Abstract
EmrE is a small H+-coupled multidrug transporter in Escherichia coli. Claims have been made for an antiparallel topology of this homodimeric protein. However, our own biochemical studies performed with detergent-solubilized purified protein support a parallel topology of the protomers. We developed an alternative approach to constrain the relative topology of the protomers within the dimer so that their activity can be assayed also in vivo before biochemical handling. Tandem EmrE was built with two identical monomers genetically fused tail to head (C-terminus of the first to N-terminus of the second monomer) with hydrophilic linkers of varying length. All the constructs conferred resistance to ethidium by actively removing it from the cytoplasm. The purified proteins bound substrate and transported methyl viologen into proteoliposomes by a proton-dependent mechanism. A tandem where one of the essential glutamates was replaced with glutamine transported only monovalent substrates and displayed a modified stoichiometry. The results support a parallel topology of the protomers in the functional dimer. The implications regarding insertion and evolution of membrane proteins are discussed.
Keywords: ion-coupled transport, membrane protein evolution, membrane protein structure, membrane protein topology, multidrug transporters
Introduction
EmrE belongs to the small multidrug resistance (SMR) family (Paulsen et al, 1996; Bay et al, 2007) of multidrug transporters and is a unique experimental paradigm for biochemical and biophysical studies of membrane-based, ion-coupled transporters due to its size, stability and its retention of function when solubilized in detergent (Yerushalmi et al, 1995; Yerushalmi and Schuldiner, 2000b; Tate et al, 2001). Study of this small, 110-residue multidrug transporter of Escherichia coli has provided valuable information for the understanding of the coupling mechanism of ion-coupled transporters (Yerushalmi and Schuldiner, 2000a, 2000b; Soskine et al, 2004; Adam et al, 2007). However, the structural information that became available for this protein in recent years has been in conflict with the existing biochemical knowledge (Ma and Chang, 2004; Pornillos et al, 2005). Even though the two X-ray crystallography papers for the protein have recently been retracted (Chang et al, 2006), their publication sparked a controversy regarding the relative topology of the protomers in the functional dimer and this controversy is still ongoing (Fleishman et al, 2006; Soskine et al, 2006; Rapp et al, 2007).
The claim for an antiparallel topology was supported by a reinterpretation of the electron density maps of 2D crystals of EmrE that showed that parts of the structure are related by quasi-symmetry (Ubarretxena-Belandia et al, 2003). A Cα-model of the transmembrane region was constructed by considering the evolutionary conservation pattern of each helix (Fleishman et al, 2006). Much of the biochemical data on EmrE seem to agree with this model. Von Heijne and co-workers designed experiments to support the concept of dual topology. They showed that the topology of the EmrE fusion proteins in the membrane is sensitive to the distribution of positive charges in the protein (Rapp et al, 2006). In addition, manipulation of the positive charges generates a set of mutants, some with Cout, others with Cin apparent topology (Rapp et al, 2006, 2007). The Cin and Cout mutants, bearing three mutations each, do not confer resistance to ethidium. The authors conclude that this is due to the modified topology. However, it was not yet tested whether the introduction of positive charges may affect homodimerization or recognition of the positive substrates. Coexpression of the inactive mutants restores the ethidium bromide resistance phenotype to the same level as seen for wild-type EmrE (Rapp et al, 2007). A possible interpretation of this finding is that coexpression results in the generation of a functional, antiparallel heterodimer. However, a more direct biochemical analysis of the mutants is needed to support it. In addition, six mutations (three in each monomer) may have indeed pushed EmrE to form a hetero-oligomer. Whether the protein that has not been mutated can form functional homodimers with an antiparallel topology still remains to be directly demonstrated. Moreover, the existence of homodimers with antiparallel orientation would pose a problem for the insertion machinery of membrane proteins. Identical protomers with Cout and Cin topologies would insert exactly at a 1:1 ratio to prevent the existence of unpaired units. How this ratio is controlled and how the assembly of such a dimer is achieved is very difficult to envisage with our present knowledge.
Since an apparent antiparallel topology of a homodimer has many intriguing implications regarding biogenesis, insertion and evolution of ion-coupled transporters, the topic has already attracted much attention (Bowie, 2006; Pornillos and Chang, 2006; Rapp et al, 2006). Our own biochemical studies support a symmetrical relationship between the monomers in the EmrE homodimer (reviewed in Schuldiner, 2007). These studies were performed with the protein after solubilization and purification. To provide an approach that combines in vivo and in vitro studies, we designed a series of genetic fusions where the monomers are connected tail to head (C-terminus of the first to N-terminus of the second monomer) by means of defined linkers that are not compatible with an antiparallel topology either because they are too hydrophilic or too short (Figure 1). The fused dimers (tandems) confer resistance to ethidium by actively removing it from the cytoplasm. The purified proteins bound the high-affinity substrate tetraphenylphosphonium (TPP+) and upon reconstitution transported methyl viologen (MV2+) into proteoliposomes by a proton-dependent mechanism. The contention that the tandem is the functional unit is supported by negative dominance studies. In these studies we also show that a single mutation changes the stoichiometry of the transporter and its ability to recognize divalent substrates.
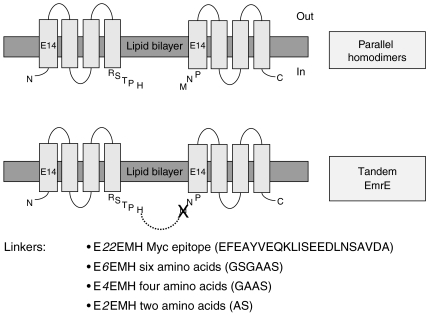
Figure 1.
Design of the EmrE tandems. Construction of EmrE to EmrE fusions is shown schematically; location of essential carboxyl residue E14 is shown; transmembrane helices are drawn as boxes. The N-terminus of the second EmrE protomer is two amino acids long (NP, the first methionine was removed) and was connected to the C-terminus (RSTPH) of the first EmrE protomer with a variety of linker sequences (displayed above). Linker sequences were chosen in a manner that enforces the parallel orientation of the two EmrE protomers (with respect to the membrane plane) within each tandem polypeptide chain. This was achieved either by very hydrophilic linker (E22EMH) or by linkers that are too short to cross the membrane plane (E2EMH, E4EMH, E6EMH). A ‘negatively dominated' tandem was built using the six-amino-acid linker and substituting the essential glutamate 14 in the first monomer with glutamine (Q6EMH).
