Abstract
Interphase phosphorylation of S10 at histone H3 is linked to transcriptional activation of a specific subset of mammalian genes like HDAC1. Recently, 14-3-3 proteins have been described as detectors for this phosphorylated histone H3 form. Here, we report that 14-3-3 binding is modulated by combinatorial modifications of histone H3. S10 phosphorylation is necessary for an interaction, but additional H3K9 or H3K14 acetylation increases the affinity of 14-3-3 for histone H3. Histone H3 phosphoacetylation occurs concomitant with K9 methylation in vivo, suggesting that histone phosphorylation and acetylation can synergize to overcome repressive histone methylation. Chromatin immunoprecipitation experiments reveal recruitment of 14-3-3 proteins to the HDAC1 gene in an H3S10ph-dependent manner. Recruitment of 14-3-3 to the promoter is enhanced by additional histone H3 acetylation and correlates with dissociation of the repressive binding module HP1γ. Finally, siRNA-mediated loss of 14-3-3 proteins abolishes the transcriptional activation of HDAC1. Together our data indicate that 14-3-3 proteins are crucial mediators of histone phosphoacetylation signals.
Keywords: 14-3-3, histone acetylation, histone code, histone phosphorylation, phosphoacetylation
Introduction
The unstructured N-terminal tails of histone proteins are targeted by various different post-translational modifications (PTMs) like acetylation, methylation, phosphorylation or ADP ribosylation. These PTMs are critical factors in the regulation of local and global chromatin function and confer distinct properties to regions of the genome that finally modulate chromatin-associated processes such as transcription. It was suggested that the specific modification make-up constitutes a ‘histone code', which is recognized via a ‘decoding machinery' comprised by modification-dependent, chromatin-associated polypeptides (Strahl and Allis, 2000; Fischle et al, 2003). One particular PTM, the phosphorylation of histone H3 at S10, emerges in two distinct phases of the cell cycle, with considerable differences in dynamics and abundance (Johansen and Johansen, 2006; McManus and Hendzel, 2006). Global mitosis-specific histone H3 phosphorylation is mediated by the Aurora B kinase (Hsu et al, 2000) and is required for the displacement of HP1 proteins (Mateescu et al, 2004; Fischle et al, 2005; Hirota et al, 2005). During interphase, H3S10 phosphorylation is targeted to only a minute fraction of nucleosomes and is tightly linked to acetylation of H3K9 and H3K14 (phosphoacetylation) (Mahadevan et al, 1991; Cheung et al, 2000b; Clayton and Mahadevan, 2003). Histone H3 phosphoacetylation can be mediated by kinases MSK1/2 that are downstream of the ERK (p42/44) or the p38 mitogen-activated protein (MAP) kinase pathways and has been correlated with transcriptional induction of the immediate-early (IE) genes c-fos and c-jun (Clayton et al, 2000; Cheung et al, 2000b; Thomson et al, 2001; Clayton and Mahadevan, 2003; Soloaga et al, 2003; Mahadevan et al, 2004).
The concept of gene activation by histone phosphorylation was extended to a variety of mammalian genes (Strelkov and Davie, 2002; Clayton and Mahadevan, 2003; Vicent et al, 2006). For instance, we reported the regulation of the HDAC1 gene by cooperative histone H3 phosphorylation and acetylation (Hauser et al, 2002). In contrast to the rapid and transient phosphoacetylation associated with IE gene activation, transcriptional induction of HDAC1 requires stable phosphoacetylation, which is achieved by stimulation of MAP kinase pathways and fine tuned via histone acetylation in an autoregulatory loop (Bartl et al, 1997; Schuettengruber et al, 2003).
In addition, detector proteins that specifically recognize PTMs on histones play a key role in the regulation of chromatin-associated events. For example, the bromodomains of GCN5 and TFIID250 have been shown to specifically associate with acetylated histones, while chromodomain proteins exemplified by the heterochromatin protein 1 (HP1) interact with specific methylated forms (Lachner et al, 2003). Recently, 14-3-3 proteins, which are well-established phospho-serine adaptor molecules (Muslin et al, 1996; Yaffe et al, 1997), have been described as detectors for phosphorylated histone H3 (Macdonald et al, 2005). However, the role of this interaction in the context of transcription is unclear.
