorylation of Fkh2p by Cdc2p results in efficient inhibition of mating, indicating that inactivation of Cdc2p is required for efficient mating. Consistent with this notion, inhibition of cyclin degradation, downregulation of a Cdk inhibitor, or overproduction of Cig2p inhibits mating (Obara-Ishihara and Okayama, 1994; Yamaguchi et al, 1997; Kitamura et al, 1998; Stern and Nurse, 1998). It has been known that cig2 cells show enhanced mating, probably because of the upregulation of Ste11p at both mRNA (this study) and protein levels (Kjaerulff et al, 2007) in addition to the enhanced G1 arrest (Obara-Ishihara and Okayama, 1994). There are therefore two distinct mechanisms by which mating is controlled by Cdc2p. First, exhaustion of nutrients, especially that of nitrogen, induces G1 arrest, which requires inactivation of Cdc2p mediated by cyclin degradation or upregulation of a Cdk inhibitor (Yamaguchi et al, 1997; Kitamura et al, 1998; Stern and Nurse, 1998). Second, during the mitotic cycle, when nutrients are available, Cdc2p phosphorylates Fkh2p and thereby inhibits both induction of ste11+ mRNA and mating. Fkh2p is not required for the G1 arrest induced by nitrogen deprivation, suggesting that it is specifically required for ste11+ induction. In other words, Cdc2p may actively inhibit mating by phosphorylating Fkh2p. We propose that these two controls ensure that mating occurs only in G1 phase when the activity of Cdc2p is low.
It has been recently shown that Cdc2p directly phosphorylates Ste11p, which inhibits its DNA binding activity (Kjaerulff et al, 2007). Therefore, Cdc2p inhibits sexual differentiation through Ste11p at both the mRNA (this study) and post-translational levels (Kjaerulff et al, 2007). It is possible that Fkh2p-dependent mechanism is less important than the more direct Cdc2 phosphorylated mechanism. However, the switch between mitosis and meiosis is vitally important to fission yeast, and it is likely that this organism having two levels of control in regulating Ste11p expression to achieve this goal. These two controls of Ste11p by Cdc2p may reinforce to repress differentiation outside G1.
ste11+ as a target gene of Fkh2p in mating
We have shown that ste11+ is a critical target of Fkh2p in the control of mating. In addition to Rst2p and Ste11p (Kunitomo et al, 2000; Higuchi et al, 2002), Fkh2p is thus required for the induction of ste11+ mRNA and mediates its effect by binding to the upstream region of the gene. Nutrient limitation therefore triggers ste11+ expression by at least two separate signaling pathways: it reduces PKA activity, thereby activates Rst2p, leading to the production of ste11+ mRNA, and it inactivates Cdc2p, thereby activates Fkh2p, again resulting in the induction of ste11+ mRNA. The latter mechanism also contributes to the coordination of cell-cycle progression and sexual development.
Regulation of Fkh2p by phosphorylation
The sequence similarity among forkhead proteins is largely limited to the DNA-binding domain. We have now detected substantial similarity of the region surrounding the T314 phosphorylation site in Fkh2p of fission yeast to FoxN3 forkhead proteins of various species. In budding yeast, Fkh2p is phosphorylated predominantly on residues in its C-terminal region by Cdk1p in vitro (Ubersax et al, 2003; Pic-Taylor et al, 2004). Phosphorylation of at least some of these residues (S683, T697 and S771) is important for recruitment of Ndd1p (Pic-Taylor et al, 2004). These Cdk1p phosphorylation sites of Fkh2p in budding yeast do not appear to be conserved in fission yeast. In mammalian cells, FoxM1 forkhead proteins are transcriptional regulators important for cell-cycle progression similar to fission yeast Mei4p (Costa et al, 2003; Laoukili et al, 2005; Murakami-Tonami et al, 2007). FoxM1B undergoes extensive phosphorylation by several kinases, with Cdks phosphorylating T596 in the activation domain of the mouse protein, a process that is essential for the recruitment of coactivator proteins and transcriptional activity (Major et al, 2004). In addition, the transcriptional activity of FoxM1C is regulated by Cdk-mediated phosphorylation (Luscher-Firzlaff et al, 2006; Wierstra and Alves, 2006). Furthermore, Cdk2 phosphorylates FoxO1 and thereby reduces its transcriptional activity (Huang et al, 2006). Regulation of forkhead proteins by Cdk-mediated phosphorylation thus appears to be evolutionarily conserved in many eukaryotes, although the number and location of phosphorylation sites appear to vary among species.