Results
Genetically fused dimers of EmrE (Tandem EmrE) confer resistance to ethidium by actively removing it from the cytoplasm
Tandem EmrE was constructed by genetic fusion of the monomers tail to head with linkers of various lengths (Figure 1). The N-terminus of the second EmrE protomer is two amino acids long (NP, the first methionine was removed) and was connected to the C-terminus (RSTPH) of the first EmrE protomer with a variety of linker sequences (Figure 1). Linker sequences were chosen in a manner that enforces the parallel orientation of the two EmrE protomers (with respect to the membrane plane) within each tandem polypeptide chain. This was achieved either by very hydrophilic linker (E22EMH) or by linkers that are too short to cross the membrane plane (E2EMH, E4EMH, E6EMH). The activity of each of the tandem proteins has been tested both in vivo and in vitro. In vivo the resistance conferred by each of the proteins was assessed by testing the ability of cells expressing them to grow under otherwise non-permissive conditions. This was achieved in solid media containing ethidium (100 μg/ml) in which 5 μl of logarithmic dilutions of an overnight culture were spotted. Cells carrying the vector plasmid without any insert cannot grow in this medium at any of the dilutions tested (Figure 2A, pT7-7). Cells expressing either EmrE or the various tandems were able to grow at each of the dilutions. While all the tandems conferred resistance to ethidium, those with shorter linkers were slightly less effective. At the high dilutions, the single colonies observed displayed a similar size indicating similar growth rates (Figure 2A). As expected, all the cells grew to a similar degree in control plates containing only ampicillin (Figure 2A, control).
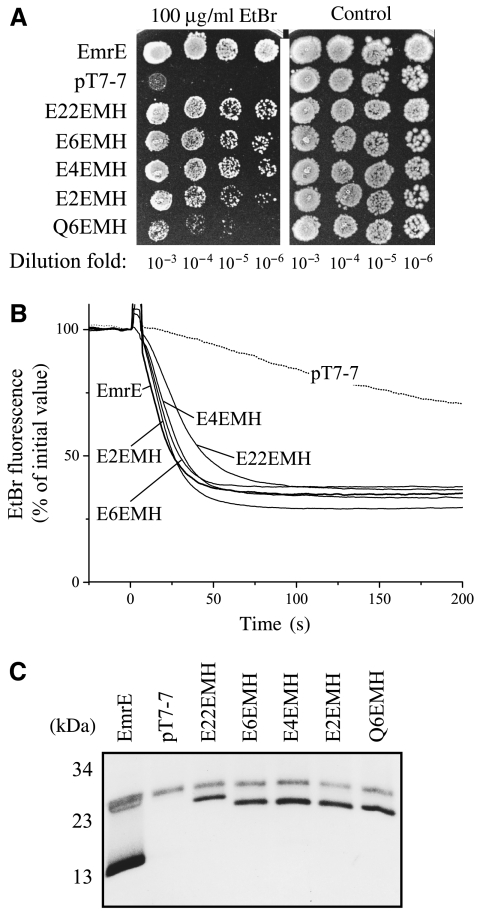
Figure 2.
EmrE tandems are functional transporters in vivo. (A) Growth phenotype of cells expressing tandems. E. coli DH5α cells transformed with either pT7-7-EmrE (EmrE), pT7-7 (vector) or with the indicated tandems were grown overnight at 37°C in LB–ampicillin medium. A 5-μl volume of 10−3–10−6 dilutions of the culture was spotted on the LB–ampicillin plates containing 30 mM BisTris propane, pH 7.0, with or without 100 μg/ml ethidium bromide. Growth was analyzed after overnight incubation at 37°C. (B) Ethidium efflux activity of the tandems. E. coli HMS 174 cells harboring the plasmid pT7-7 containing EmrE or tandems with varying linker length, or pT7-7 alone were grown in minimal medium A to mid-logarithmic phase and then induced with 0.5 mM IPTG for 2 h. The cells were then transferred to minimal medium containing ethidium bromide and CCCP, as described under Materials and methods. After 60 min of incubation at 37°C, the cells were washed quickly in CCCP-free medium and monitored for fluorescence (excitation at 525 nm, emission at 585 nm). Glucose (20 mM) was added to initiate the active efflux of ethidium. (C) Comparison of the expression levels of the EmrE and tandems in HMS 174 cells after induction. His-tagged proteins were purified as described under Materials and methods and analyzed by SDS–PAGE.
To further evaluate the ability of the various constructs to actively remove ethidium from the cytoplasm, we assayed active transport of ethidium in intact cells. In these studies, after induction of expression of the various constructs, cells were starved in the presence of ethidium by incubation in the absence of an energy source and in the presence of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP). After removal of the uncoupler, transport was started by addition of glucose. Although ethidium is present both in the medium and the cells, the fluorescence observed originates almost exclusively from the intracellular ethidium bound to nucleic acid. Upon addition of glucose, a quick decrease in the fluorescence is observed, which represents removal of ethidium from the cell against its concentration gradient (Figure 2B). This decrease was completely prevented when the cells were resuspended in a medium with CCCP (not shown). The rate of extrusion in the cells bearing wild-type EmrE or any of the four tandems tested was very similar and reached very similar equilibrium after about 2 min. In cells transformed with empty vector, removal of ethidium is much slower and is most likely mediated by multidrug transporters other than EmrE. Expression levels of the tandems were assessed after purification and were found to be very similar (Figure 2C). No proteolytic products were detectable and a visible impurity in all cells is due to the metal-binding protein YodA (David et al, 2002, 2003). Taken as a whole the results described demonstrate that all the tandems are capable of active extrusion of ethidium.
Tandem EmrE binds the high-affinity substrate TPP+ and transports MV2+ into proteoliposomes
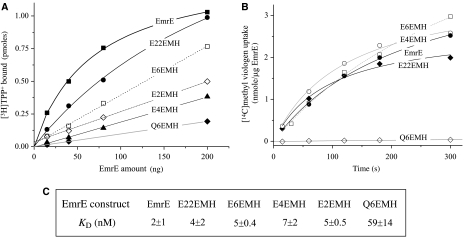
The tandem proteins were purified in order to further characterize their activity and to rule out the possibility that the transport observed in vivo is due to proteolytic products. The results of the binding experiments are summarized in Figure 3A. All the EmrE tandem fusions bound TPP+, although to somewhat lesser degrees than the wild-type EmrE due to some decrease of the affinities from 2 to 4–7 nM (Figure 3C). Tandem EmrE contains two essential glutamates (one in each protomer) that correspond to the membrane-embedded glutamate 14 in wild-type EmrE. EmrE tandem where one of the essential glutamates was substituted by glutamine (Q6EMH) displays a profoundly lower binding affinity to TPP+ (59 nM; Figure 3C). The results are in agreement with previous findings where it was shown that both Glu14 within the EmrE dimer are required for assembly of the high-affinity substrate-binding site (Rotem et al, 2001).