Here, we report that interaction of 14-3-3ɛ and ζ with histone H3 is modulated by combinatorial PTM patterns. Binding of these proteins to phosphorylated H3S10 is stabilized by additional lysine acetylation. Phosphoacetylation of histone H3 at the HDAC1 promoter leads to the recruitment of 14-3-3 proteins concomitant with dissociation of HP1. As detector for specific active histone marks, we found that 14-3-3ζ is necessary for activation of the HDAC1 gene. Finally, we identify 14-3-3 as counterpart of the repressive binding module HP1, with reciprocal binding affinities for the modified histone H3 tail.
Results
Purification of 14-3-3 proteins as phosphoacetylated histone H3-binding factors
Histone H3 phosphorylation and acetylation synergize in transcriptional activation of the late inducible HDAC1 gene (Hauser et al, 2002), implying that phosphoacetylation is a biologically relevant PTM pattern. Therefore, we asked whether the phosphoacetylation mark is recognized by specific cellular factors. To this end, differentially modified matrix-coupled histone H3 peptides (Table I) were incubated with nuclear extracts from HeLa cells that were either left untreated or treated with the p38–MAP kinase activator anisomycin and the HDAC inhibitor trichostatin A (TSA). This combinatorial treatment was previously shown to stimulate histone H3 phosphoacetylation and HDAC1 gene expression (Hauser et al, 2002). Two modification-dependent factors of approximately 30 and 27 kDa were found that specifically interacted with the S10phK14ac (ph/ac)–histone H3 peptide (Supplementary Figure S1). These proteins were identified as the epsilon (ɛ) and zeta (ζ) isoforms of the 14-3-3 protein family via mass spectrometry. The presence of the two isoforms in HeLa nuclear extracts was verified by immunoblotting with isoform-specific 14-3-3 antibodies (Supplementary Figure S1B). In in vitro binding assays, 14-3-3ζ extracted either from untreated or anisomycin/TSA treated HeLa cells bound equally well to S10phK14ac H3 peptide, suggesting that activation of the MAP kinase pathway or HDAC inhibition does not alter the affinity of 14-3-3 for the modified histone H3 tail (Supplementary Figure S1C). Although 14-3-3 proteins have been recently shown to interact with phosphorylated histone H3 (Macdonald et al, 2005), the importance of this interaction for gene regulation is not yet clarified. We therefore examined the role of 14-3-3 proteins as detectors for ph/ac histone H3 and studied their role in the activation of transcription.
Table 1.
Histone H3 peptides used in this study
| Peptide | Sequence N–C |
|---|---|
| um | ARK STG GKA PRK QLC |
| K14ac | ARK STG GacKA PRK QLC |
| S10ph | ARK phSTG GKA PRK QLC |
| K9ac/S10ph | ARacK phSTG GKA PRK QLC |
| S10ph/K14ac | ARK phSTG GacKA PRK QLC |
| K9me2/S10ph | Arme2K phSTG GKA PRK QLC |
| K9me2/S10ph/K14ac | ARme2K phSTG GacKA PRK QLC |
| H3(1–20) um | ART KQT ARK STG GKA PRQ LC |
| H3(1–20) S10ph | ART KQT ARK phSTG GKA PRQ LC |
| H3(1–20) K9ac/S10ph | ART KQT ARacK phSTG GKA PRQ LC |
| H3(1–20) S10ph/K14ac | ART KQT ARK phSTG GacKA PRQ LC |
| H3(1–20) K9me2/S10ph | ART KQT AR me2K phSTG GKA PRQ LC |
| H3(1–20) K9me2/S10ph/ K14ac | ART QT AR me2K phSTG GacKA PRQ LC |
| H3 um (25–38) | ARK SAP ATG GVK KPC |
| H3 S28ph (25–38) | ARK phSAP ATG GVK KPC |
| acK, acetylated lysine; me2K, dimethylated lysine; pS, phosphoserine. | |
14-3-3 Proteins interact with histone H3 in a modification-dependent manner
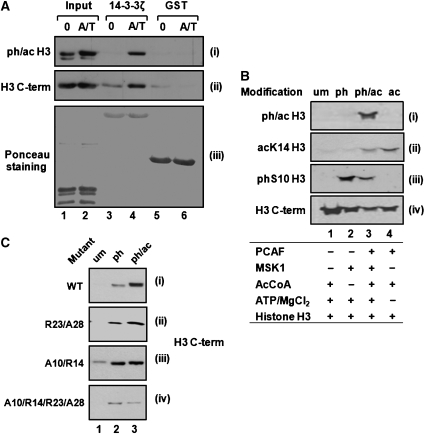