Materials and methods
Yeast strains, media, and genetic methods
All media and standard methods were as described previously (Moreno et al, 1991). The procedures for gene disruption and N-terminal or C-terminal tagging of proteins were also as described previously (Bahler et al, 1998). The S. pombe strains used in this study are listed in Supplementary Table 1.
Primers and probes
Oligonucleotide primers and probes used in this study are listed in Supplementary Table 2.
Mating assay
After exponential growth in YE4S medium, cells were cultured in EMM2 medium for 12–16 h at 30°C to a density of 1 × 107–2 × 107 cells/ml, washed several times with EMM2 medium without a nitrogen source, resuspended at a density of 2 × 107 cells/ml, and shook gently. After the incubation for the indicated times at 30°C, samples of the cell suspension were collected and the number of zygotes was counted with the use of a light microscope. The percent mating frequencies were calculated by dividing the number of zygotes (one zygote counted as two cells) by the number of total cells.
Construction of fkh2 mutants
To construct a mutant lacking the 7-bp core sequence of FLEX1, we performed the polymerase chain reaction (PCR) with genomic DNA and the primers 1088 and 1089. The amplified DNA fragment was introduced by transformation into a strain in which the FLEX1 region was replaced with ura4+. Colonies resistant to 5-fluoroorotic acid (5-FOA) were selected, and the 7-bp deletion of FLEX1 was verified by sequencing.
To construct strains with mutations of the putative Cdc2p phosphorylation sites of Fkh2p, we performed site-directed mutagenesis with the fkh2+ coding region amplified by PCR with the primers 1167 and 1172. The primers used for mutagenesis were as follows: 1167, 1168, 1169, 1170, 1171, and 1172 for fkh2-T314A S462A S481A; 1167, 1270, 1271, 1272, 1273, and 1172 for fkh2-T314E S462E S481E; 1167, 1270, 1271, and 1172 for fkh2-T314E; 1167, 1272, 1373, and 1172 for fkh2-S462E; and 1167, 1038, 1273, and 1172 for fkh2-S481E. The plasmids were then used as templates for PCR with primers 1167 and 1172, and the resulting DNA fragments were introduced into fkh2∷ura4+ cells (HM5657) by transformation. Colonies resistant to 5-FOA were selected, and the mutations were confirmed by PCR and sequencing.
Protein extraction, immunoprecipitation, and immunoblot analysis
Protein extracts were prepared and immunoblot analysis was performed as described previously (Shimada et al, 2005). Immunoprecipitation was also performed as previously described (Shimada et al, 1999) with the exception described below. Mouse monoclonal antibody to HA (1:1000) was obtained from Roche. Polyclonal antibodies specific for phosphorylated forms of Fkh2p were generated in rabbits with the keyhole limpet hemocyanin-conjugated peptides AKTRKpTPRKRS (residues 309–319) for phospho-T314 and GSYDTIpSPYRN (residues 456–466) for phospho-S462 as antigens. Immunoblot analysis was performed with affinity-purified anti-pT314 (1:100 dilution) or anti-pS462 (1:50 dilution) after immunoprecipitation of HA-tagged Fkh2p with anti-HA antibody. Immune complexes were detected with horseradish peroxidase-conjugated goat antibodies to mouse or rabbit immunoglobulin G (both at 1:1000 dilution, Amersham) and ECL reagents (Amersham).
Isolation of RNA and northern blot analysis
Total RNA was extracted and northern blot analysis was performed as described previously (Shimada et al, 2005). The probe used for northern analysis was a 32P-labeled 1.3-kb PvuII fragment of ste11+.
Production of recombinant Fkh2p proteins
To express GST-tagged versions of Fkh2p in Escherichia coli, we amplified DNA fragments encoding Fkh2p (amino acids 305–492 containing the wt sequence or T314A, S462A, and S481A mutations; amino acids 216–330 containing the wt sequence or T314A or T314E mutations; amino acids 317–479 containing the wt sequence or the S462A mutation) by PCR using the constructs mentioned above as templates. The primers used here were as follows: 1367 and 1368 for amino acids 305–492; 1002 and 1003 for amino acids 216–330; 1411 and 1412 for amino acids 317–479. The PCR products were digested and cloned them into the corresponding sites of pGEX5X-1 (Pharmacia Biotech). The expression and purification of the GST fusion proteins were performed as described previously (Smith and Johnson, 1988).