Figure 3.
Substrate binding and transport activity of the tandems. (A) [3H]TPP+-binding activity of the purified tandems. EmrE and tandems were purified as described under Materials and methods and assayed for the ability to bind the high-affinity substrate TPP+ with 2.5 nM of [3H]TPP+ in 0.08% DDM Na buffer. (B) [14C]MV2+ uptake activity of the reconstituted tandems. EmrE (filled circles), E22EMH (filled diamonds), E6EMH (hollow squares), E4EMH (hollow circles) and Q6EMH (hollow diamonds) were purified, reconstituted and assayed as described under Materials and methods. (C) TPP+ binding affinities of the tandems used in this study, assayed by [3H]TPP+ binding and determined as previously described (Rotem et al, 2001).
To further test the functionality of the purified tandem proteins, we reconstituted them into proteoliposomes and assayed their ability to catalyze proton gradient-driven uptake of MV2+. Upon generation of a pH gradient, the rates of uptake and the level of accumulation of MV2+ detected in proteoliposomes reconstituted with either one of the ‘wild-type' tandems were very similar to each other and even slightly higher than the rates observed for wild-type EmrE proteoliposomes (Figure 3B). These findings allow us to conclude that tandem EmrE is a functional transporter.
Probing packing and topology of the protomers in tandem EmrE
The linkers were designed to be either short enough or hydrophilic enough in order not to cross the membrane plane (Figure 1). The results described above demonstrate that the linkers have no significant effect on the activity of the protein and therefore most likely do not affect the overall packing of the dimer, a prerequisite for function. However, since manipulation of charge bias (Gafvelin and von Heijne, 1994) or lipid composition (Bogdanov et al, 2002; Zhang et al, 2003) may affect the integration of transmembrane segments and may induce generation of semi-inverted topologies, we tested whether the insertion of the linkers affected the packing and the relative topology of the protomers in tandem EmrE. The membrane domain of EmrE is completely resistant to a battery of proteases, including proteinase K. After exposure of EmrE to either chymotrypsin or proteinase K, only the C-terminal tag is digested (Figure 4A, lanes 1–3), even at prolonged overnight digestions. When E22EMH is treated with either one of the enzymes, the linker and the tag are digested by the protease treatment (Figure 4A, lanes 4–6) supporting the contention that the packing in the membrane is undistinguishable from that of EmrE and that the linker is completely exposed to the protease. As expected, only proteinase K digests E6EMH since there are no sites for either one of the other enzymes (not shown). In addition, to support the contention that the protomers in E22EMH are in a parallel orientation, we treated the protein with hexamethylene diisocyanate (HMDC), a reagent previously shown to react with Lys22 in the first loop of EmrE (Soskine et al, 2002). The cross-linking is dependent on the proximity of the two monomers, since it is not observed after SDS treatment that induces dissociation of the dimer (Soskine et al, 2002). The results of this experiment are shown in Figure 4B. EmrE (lanes 4–6) and E22EMH (lanes 1–3) were digested with proteinase K as above so that the tags and the linker were digested and yielded a construct shorter than the tagged EmrE (lanes 5 and 2, respectively). After digestion, cross-linking resulted in both cases in the production of a polypeptide with an apparent molecular mass of about 22 kDa, smaller than the tagged dimer (lanes 3 and 6). These experiments support the contention that the relative topology of the protomers in tandem EmrE is parallel and that a dimer with parallel topology is functional.
Figure 4.
Probing packing and topology of the protomers in E22EMH. (A) Membranes bearing radiolabeled EmrE-His tagged (EmrE, lanes 1–3) and E22EMH (lanes 4–6) were incubated with the corresponding proteases as described under Materials and methods. In EmrE, the MycHis tag is digested, producing a polypeptide of about 10 kDa (lanes 2 and 3); E22EMH is digested to produce polypeptides with a similar molecular mass as a result of the digestion of the tag and the linker. (B) After digestion with proteinase K, both proteins were treated with the cross-linker HMDC as described under Materials and methods. In both cases, cross-linking is very effective (lanes 3 and 6), producing a polypeptide slightly smaller that the original tandem because the tag has been digested.
Probing interdimeric interaction by ‘monomer swapping' with E14C, an inactive EmrE mutant
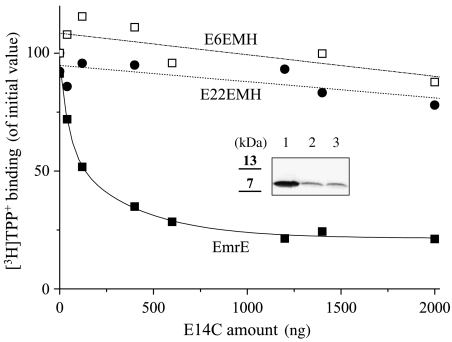
EmrE dimers solubilized with DDM (n-dodecyl-β-maltoside) may be reversibly dissociated by a short heat treatment. If a mixture of two mutant proteins is subjected to this procedure, mixed dimers are formed upon cooling (‘monomer swapping') (Rotem et al, 2001; Sharoni et al, 2005). Replacement of Glu14 with an uncharged amino acid completely abolishes binding, but the mutants can form mixed dimers with other functional EmrE protomers. The mixed dimers display more than 20-fold lower affinity toward TPP+, a phenomenon defined as a negative dominant effect of the inactive mutant on the functional one (Rotem et al, 2001; Elbaz et al, 2004). We used this experimental paradigm to test the contention that the functional unit is within the tandem rather than derived from interdimeric interaction. Increasing amounts of the well-characterized mutant EmrE E14C were added to wild-type EmrE and to two tandems with the longest linkers, E22EMH and E6EMH, and the mixtures were subjected to the monomer swapping procedure and assayed for TPP+-binding activity. The TPP+ concentration used in this assay, 2.5 nM, equals the KD of EmrE. Therefore, inhibition of binding reflects the formation of mixed dimmers, since the contribution of heterodimers EmrE/E14C is insignificant due to their lower affinity (Figure 5). After monomer swapping with a 50-fold excess of the E14C mutant, the binding activity of the wild-type protein was inhibited by more than 80% (Figure 5, closed squares). Under the same conditions, binding activity of both tandems is only barely affected (∼15%; Figure 5, closed circles, open squares).
Figure 5.