To verify that 14-3-3 proteins bind indeed to ph/ac histone H3, we performed GST pull-down assays with histones isolated from 3T3 mouse fibroblasts. To avoid mitotic S10 phosphorylation, we used resting cells, which show only low levels of ph/ac histone H3 and cells that were simultaneously treated with anisomycin and TSA to stimulate H3 phosphoacetylation (Figure 1A, panel i, lanes 1 and 2). After incubation with GST–14-3-3ζ or GST as control, bound histones were analyzed by immunoblotting. Probing of the blot with antibodies specific for S10phK14ac H3 revealed that this modification form could interact with GST–14-3-3ζ but not with GST (Figure 1A, panel i). The total amount of H3 associated with 14-3-3 proteins was increased upon TSA/anisomycin treatment as displayed with modification independent C-terminal H3 antibodies (Figure 1A, panel ii). A similar in vitro interaction was also found for 14-3-3ɛ (data not shown). These results indicate that 14-3-3ζ and ɛ bind to histone H3 in a modification-dependent manner.
Figure 1.
14-3-3 Binding to histone H3 is dependent on H3 phosphorylation and stabilized by additional acetylation. (A) Induction of phosphoacetylation increases histone H3 interaction with 14-3-3. Histones were isolated from resting 3T3 fibroblasts that were either left untreated (0) or stimulated for 1 h with anisomycin and TSA (A/T) and incubated with GST or GST–14-3-3ζ. Bound histones were analyzed by immunoblotting with antibodies against ph/ac histone H3 (panel i) and C-terminal histone H3 (H3 C-term) (panel ii). Loading of GST and GST–14-3-3 was monitored by Ponceau staining (panel iii). (B) In vitro modification of histone H3. Recombinant histone H3 was phosphorylated by MSK1 (lane2), acetylated by PCAF (lane 4) or phosphoacetylated with both enzymes (lane 3). Enzymes were omitted in control reactions (lane 1). The modification status was analyzed by sequential immunoblotting with the indicated antibodies. Corresponding modifications are denoted at the top. (C) Acetylation effects on the 14-3-3/histone H3 interaction are more dominant for the R23A28 mutant than for the A10R14 mutant. The indicated histone H3 mutants were in vitro modified as indicated and incubated with GST–14-3-3ζ proteins. Bound histone H3 proteins were analyzed by immunoblotting with C-terminal histone H3 antibodies. The panel shows one representative experiment for each mutant or WT histone H3.
Additional acetylation stabilizes the interaction between S10 phosphorylated histone H3 and 14-3-3 proteins
Histone proteins extracted from mammalian cells may carry in addition to phosphorylation and acetylation various other PTMs. To utilize a more defined set of modifications, we modified recombinant histone H3 in vitro. Phosphorylation or acetylation reactions were performed using MSK1 (Figure 1B, panel iii, lane 2) or the histone acetyltransferase PCAF, respectively (Figure 1B, panel ii, lane 4). Initial phosphorylation and subsequent acetylation reactions generated ph/ac histone H3 (Figure 1B, panel i, lane 3). As control, enzymes were omitted from reactions (Figure 1B, lane 1) and total amounts of H3 in the different modification reactions were visualized with the C-terminal H3 antibodies (Figure 1B, panel iv).
The interaction of in vitro modified H3 with 14-3-3ζ was analyzed in GST pull-down assays. As expected, phosphorylation led to association with GST–14-3-3ζ (Figure 1C, panel i), whereas acetylation by PCAF alone did not mediate any binding (Supplementary Figure S2C, and data not shown). Strikingly, 14-3-3ζ binding was stronger for phosphoacetylated than for phosphorylated H3, indicating that in the context of S10 phosphorylation acetylation exerts a stabilizing effect (Figure 1C, panel i).