EMSA analysis
Double-stranded oligonucleotide probes used are as follows: the primers 1141 and 1142 (FLEX1), 1179 and 1180 (FLEXL1), 1181 and 1182 (FLEXL2), 1183 and 1184 (FLEXL3), or 1143 and 1144 (TR). Purified GST–Fkh2p(216–330) (0.2 μg) was incubated for 30 min at room temperature with 20 μg of 32P-labeled probe in a total volume of 5 μl containing 50 mM Tris–HCl (pH 7.5), 10 mM KCl, 5 mM MgCl2, 15% glycerol, 1 mM dithiothreitol, and poly(dI-dC) (0.1 mg/ml). In some instances, samples were incubated for 15 min at room temperature either with a 100-fold molar excess of nonradioactive competitor oligonucleotide or with 0.9 μg of antibodies to GST (Santa Cruz Biotechnology) before addition of the probe. DNA–protein complexes were resolved by nondenaturing electrophoresis on a 5% polyacrylamide gel, which was then dried and subjected to autoradiography.
ChIP analysis
ChIP was performed as described previously (Saitoh et al, 1997) with some modifications. Immunoprecipitation was performed with anti-GFP conjugated to magnetic beads (Dynal). Primer set A comprised 1325 and 1326, and primer set B consisted of 1327 and 1328. For detection of the dis3+ region as a negative control, the primers 1329 and 1330 were used. For detection of the cdc15+ upstream region, the primers 1569 and 1570 were used. PCR amplification was performed with SYBR Green PCR Master Mix containing immunoprecipitated DNA (or total DNA) and a mixture of the three sets of primers. The amounts of the DNA were determined by the ΔΔCT method with the use of an ABI prism 7700 instrument and the primers described above.
Preparation of recombinant Cdc2/CyclinB1 complex
Baculoviruses expressing Myc- and His × 6-tagged human Cdc2 wild type or kinase dead (K33M) and CyclinB1 were generated by cotransfection of pVL1392-Cdc2 or pVL1392-CyclinB1 with the linearized baculovirus DNA (BaculoGold; BD biosciences) into Sf9 cells. After amplification of the virus, Sf9 were infected with both Cdc2 wild type and CyclinB1 viruses or Cdc2 kinase dead and CyclinB1 viruses for 72 h. Cell lysates were prepared and 0.1 mg of protein immunoprecipitated with anti-Myc antibody (Santa Cruz) was used for kinase assay.
Kinase assay
Protein extracts were prepared with glass beads in lysis buffer as described previously (Shimada et al, 1999). The kinase assay was performed as described previously (Murakami and Nurse, 1999).
Statistics
Experiments with at least three replicates were carried out and statistical analyses were performed by Student's t-test. Values of P<0.05 are considered significant.
Supplementary Material
Supplementary Information
Acknowledgments
We thank P Nurse and K Kitamura for yeast strains and K Okazaki for plasmids and helpful discussion. We also thank H Kojima and Y Katsuno for technical assistance. This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to HM).
References
- Bahler J (2005) Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet 39: 69–94 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Buck V, Ng SS, Ruiz-Garcia AB, Papadopoulou K, Bhatti S, Samuel JM, Anderson M, Millar JB, McInerny CJ (2004) Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J Cell Sci 117: 5623–5632 [DOI] [PubMed] [Google Scholar]
- Bulmer R, Pic-Taylor A, Whitehall SK, Martin KA, Millar JB, Quinn J, Morgan BA (2004) The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe. Eukaryot Cell 3: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RH (2005) FoxM1 dances with mitosis. Nat Cell Biol 7: 108–110 [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38: 1331–1347 [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P (2005) New and unexpected: forkhead meets ARF. Curr Opin Genet Dev 15: 42–48 [DOI] [PubMed] [Google Scholar]
- Higuchi T, Watanabe Y, Yamamoto M (2002) Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol 22: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ (2006) CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314: 294–297 [DOI] [PubMed] [Google Scholar]
- Kato T Jr, Okazaki K, Murakami H, Stettler S, Fantes PA, Okayama H (1996) Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett 378: 207–212 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, Toda T (2001) Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev Cell 1: 389–399 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Maekawa H, Shimoda C (1998) Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol Biol Cell 9: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S, Andersen NR, Borup MT, Nielsen O (2007) Cdk phosphorylation of the Ste11 transcription factor constrains differentiation-specific transcription to G1. Genes Dev 21: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S, Lautrup-Larsen I, Truelsen S, Pedersen M, Nielsen O (2005) Constitutive activation of the fission yeast pheromone-responsive pathway induces ectopic meiosis and reveals ste11 as a mitogen-activated protein kinase target. Mol Cell Biol 25: 2045–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Higuchi T, Iino Y, Yamamoto M (2000) A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol Biol Cell 11: 3205–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136 [DOI] [PubMed] [Google Scholar]
- Li P, McLeod M (1996) Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87: 869–880 [DOI] [PubMed] [Google Scholar]
- Luscher-Firzlaff JM, Lilischkis R, Luscher B (2006) Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett 580: 1716–1722 [DOI] [PubMed] [Google Scholar]