The tandem as the functional unit: it does not take part in the intermolecular interactions with other EmrE molecules. Increasing amounts of the membranes containing the indicated amounts of inactive EmrE mutant E14C were solubilized in 1% DDM Na-buffer and mixed with the solubilized membranes containing ∼40 ng of EmrE (WT), E22EMH or E6EMH. After incubation at 80°C (10 min), the mixture was transferred to 4°C and assayed for [3H]TPP+ binding as described under Materials and methods. Inset: about 100 ng of His-tagged EmrE (1), E22EMH (2) and E6EMH (3) were mixed with [35S]methionine-labeled, cysteine-less untagged EmrE solubilized as above, mixture was treated as described under Materials and methods, after pull down with Ni-NTA, radiolabeled untagged protein was analyzed by SDS–PAGE and visualized with a Fujifilm FLA-3000 imaging system.
Interdimeric interaction that does not result in functional changes was also ruled out in a parallel experiment where unlabeled tagged proteins were used as ‘bait' to pull down untagged radiolabeled wild-type protein (Figure 5, inset). The generation of heterodimers between tagged and radiolabeled untagged EmrE is evidenced by the fact that after the swapping procedure, most of the untagged protein is associated with the Ni–NTA (Ni2+–nitrilotriacetic acid) beads. Neither E22EMH nor E6EMH pull down more than one-tenth of the radiolabeled protein.
Tandem EmrE with a single Glu14: modified stoichiometry and specificity
EmrE transports monovalent and divalent substrates with the same stoichiometry of 2H+/substrate. Thus, transport of monovalent substrates involves charge movement (i.e., electrogenic), whereas transport of divalent substrate does not. Both protons are released from Glu residues at position 14. Heterodimers were generated by monomer swapping to test whether two glutamyl residues are required for substrate recognition and binding (Rotem et al, 2001). Heterodimers with only one Glu residue bind substrate with a lower affinity (Rotem et al, 2001). Previously, reconstitution of these heterodimers could not yield reliable results, since our protocol includes a step where the proteins are treated with octyl glucoside, a detergent that increases the fraction of monomeric protein and allows reshuffling of the preformed heterodimers (Soskine et al, 2006).
Therefore, to further study this question, a tandem was generated in which one of the Glu14 was replaced with glutamine (Q6EMH). The tandem also allowed testing the effect of such replacement in vivo. It confers decreased but significant resistance to ethidium (Figure 2A). In addition, it actively removes ethidium albeit at rates slower than those of E6EMH or any of the other tandems (Figures 2B and 6A). Furthermore, the level of removal as judged from the equilibrium value is lower than that achieved by the other tandems. The results demonstrate that a tandem protein with only one carboxyl is capable of catalyzing the transport of ethidium. The lower levels of removal and resistance may be due to the different stoichiometry dictated by the fact that since there is only one Glu14 per dimer, we may expect that only 1H+ is exchanged per substrate molecule rather than two for the protein with two carboxyls. To test this contention, we purified Q6EMH and characterized its activity in vitro. As expected from monomer swapping experiments, the purified protein binds TPP+ (Figure 3A) with an affinity significantly lower than the wild-type or the other tandems (KD=59 nM).
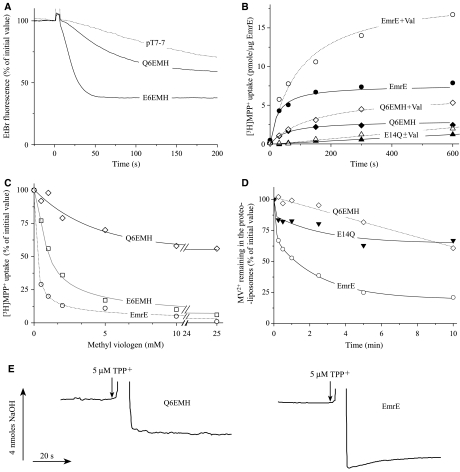
Figure 6.
Negative dominance by design: a tandem with the single Glu 14. (A) Ethidium efflux activity in whole cells expressing Q6EMH and E6EMH was measured as described in Figure 2. (B) [3H]MPP+ uptake by EmrE, EmrE E14Q and Q6EMH was tested with ammonium-loaded proteoliposomes (2 μl, containing ∼0.5 μg of protein). They were diluted into an ammonium-free medium with pH 8.5 containing 1 μM [3H]MPP+, 140 mM K2SO4 with or without addition of 100 nM valinomycin. Hollow triangles display MPP+ incorporated into proteoliposomes containing E14Q in the presence of 100 nM valinomycin. (C) Inhibition of [3H]MPP+ uptake by MV2+. Uptake was measured as above in the presence of 100 nM valinomycin and the indicated amounts of MV2+. (D) [14C]MV2+ efflux activity of EmrE, EmrE E14Q and Q6EMH. K2SO4-containing proteoliposomes were assayed as described under Materials and methods. (E) Substrate induced proton release from EmrE and Q6EMH. The pH of the unbuffered solution containing 105 μg Q6EMH or EmrE was monitored by absorbance of Phenol Red as described under Materials and methods. Addition of 5 μM TPP+ is indicated by the arrow. Addition of 4 nmol of NaOH was to estimate the amount of protons released from the protein upon substrate binding.
The reconstituted proteoliposomes were assayed for transport of two different substrates: the monovalent 1-methyl-4-phenylpyridinium (MPP+) (Figure 6B) and a divalent one, MV2+ (Figure 3B). Wild-type EmrE and E6EMH accumulate both the divalent (Figure 3B) and the monovalent substrates (Figures 3B and 6B). As expected from an electrogenic exchange of 2H+/MPP+, transport of the latter is stimulated by a membrane potential generated by valinomycin in the presence of external potassium (Figure 6B). Strikingly, Q6EMH transports MPP+ but does not catalyze accumulation of MV2+ (Figures 3B and 6B, respectively). To distinguish between the effect of a modified stoichiometry and loss of recognition of the divalent substrate by a protein with a single Glu14, we assessed its ability to inhibit transport of MPP+ (Figure 6C). While 10 mM MV2+ significantly inhibits MPP+ accumulation in EmrE and in E6EMH (IC50 <1 mM), it only partially inhibits transport catalyzed by Q6EMH and inhibition plateaus above 10 mM to a level of about 40%. To further test the finding that the tandems with only one Glu14 are impaired in the recognition of divalent substrates, we assayed downhill efflux of MV2+ from proteoliposomes preloaded with the substrate, and diluted to a medium without it. Wild type (Figure 6D) and E6EMH (not shown) rapidly catalyze equilibration, while Q6EMH or E14Q, a mutant with no Glu14, do not facilitate the downhill movement (Figure 6D).