Mass spectrometry analysis of MSK1-modified histone H3 revealed that not only S10 but also S28 was phosphorylated (Supplementary Figure S2D). 14-3-3 Proteins were previously shown to interact not only with H3S10ph but also with H3S28ph peptides (Macdonald et al, 2005). Furthermore, acetylation of the neighboring K23 residue was reported (Daujat et al, 2002). Therefore, we performed binding assays with different mutated histone H3 proteins. The efficiency of phosphorylation by MSK1 and acetylation by PCAF was monitored by immunoblot analysis and kinase assays with γ-32P-ATP (Supplementary Figures S2A and B). Mutation of either S10 in combination with K14 or S28 and K23 led to 55–60% reduction in 32P incorporation, while the quadruple mutant displayed about 20% residual phosphorylation (Supplementary Figure S2B). According to the mass spectrometry analysis, histones H3 becomes phosphorylated at T45 and S57 in the absence of both serines. Surprisingly, a K9R mutation significantly increased 32P incorporation by MSK1 and therefore these mutants were omitted from further analysis.
Pull-down assays with histone H3 bearing K23R/S28A double mutations (R23/A28) revealed an increased interaction of 14-3-3ζ with the phosphoacetylated than to the phosphorylated form (Figure 1C, panel ii). Mass spectrometry analysis confirmed that this mutant was predominantly phosphorylated on S10. In contrast, 14-3-3ζ binding to the S10A/K14R mutant (A10/R14) was similar in the presence and absence of acetylation (Figure 1C, panel iii), and loss of both serines in the quadruple mutant resulted in only weak interaction with 14-3-3ζ. Taken together, these data indicate that the binding affinity of 14-3-3ζ for S10-phosphorylated histone H3 is significantly enhanced by additional lysine acetylation. Since combinatorial binding of 14-3-3 to target factors has not been reported, we decided to investigate this effect in more detail.
Interphase phosphorylation of histone H3 occurs in the context of additional PTMs
The efficiency of phosphorylation-mediated binding of 14-3-3 to target proteins is strongly dependent on the amino-acid environment around the phosphorylated residue (Yaffe et al, 1997; Uchida et al, 2006). As a basis for studying the impact of additional modifications on the interaction with 14-3-3, we determined PTM patterns present on S10-phosphorylated N-terminal tails of histone H3 via a mass spectrometry approach. As already mentioned, interphase phosphorylation of histone H3 affects only a small sub-fraction of all nucleosomes (Barratt et al, 1994). Our mass spectrometry analysis clearly indicates the presence of various different modifications like lysine methylation and acetylation in addition to H3S10ph (Table II). In agreement with previously published data (Dyson et al, 2005), we observed some residual histone H3 phosphorylation in samples derived from resting and untreated cells.
Table 2.
PTMs on the S10-phosphorylated tryptic histone H3 peptide K9STGGKAPR17
| Condition | Peptide sequence | MH+ | LTQ-FT | LTQ |
|---|---|---|---|---|
| Resting | R.KpSTGGKAPR.K | 1093.5 | 0/3 | 1/1 |
| R.me1KpSTGGKAPR.K | 1107.556 | 1/3 | 0/1 | |
| R.me2KpSTGGKAPR.K | 1065.545 | 2/3 | 0/1 | |
| R.me3KpSTGGKAPR.K | 1079.561 | 2/3 | 1/1 | |
| R.KpSTGGacKAPR.K | 1079.524 | 1/3 | 0/1 | |
| R.acKpSTGGme3KAPR.K | 1065.545 | 1/3 | 0/1 | |
| R.me3KpSTGGacKAPR.K | ||||
| sAn | R.KpSTGGKAPR.K | 1093.540 | 1/3 | 1/1 |
| R.me1KpSTGGKAPR.K | 1107.556 | 2/3 | 1/1 | |
| R.me2KpSTGGKAPR.K | 1065.545 | 3/3 | 1/1 | |
| R.me3KpSTGGKAPR.K | 1079.561 | 3/3 | 1/1 | |
| R.KpSTGGacKAPR.K | 1079.525 | 2/3 | 1/1 | |