- MacNeill SA, Nurse P (1997) Cell cycle control in fission yeast, Schizosaccharomyces pombe. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology, Pringle JR, Broach JR, Jones EW (eds), pp 697–763. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory [Google Scholar]
- Major ML, Lepe R, Costa RH (2004) Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol 24: 2649–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Bahler J (2006) Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci USA 103: 15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM (2003) Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 316: 79–89 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P (1999) Meiotic DNA replication checkpoint control in fission yeast. Genes Dev 13: 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami-Tonami Y, Yamada-Namikawa C, Tochigi A, Hasegawa N, Kojima H, Kunimatsu M, Nakanishi M, Murakami H (2007) Mei4p coordinates the onset of meiosis I by regulating cdc25+ in fission yeast. Proc Natl Acad Sci USA 104: 14688–14693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA (1993) Targets of cyclin-dependent protein kinases. Curr Opin Cell Biol 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Obara-Ishihara T, Okayama H (1994) A B-type cyclin negatively regulates conjugation via interacting with cell cycle ‘start' genes in fission yeast. EMBO J 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD (2004) Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol Cell Biol 24: 10036–10046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P (1994) Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J 13: 5002–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Kang W, Leung B, McLeod M (2003) Ste11p, a high-mobility-group box DNA-binding protein, undergoes pheromone- and nutrient-regulated nuclear-cytoplasmic shuttling. Mol Cell Biol 23: 3253–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar B, Grallert A, Olah E, Szallasi Z (1999) Deletion of the sep1(+) forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem Biophys Res Commun 263: 465–474 [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J (2004) Periodic gene expression program of the fission yeast cell cycle. Nat Genet 36: 809–817 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M (1997) Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90: 131–143 [DOI] [PubMed] [Google Scholar]
- Scott KL, Plon SE (2003) Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol Cell Biol 23: 4522–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Namikawa-Yamada C, Nakanishi M, Murakami H (2005) Regulation of Cdc2p and Cdc13p is required for cell cycle arrest induced by defective RNA splicing in fission yeast. J Biol Chem 280: 32640–32648 [DOI] [PubMed] [Google Scholar]
- Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai KK, Shimoda C, Nojima H (1999) Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell 10: 3991–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P (1996) Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10: 2276–2288 [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- Stern B, Nurse P (1997) Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B- p34cdc2 kinase. EMBO J 16: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B, Nurse P (1998) Cyclin B proteolysis and the cyclin-dependent kinase inhibitor rum1p are required for pheromone-induced G1 arrest in fission yeast. Mol Biol Cell 9: 1309–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A, Iino Y, Wantanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11 encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 5: 1990–1999 [DOI] [PubMed] [Google Scholar]
- Szilagyi Z, Batta G, Enczi K, Sipiczki M (2005) Characterisation of two novel fork-head gene homologues of Schizosaccharomyces pombe: their involvement in cell cycle and sexual differentiation. Gene 348: 101–109 [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N (1995) Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J 14: 6193–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 25: 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J (2006) FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3alpha. Biochem Biophys Res Commun 348: 99–108 [DOI] [PubMed] [Google Scholar]
- Xue-Franzen Y, Kjaerulff S, Holmberg C, Wright A, Nielsen O (2006) Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genomics 7: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Murakami H, Okayama H (1997) A WD repeat protein controls the cell cycle and differentiation by negatively regulating Cdc2/B-type cyclin complexes. Mol Biol Cell 8: 2475–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M (1996) The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci 21: 18–22 [PubMed] [Google Scholar]
- Yamamoto M, Imai Y, Watanabe Y (1997) Mating and sporulation in scizosaccharomyces pombe. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology, Pringle JR, Broach JR, Jones EW (eds), pp 1037–1106. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory [Google Scholar]
- Yamano H, Kominami K, Harrison C, Kitamura K, Katayama S, Dhut S, Hunt T, Toda T (2004) Requirement of the SCFPop1/Pop2 ubiquitin ligase for degradation of the fission yeast S Phase cyclin Cig2. J Biol Chem 279: 18974–18980 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information