We conclude that two carboxyls are needed for high-affinity binding of TPP+ and transport of the divalent substrate MV2+, but one is enough to bind substrate with a lower affinity, to transport ethidium and to confer limited resistance to it. Valinomycin had a small but reproducible effect on transport of MPP+ in Q6EMH proteoliposomes (Figure 6B). Since such stimulation is not expected in an electroneutral process, we further tested the exchange stoichiometry by measuring directly substrate-induced release of H+ from the detergent-solubilized transporter. In such experiments, purified protein has been shown to release 2H+/dimer (Soskine et al, 2004). In Figure 6E the substrate-induced H+ release was assayed with identical dimer concentration of EmrE and Q6EMH. As expected, only about half of the amount of protons is released from Q6EMH upon substrate addition. The average release as determined using six different concentrations of proteins between 30 and 105 μg was 1.9±0.5 nmol for EmrE and 1.2±0.4 for Q6EMH.
Discussion
The functional properties of EmrE were extensively studied by biochemical methods (Schuldiner, 2007), but the structural information available for this protein thus far is derived largely from modeling studies. X-ray crystallography studies (Ma and Chang, 2004; Pornillos et al, 2005), since retracted (Chang et al, 2006), sparked a controversy about the orientation of the protomers in the EmrE dimer. Even after the retraction, the concept of the antiparallel topology has survived, probably because of its appeal to explain evolution of membrane protein topology. Nevertheless, numerous experimental lines of evidence such as several biochemical studies, direct topology assessment with impermeable thiol reagents or antibodies and comprehensive cross-linking studies strongly suggest that EmrE homodimer has the parallel orientation of the monomers with the N- and C-termini of the protein exposed to the cytoplasm milieu (reviewed in Schuldiner, 2007). In addition, a question that has not been addressed in the papers that suggest antiparallel topology of two protomers in a homodimer is how the insertion mechanism of membrane proteins deals with the uncertainty of the topology. Is this a stochastic process where some of the units will remain unpaired while others find the ‘wrong' partner?
Our approach to this controversy has been to enforce the parallel orientation on the two EmrE monomers within the dimer and observe the functional consequences of this constraint. Previously we showed that EmrE, cross-linked in vitro in a manner that requires the parallel orientation of the monomers, is functional (Soskine et al, 2006). In this work we report the properties of parallel EmrE dimers fused genetically with linkers of various lengths between the C-terminus of the first EmrE protomer and the N-terminus of the second protomer (tail to head). The linkers were designed to be either short enough or hydrophilic enough in order not to cross the membrane plane (Figure 1). The generation of a set of different linkers allowed us to separate between their impact on activity derived from imposed parallel orientation of the monomers (effect should be prominent in all of the tandems constructed) and the distance constrain caused by linker length (should be more pronounced in the shorter linker tandems). The genetic fusions are not simply an extension of the cross-linking studies. They provide a unique and powerful tool for studying activity in vivo, to avoid possible artifacts of detergents and to construct mutants with well-controlled and defined genotypes.
All the tandems tested are functional transporters: they rendered bacteria resistant to the non-permissive toxic environment and catalyzed ethidium efflux in the whole cell. The purified, detergent-solubilized tandem proteins bind the high-affinity substrate TPP+ and display active uptake of MV2+ when reconstituted in proteoliposomes. A slight decrease in the affinity to TPP+ is the only significant change observed in the different constructs: shorter linkers displayed lower affinities. Purification of the functional proteins eliminated the possibility that the in vivo phenomena arise from proteolysis of the linker between the two protomers, since our preparations are free of any visible monomeric EmrE.
Although EmrE conferred slightly better resistance than the tandem EmrE fusions, the positive phenotype provides a strong indication that the tandem is functional in vivo. Even though a positive phenotype is a powerful tool, we performed a more extensive study both in vivo and in vitro in order to rule out effects of expression levels or proteolysis. Upon induction it is possible to detect and quantitate expression and assay the ability of the tandems to catalyze ethidium efflux, and this proved to be practically identical to that of EmrE. In our work with the purified proteins, the only detectable functional effect of the genetic fusion was a moderately negative impact on the affinity to TPP+.
The simplest interpretation of our results is that the parallel orientation of the monomers within the EmrE dimer describes the functionally and physiologically relevant state. To support this interpretation, we tested several possible scenarios. Manipulation of charge bias (Gafvelin and von Heijne, 1994) or lipid composition (Bogdanov et al, 2002; Zhang et al, 2003) may affect the integration of transmembrane segments and may induce generation of semi-inverted topologies. However, when activity was tested, the inversions have a severe effect on activity so that at least the coupling mechanism is affected (Bogdanov et al, 2002; Zhang et al, 2003). To test whether the insertion of the linkers affected the packing of the tandem proteins, we showed that their membrane domain is, like in native EmrE, completely resistant to a battery of proteases, including proteinase K. Only the C-terminal tag and the linker are digested by the protease treatment, supporting the contention that the packing in the membrane is undistinguishable from that of EmrE, and that the linker is completely exposed to the protease. In addition, after digestion, the protomers are cross-linked with HMDC, a reagent we previously showed to react with Lys22. These experiments support the contention that the relative topology of the protomers in tandem EmrE is parallel and that a dimer with parallel topology is functional.
An unlikely but still possible scenario would include formation of a dimer of tandems—that would require ‘protomer swapping' between the two EmrE tandems that have been inserted into bacterial membrane with the opposite orientations, forming antiparallel functional contact. In order to rule out this possibility, we showed the lack of biochemical negative dominance and no ‘pull down' using the [35S]Met-labeled untagged EmrE as prey. A higher organized oligomeric form was shown to have a very low affinity of interaction and could not be detected directly (Elbaz et al, 2004). In other words, the dimer–dimer affinity is so low that it cannot be responsible for activity and would certainly not survive the heat treatment used in the negative dominance and pull-down experiments. In addition, the possible dimer–dimer interaction was a parallel one (Elbaz et al, 2004). Furthermore, we designed a tandem with negative dominance within the dimer: one of the essential glutamates (corresponding to glutamate 14 in the EmrE wild type) was substituted for glutamine. Functional characterization of the tandem with a single essential glutamate Q6EMH revealed that it renders the bacteria resistant to ethidium much less effectively than any of the ‘wild-type' tandems that have two intact essential glutamates, but the low resistance displayed is higher than observed in cells carrying the vector alone. Similarly, in the ethidium efflux experiments, Q6EMH displays a behavior intermediate between the vector control and the E6EMH. In vitro characterization of Q6EMH implied that the protein with one Glu14 has a decreased affinity to TPP+, does not recognize divalent substrates and displays a modified stoichiometry of 1H+/substrate. Our findings suggest that both slower rates and smaller gradients are related to the change in the stoichiometry of the transport. If a tandem with the single essential glutamate per functional unit exchanges one substrate per H+ during the catalytic cycle, as opposed to wild-type EmrE that transports 2H+/substrate, the expected gradients will be 10-fold lower. In addition, the protein with a single Glu14 has an impaired recognition of substrates with two charges, since MV2+ does not inhibit transport of singly charged substrates and it is not transported downhill in efflux experiments. If tandem recombination were to happen, Q6EMH would give rise to a mixture of the protomer dimers (QQ, QE and EE) with different functional properties reflecting constructs with two, one or none of the essential glutamates per functional unit. The properties of this mutant strongly support the contention that the functional unit is the genetically fused dimer.