| R.me1KpSTGGacKAPR.K | 1093.540 | 1/3 | 0/1 | |
| R.me2KpSTGGKacAPR.K | 1051.530 | 1/3 | 1/1 | |
| R.acKpSTGGme3KAPR.K | ||||
| R.me3KpSTGGacKAPR.K | 1065.5 | 0/3 | 1/1 | |
| R.me3KpSTGGme3KAPR.K | ||||
| SAn/TSA | R.KpSTGGKAPR.K | 1093.540 | 1/3 | 1/1 |
| R.me1KpSTGGKAPR.K | 1107.556 | 1/3 | 0/1 | |
| R.me2KpSTGGKAPR.K | 1065.545 | 2/3 | 1/1 | |
| R.me3KpSTGGKAPR.K | 1079.561 | 3/3 | 1/1 | |
| R.KpSTGGacKAPR.K | 1079.524 | 2/3 | 0/1 | |
| R.acKpSTGGacKAPR.K | 1065.509 | 2/3 | 1/1 | |
| R.me1KpSTGGacKAPR.K | 1093.540 | 3/3 | 1/1 | |
| R.me2KpSTGGacKAPR.K | 1051.530 | 2/3 | 1/1 | |
| R.acKpSTGGme3KAPR.K | 1065.545 | 1/3 | 1/1 | |
| R.me3KpSTGGacKAPR.K | ||||
| acK, acetylated lysine; LTQ-FT/LTQ, the value indicates the frequency of peptide recovery either on the LTQ-FTICR hybrid instrument or the LTQ mass spectrometer (e.g., 2/3 meaning two times out of three experiments); me1K, monomethylated lysine; me2K, dimethylated lysine; me3K, trimethylated lysine; MH+, mono-protonated mass; pS, phosphorylated serine; PTM, post-translational modification; sAn, anisomycin; TSA, trichostatin A. Vertical lines indicate one peptide species where modifications were not unequivocally assigned to a particular position. | ||||
Anisomycin treatment increased the S10ph histone H3 pool, as well as the complexity of modification patterns (Table II). Besides single phosphorylated histone H3 and H3K9me1/2/3/S10ph forms, we identified a phosphoacetylated species with the acetyl moiety at position 14 (S10phK14ac). In addition, we identified a triple modified H3 peptide with the modification status K9me2S10phK14ac. This form is particular interesting as it carries an active and a repressive modification in addition to H3S10ph.
Additional treatment with TSA led to further changes in the phospho-form composition and gave also rise to a K9K14 diacetylated ph/ac form (K9acS10phK14ac). While TSA treatment had no effect on overall S10 phosphorylation (Supplementary Figure S3A, panel i, lanes 5 and 6 and Supplementary Figure S3B), the abundance of the S10phK14ac epitope was almost doubled (Supplementary Figure S3A, panel i, compare lanes 2 and 3). These results suggest that p38 MAP kinase activation leads to the formation of several different phospho-histone H3 forms and the composition of this pool is altered by additional TSA treatment. Taken together, interphase H3S10 phosphorylation occurs as single modification, but frequently coincides with lysine methylation and acetylation on the same histone H3 tail.
Additional histone modifications affect the 14-3-3/histone H3 interaction
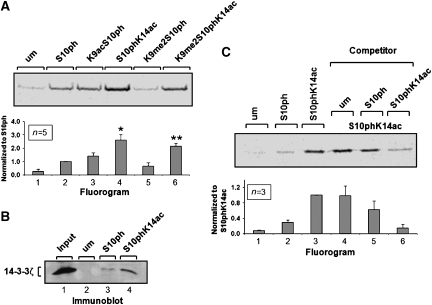
To investigate the impact of lysine acetylation and methylation on the interaction between 14-3-3 and histone H3, we performed in vitro peptide pull-down assays. This experimental setup also allowed us to use a homogenously modified system for the interaction studies. Differentially modified histone H3 peptides were synthesized on the basis of the mass spectrometry results (Table I). Since we are interested in the role of H3S10ph during transcriptional activation, we focused on modifications that are known to prevalently reside in euchromatin and excluded H3K9me3, the archetype of heterochromatic histone modifications. Equal amounts of the differentially modified immobilized H3 peptides were incubated with in vitro translated (IVT) 14-3-3ζ protein. Phosphorylation of H3S10 was required for significant interaction with 14-3-3 (Figure 2A, lane 2), whereas only slight background signals were observed for the unmodified or the H3K14ac peptide (Figure 2A, lane 1, and data not shown).
Figure 2.