EmrE may have been in an evolutionary junction where the need to expand the range of substrates of this multidrug transporter could only be met with the larger number of combinations possible in heterodimeric proteins. The evolutionary pressure may have selected for SMR heterodimers that can yield a larger number of permutations and originated from gene duplication of the more ancient homodimers. In this manner, one protein with only a slightly modified sequence may extend the range of the substrate specificity. A bioinformatic analysis of SMR heterodimers suggests that in most of them the distribution of positive charges is different in a way that would predict a topology of opposite direction for each protomer, that is, antiparallel (Rapp et al, 2006). After gene duplication, a relatively small number of mutations would allow them to assume either parallel or antiparallel orientation of the monomers within the heterodimer (Kikukawa et al, 2006; Rapp et al, 2007). Topology evolution of larger proteins with two oppositely oriented membrane domains can now be visualized starting from gene duplication, mutations and then fusion of SMR heterodimers. The case for antiparallel topology is suggested by the studies from von Heijne's laboratory (Rapp et al, 2006, 2007) and by the low-resolution CryoEM structure that was recently used to derive a Cα-model structure (Fleishman et al, 2006). The Cα-model agrees with much of the biochemical data and indeed most of the positions that were identified as affecting substrate translocation are located around the substrate-binding cavity. However, the functionality of EmrE with an antiparallel orientation of the monomers has not yet been biochemically demonstrated.
If antiparallel homodimers were to exist, this would pose intriguing questions about the insertion and assembly of these proteins in the membrane. In addition many experimental findings are consistent with the fact that EmrE with a parallel arrangement of the protomers in the dimer is fully functional. Some experimental findings are suggestive of the possibility that a few mutations in the hydrophilic loops transform a functional parallel homodimer to a functional antiparallel heterodimer (Rapp et al, 2007) and vice versa (Kikukawa et al, 2006). If this indeed will be supported by further biochemical work, it will open a fascinating question of what is the minimal requirement for catalysis of ion-coupled transport and for interaction of the protomers. In such a case, the binding cavity of the parallel and antiparallel dimer would be very different but still would keep one basic component: two charges in a highly hydrophobic environment formed by, in the case of EmrE, six aromatic residues. Is this enough to ensure the vectorial movements of protons and substrates? This is an intriguing question that awaits more detailed studies.
Materials and methods
Bacterial strains, plasmids and mutagenesis
E. coli DH5α (Invitrogene Inc.), HMS 174 (Novagen) and TA15 (Goldberg et al, 1987) strains were used throughout this work. The TA15 strain was previously transformed with plasmid pGP1-2, which codes for the T7 polymerase under the inducible control of the λ PL promoter (Tabor and Richardson, 1985). The plasmids used for EmrE gene expression are pT7-7 (Tabor and Richardson, 1985) derivatives with an MycHis tag (for simplicity it will be called EmrE throughout this paper unless otherwise indicated) (Muth and Schuldiner, 2000).
The genetic fusions were constructed in two steps: first the pT7-7 plasmid was digested by BamHI and HindIII and ligated to the product of a PCR reaction where MycHis-tagged EmrE (EMH) was used as a template, with primers bearing the same sites at the ends. The resulting plasmid was then digested with NdeI and BamHI and ligated to the product of a PCR reaction where untagged EmrE was used as a template, with primers designed to eliminate termination and bearing the same sites at the ends. This manipulation yields a construct bearing the EmrE gene, without termination, followed by a linker with the sequence as indicated in Figure 1, and a second EmrE with the MycHis tag. The cloning sites for the whole construct are NdeI and HindIII at the ends. Sites for NheI (all the tandems), KasI (all the tandems except E2EMH) and BamHI (E6EMH and Q6EMH) were created between the two genes. E22EMH was constructed as follows: pT7-7 plasmid bearing EmrE with the MycHis tag was digested by SalI (at the end of the Myc tag) and PstI (after the end of the gene). EmrE with MycHis was created by PCR with sites for XhoI and PstI and ligated to the above vector. The identity of all the constructs was verified by sequencing.
Resistance to toxic compounds
E. coli DH5α cells transformed with pT7-7-EmrE, pT7-7 (vector), or pT7-7 with the various EmrE fusions were grown overnight at 37°C in LB–ampicillin medium. A 5-μl volume of serial dilutions of the culture was spotted on LB–ampicillin plates containing 30 mM BisTris propane, pH 7.0, with or without addition of 100 μg/ml ethidium bromide. Growth was analyzed after overnight incubation at 37°C.
Transport of ethidium in whole cells
Transport was assayed essentially as described (Yerushalmi et al, 1995). E. coli HMS 174 cells bearing the appropriate plasmids were grown in minimal medium A (Davies and Mingioli, 1950) supplemented with 20 mM glucose at 37°C to A600=0.5. EmrE expression was induced by 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside), and 2 h later cells were collected by centrifugation and resuspended to A600=0.5 in minimal medium A with no glucose. Then, ethidium and carbonyl cyanide m-chlorophenylhydrazone (CCCP) were added to a final concentration of 5 and 40 μM, respectively, and the cells were incubated for 60 min at 37°C. The cells were collected by centrifugation and resuspended in medium containing 5 μM ethidium without CCCP, and the reaction was initiated by addition of 20 mM glucose. Fluorescence was measured at 37°C with a Perkin Elmer fluorometer (LS 50 B luminescence spectrometer) using FL WinLab software with exciting wavelength at 525 nm and emission at 585 nm. To assess expression, membranes were prepared by sonication and the His-tagged proteins were extracted with 2% SDS, purified on Ni–NTA (Qiagen, Hilden, Germany) and analyzed by SDS–PAGE.
Overexpression and purification
TA15 cells bearing plasmids pGP1-2 and His-tagged EmrE constructs (cloned into pT7-7 expression vector) were used for overexpression. Purification was performed essentially as described in Soskine et al (2006), except that metal chelate chromatography was performed on the bench with 1-ml columns using Ni–NTA (Qiagen, Hilden, Germany), and the eluted protein was further purified on a Superdex™ 200HR column (Amersham Biosciences) equilibrated with 0.08% DDM Na-buffer (150 mM NaCl, 15 mM Tris–Cl, pH 7.5) and mounted on Akta Explorer (Amersham Biosciences). Major peak fractions were pooled and the protein solution was brought to ∼0.25 mg/ml EmrE. The protein stock was aliquoted and stored at −70°C.
Reconstitution
Reconstitution was performed essentially as described (Yerushalmi et al, 2001), except that proteoliposomes were prepared in buffer containing 0.15 M (NH4)2SO4, 15 mM Tris, pH 7.5, and 1 mM dithiothreitol. To determine the protein concentration in the proteoliposomes, the proteoliposomes were solubilized in SDS and His-tagged proteins were purified with Ni–NTA (Qiagen, Hilden, Germany) and analyzed by SDS–PAGE. The intensity of the staining was analyzed using Gauge 3.46 Fujifilm software and compared with the intensity of samples with known amounts of EmrE (range 0.5–5 μg).
[3H]TPP+-binding assay
TPP+ binding was assayed essentially as described (Muth and Schuldiner, 2000). Amounts of purified EmrE and tandems were determined according to A280. All binding reactions were performed in duplicates and in each experiment the values obtained in a control reaction with 25 μM unlabeled TPP+ were subtracted. All the experiments were repeated at least twice.
[14C]MV2+ and [3H]MPP+ uptake assay
Uptake of [14C]MV2+ or [3H]MPP+ into proteoliposomes was assayed at 25°C by dilution of 2 μl of the (NH4)2SO4-containing proteoliposomes into 200 μl of an ammonium-free solution (Yerushalmi et al, 1995, 2001). The latter contained 20 μM [14C]MV2+ (8.1 mCi/mmol; Sigma-Aldrich, St Louis, MO) or 1 μM [3H]MPP (0.566 Ci/mol), 140 mM K2SO4, 10 mM tricine, 5 mM MgCl2 and 10 mM Tris, pH was 8.5. Where indicated, valinomycin was added to 100 nM. At the given times, the reaction was stopped by dilution with 2 ml of the same ice-cold solution, filtration through Millipore GSWP (MV2+) or Supor®-200 filters (MPP+) (0.22 and 0.2 μm pore size respectively) and washing with additional 2 ml of solution. The radioactivity on the filters was estimated by liquid scintillation. Values obtained in a control reaction, with 15 μM Nigericin, were subtracted from all experimental points. Each experiment was performed at least twice.
[14C]MV2+ efflux assay
Proteoliposomes were prepared as described above, except that buffer contained K2SO4 instead of (NH4)2SO4. After thawing the proteoliposomes, [14C]MV2+ was added to a final concentration of 2.1 mM (8.1 mCi/mmol; Sigma-Aldrich, St Louis, MO) and the proteoliposomes were sonicated to clarity. Efflux was assayed at 15°C by dilution of 2 μl of the [14C]MV2+-loaded proteoliposomes into 200 μl of solution containing 140 mM K2SO4, 5 mM MgCl2, 1 μM valinomycin and 20 mM K-Hepes, pH 8.5. At the given times, the reaction was stopped by dilution with 2 ml of the same ice-cold solution, filtration through Millipore GSWP 0.22 μm pore size and washing with an additional 2 ml of solution. The radioactivity on the filters was estimated by liquid scintillation. Values obtained with proteoliposomes diluted into a medium containing 0.5% DDM were subtracted from each point. Each experiment was performed at least twice.
Substrate-induced proton release measurements
Substrate-induced proton release was measured as previously described (Adam et al, 2007). A 30–105-μg weight of unbuffered protein in a solution of 150 mM NaCl, 0.08% DDM and 100 μM Phenol Red was titrated to pH ∼7.0 (according to OD using a pH calibration curve). Reaction was started by addition of TPP+ to a concentration of 5 μM. To calculate the amount of protons released, 4 nmol NaOH were added at the end of the reaction and the absorption recorded.
Protease treatment and cross-linking experiments
Membranes from cells expressing EmrE and E22EMH were prepared as described (Soskine et al, 2002). The proteins were specifically radiolabeled with [35S]methionine (Soskine et al, 2002) and were incubated with the corresponding protease for 1 h at 37°C in a final volume of 30 μl of 150 mM NaCl, 15 mM Tris–Cl, pH 8.0, and 10 mM CaCl2. Chymotrypsin (Sigma-Aldrich, St Louis, MO) concentration was 1.9 U/ml, and of proteinase K (Sigma-Aldrich, St Louis, MO) was 0.26 U/ml. After digestion, 1 ml of 150 mM NaCl, 15 mM Tris–Cl, pH 8.0, was added and the membranes were collected by centrifugation, solubilized in 30 μl of a buffer containing 200 mM β-mercaptoethanol, 100 mM Tris–HCl, pH 6.8, 4% SDS, 40% glycerol and 0.2% bromophenol blue, and were run on SDS–PAGE gels, visualized with a Fujifilm FLA-3000 imaging system and digitally analyzed with Image Gauge 3.46 Fujifilm software.
When cross-linking was performed, digestion was performed with 1.52 U/ml proteinase K–agarose (Sigma-Aldrich, St Louis, MO) and after removal of the beads by centrifugation, the volume was adjusted to 100 μl with 150 mM NaCl, 15 mM Tris–Cl, pH 7.5, 1.5% DDM and HMDC (1:500). After 20 min at room temperature, the preparation was analyzed as above. Identical results were obtained whether DDM was added before or after proteolysis, suggesting that the same overall packing is maintained after solubilization.
Negative dominance and pull-down experiments
For activity measurements, membrane aliquots containing ∼40 ng EmrE or EmrE tandem per assay were solubilized in 1 ml of Na-buffer containing 1% DDM, 0.5 mM phenylmethylsulfonyl fluoride and 15 mM β-mercaptoethanol for 30 min at room temperature. After removal of unsolubilized material by centrifugation (20 000 g for 30 min), the supernatant was mixed with increasing amounts of membranes solubilized as above and containing the indicated amounts of untagged EmrE E14C protein in 110 μl of 0.08% DDM Na buffer. After 15 min at 80°C, the samples were allowed to cool down and subjected to pulse centrifugation. [3H]TPP+ binding was measured as described above. For pull-down experiments, the untagged protein was radiolabeled with [35S]methionine (Soskine et al, 2002), and after cooling the mixture was subjected to pulse centrifugation and immobilized on Ni–NTA beads as described above. The proteins were eluted using a buffer containing 200 mM β-mercaptoethanol, 100 mM Tris–HCl, pH 6.8, 4% SDS, 40% glycerol, 0.2% bromophenol blue and 450 mM imidazole, and were run on SDS–PAGE gels, visualized with a Fujifilm FLA-3000 imaging system and digitally analyzed with Image Gauge 3.46 Fujifilm software.