Modulation of the histone H3/14-3-3 interaction by additional modifications. (A) Histone H3/14-3-3 interaction is modulated by additional lysine acetylation. IVT 35S-methionine-labeled 14-3-3ζ was incubated with differentially modified gel-coupled histone H3 peptides. Bound proteins were analyzed by SDS–PAGE and fluorography. The panel shows one representative experiment. The signal intensity for each band was quantified and is depicted as summary of five independent measurements (mean±s.d.). Values were normalized relative to H3S10ph peptide-bound fraction (lane 2). Additional acetylation increased the association with 14-3-3 proteins (lane 4, *P=0.001, t-test). A similar effect of H3K14 acetylation was observed for the H3K9me2/S10ph peptide (lane 7, **P<0.001, t-test). (B) Nuclear 14-3-3 proteins preferentially bind to the S10phK14ac histone H3 peptide. Nuclear extracts were incubated with unmodified, S10ph or S10phK14ac H3 peptides. An aliquot of the nuclear extracts was used as input control. Bound proteins were analyzed on immunoblots with 14-3-3ζ antibodies. (C) Additional K14 acetylation increases the competitor potential of the S10ph histone H3 peptide. Binding reactions were performed as described for panel A (lanes 1–3). In addition, binding reactions on the ph/ac peptide were performed in the presence of a 20-fold molar excess of unmodified, H3S10ph or H3S10phK14ac free competitor peptides. Bound 14-3-3ζ proteins were analyzed by SDS–PAGE and fluorography. Each signal was normalized to the non-competed H3S10phK14ac peptide binding (lane 3) and is depicted as histogram showing the average of three independent experiments (mean±s.d.).
Acetylation of H3K9 caused a moderate but reproducible reinforcement of the interaction (Figure 2A, lane 3). Remarkably, additional acetylation of H3K14 strongly increased the interaction, supporting the results from the GST pull-down experiments (Figure 2A, lane 4 and Figure 1C). To rule out the possibility that the increased binding of 14-3-3ζ to the H3S10phK14ac peptide is a unique property of IVT or recombinant proteins, we confirmed this effect with endogenous 14-3-3 present in HeLa nuclear extracts (Figure 2B).
In contrast, binding to the H3K9me2S10ph peptide was slightly reduced compared with the S10ph peptide (Figure 2A, compare lanes 2 and 5). Our mass spectrometry analysis revealed the presence of a K9me2S10phK14ac histone H3 form (Table II). Binding studies with the corresponding triple modified peptide (K9me2S10phK14ac) demonstrated that additional acetylation of K14 also increased binding of 14-3-3ζ to the phosphomethylated H3 peptide (Figure 2A, lane 6).
To further confirm the stabilizing effect exerted by H3K14 acetylation, we performed competition assays using the H3S10phK14ac peptide as bait and free unmodified, H3S10ph and H3S10phK14ac peptides as competitors (Figure 2C, lanes 4–6). Addition of the H3S10ph peptide reduced binding of 14-3-3ζ to the H3S10phK14ac peptide to approximately 65% compared with non-competed binding, whereas the unmodified peptide had no effect (Figure 2C, lanes 4 and 5). Importantly, the H3S10phK14ac peptide was found to be a much more potent competitor with an average reduction of binding to about 15% of non-competed assays (Figure 2C, lane 6). These data also demonstrate that the binding of 14-3-3ζ to the phosphoacetylated histone H3 tail is highly dynamic and reversible.
Together, our data suggest that significant binding of 14-3-3 to the histone H3 tail requires initial phosphorylation but is susceptible to the presence of additional PTMs.