Acknowledgments
This work was supported by Grant NS16708 from the National Institutes of Health and Grant 119/04 from the Israel Science Foundation. SS is Mathilda Marks-Kennedy Professor of Biochemistry at the Hebrew University of Jerusalem. We thank Dr Mario Lebendiker from The Protein Purification Facility, Wolfson Center for Applied Structural Biology (Life Sciences Institute, Hebrew University of Jerusalem), valuable technical assistance and helpful advice.
References
- Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S (2007) The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc Natl Acad Sci USA 104: 17989–17994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay DC, Rommens KL, Turner RJ (2007) Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta (doi:10.1016/j.bbamem.2007.08.015) [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W (2002) A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J 21: 2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU (2006) Flip-flopping membrane proteins. Nat Struct Mol Biol 13: 94–96 [DOI] [PubMed] [Google Scholar]
- Chang G, Roth CB, Reyes CL, Pornillos O, Chen Y-J, Chen AP (2006) Retraction. Science 314: 1875. [DOI] [PubMed] [Google Scholar]
- David G, Blondeau K, Renouard M, Lewit-Bentley A (2002) Crystallization and preliminary analysis of Escherichia coli YodA. Acta Crystallogr D 58: 1243–1245 [DOI] [PubMed] [Google Scholar]
- David G, Blondeau K, Schiltz M, Penel S, Lewit-Bentley A (2003) YodA from Escherichia coli is a metal-binding, lipocalin-like protein. J Biol Chem 278: 43728–43735 [DOI] [PubMed] [Google Scholar]
- Davies B, Mingioli E (1950) Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol 60: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz Y, Steiner-Mordoch S, Danieli T, Schuldiner S (2004) In vitro synthesis of fully functional EmrE, a multidrug transporter, and study of its oligomeric state. Proc Natl Acad Sci USA 101: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N (2006) Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J Mol Biol 364: 54–67 [DOI] [PubMed] [Google Scholar]
- Gafvelin G, von Heijne G (1994) Topological ‘frustration' in multispanning E. coli inner membrane proteins. Cell 77: 401–412 [DOI] [PubMed] [Google Scholar]
- Goldberg EB, Arbel T, Chen J, Karpel R, Mackie GA, Schuldiner S, Padan E (1987) Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA 84: 2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikukawa T, Nara T, Araiso T, Miyauchi S, Kamo N (2006) Two-component bacterial multidrug transporter, EbrAB: mutations making each component solely functional. Biochim Biophys Acta 1758: 673–679 [DOI] [PubMed] [Google Scholar]
- Ma C, Chang G (2004) Structure of the multidrug resistance efflux transporter EmrE from Escherichia coli. Proc Natl Acad Sci USA 101: 2852–2857 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Muth TR, Schuldiner S (2000) A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J 19: 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I, Skurray R, Tam R, Saier M, Turner R, Weiner J, Goldberg E, Grinius L (1996) The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol 19: 1167–1175 [DOI] [PubMed] [Google Scholar]
- Pornillos O, Chang G (2006) Inverted repeat domains in membrane proteins. FEBS Lett 580: 358–362 [DOI] [PubMed] [Google Scholar]
- Pornillos O, Chen YJ, Chen AP, Chang G (2005) X-ray structure of the EmrE multidrug transporter in complex with a substrate. Science 310: 1950–1953 [DOI] [PubMed] [Google Scholar]
- Rapp M, Granseth E, Seppala S, von Heijne G (2006) Identification and evolution of dual-topology membrane proteins. Nat Struct Mol Biol 13: 112–116 [DOI] [PubMed] [Google Scholar]
- Rapp M, Seppala S, Granseth E, von Heijne G (2007) Emulating membrane protein evolution by rational design. Science 315: 1282–1284 [DOI] [PubMed] [Google Scholar]
- Rotem D, Sal-man N, Schuldiner S (2001) In vitro monomer swapping in EmrE, a multidrug transporter from Escherichia coli, reveals that the oligomer is the functional unit. J Biol Chem 276: 48243–48249 [DOI] [PubMed] [Google Scholar]
- Schuldiner S (2007) When biochemistry meets structural biology: the cautionary tale of EmrE. Trends Biochem Sci 32: 252–258 [DOI] [PubMed] [Google Scholar]
- Sharoni M, Steiner-Mordoch S, Schuldiner S (2005) Exploring the binding domain of EmrE, the smallest multidrug transporter. J Biol Chem 280: 32849–32855 [DOI] [PubMed] [Google Scholar]
- Soskine M, Adam Y, Schuldiner S (2004) Direct evidence for substrate induced proton release in detergent solubilized EmrE, a multidrug transporter. J Biol Chem 279: 9951–9955 [DOI] [PubMed] [Google Scholar]
- Soskine M, Mark S, Tayer N, Mizrachi R, Schuldiner S (2006) On parallel and antiparallel topology of an homodimeric multidrug transporter. J Biol Chem 281: 36205–36212 [DOI] [PubMed] [Google Scholar]
- Soskine M, Steiner-Mordoch S, Schuldiner S (2002) Crosslinking of membrane-embedded cysteines reveals contact points in the EmrE oligomer. Proc Natl Acad Sci USA 99: 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S, Richardson C (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82: 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C, Kunji E, Lebendiker M, Schuldiner S (2001) The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 Å resolution. EMBO J 20: 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG (2003) Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J 22: 6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi H, Lebendiker M, Schuldiner S (1995) EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem 270: 6856–6863 [DOI] [PubMed] [Google Scholar]
- Yerushalmi H, Mordoch SS, Schuldiner S (2001) A single carboxyl mutant of the multidrug transporter EmrE is fully functional. J Biol Chem 276: 12744–12748 [DOI] [PubMed] [Google Scholar]
- Yerushalmi H, Schuldiner S (2000a) A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett 476: 93–97 [DOI] [PubMed] [Google Scholar]
- Yerushalmi H, Schuldiner S (2000b) A model for coupling of H(+) and substrate fluxes based on ‘time-sharing' of a common binding site. Biochemistry 39: 14711–14719 [DOI] [PubMed] [Google Scholar]
- Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W (2003) Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem 278: 50128–50135 [DOI] [PubMed] [Google Scholar]