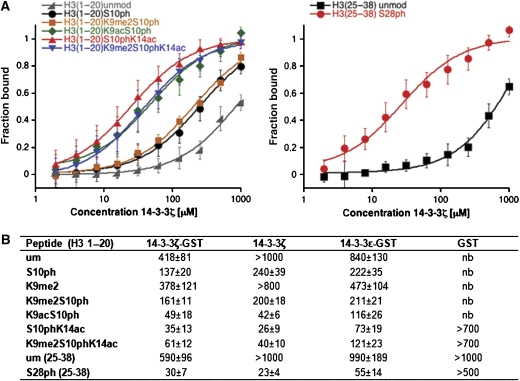
To quantify the binding of 14-3-3 to various combinations of H3 modifications, we determined dissociation constants of the interactions. Therefore, we performed fluorescence polarization measurements using recombinant 14-3-3 in combination with fluorescinated, differentially modified H3 peptides (Figure 3). In concordance with the peptide pull-down assays, we detected strong interaction of 14-3-3 with the histone H3 peptide upon phosphorylation of S10, whereas only very weak interaction with the unmodified peptide was observed. The H3K9me2S10ph peptide was bound with comparable strength as the H3S10ph peptide, suggesting that K9me2 does not significantly impair the binding of 14-3-3. The affinity of 14-3-3 for the H3S10ph and the H3K9me2S10ph peptide was further enhanced by additional acetylation. This effect was slightly more pronounced for K14ac (Kd=49 μM for K9ac/S10ph and Kd=35 μM for S10ph/K14ac). The H3S28ph peptide was bound with much higher initial affinity than the H3S10ph peptide (Kd=30 μM), which may possibly be attributed to the proline at position 30 that can also be found in one of the high-affinity 14-3-3-binding motifs RSXSpXP, where a proline is located at position n+2 from the phosphorylated serine (Yaffe et al, 1997). Importantly, similar results were also obtained with 14-3-3ζ without GST-tag and 14-3-3ɛ (Figure 3B).
Figure 3.
Binding of 14-3-3 to an H3S10ph peptide is enhanced by additional lysine acetylation. (A) Lysine acetylation increases the affinity of 14-3-3 for the phosphorylated H3 peptide. Binding of 14-3-3ζ to the indicated H3 peptides was analyzed by fluorescence polarization measurements. The panel shows the average of at least three independent measurements (mean±s.d.). (B) Dissociation constants (Kd in μM) for the interaction of different 14-3-3 isoforms with the indicated histone H3 peptides determined by fluorescence polarization measurements. Values are average (mean±s.d.) of at least three independent measurements.
In conclusion, our biochemical and biophysical studies indicate a function of the double and triple modified histone H3 forms in the recruitment of 14-3-3 proteins: interaction between histone H3 and 14-3-3 is mediated by S10 phosphorylation and acetylation of K9 or K14 significantly increases the affinity of 14-3-3 for the histone H3 tail.
14-3-3 Proteins are recruited to the promoter region of the HDAC1 gene by histone H3 phosphoacetylation
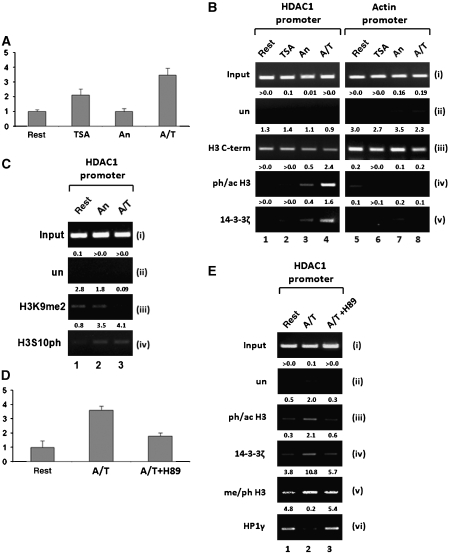
Next, we sought to determine whether 14-3-3 proteins associate with the HDAC1 gene in a histone H3 phosphoacetylation-dependent manner. As shown previously (Hauser et al, 2002), HDAC1 expression in resting 3T3 fibroblasts was low but could be efficiently stimulated by combinatorial treatment with anisomycin and TSA (Figure 4A). In contrast, anisomycin alone did not induce HDAC1 transcription and TSA had an intermediate effect. A possible recruitment of 14-3-3 proteins to the HDAC1 promoter region was investigated by ChIP assays of resting and stimulated 3T3 fibroblasts (Figure 4B; Supplementary Figure S4B). Histone H3 phosphoacetylation was absent from the HDAC1 promoter region in resting cells, both in the presence and absence of TSA (Figure 4B, panel iv). Anisomycin treatment moderately elevated the levels of ph/ac histone H3, whereas additional treatment with TSA led to high levels of phosphoacetylation that are linked to transcriptional induction of the HDAC1 gene (Figure 4A and B; Hauser et al, 2002). Interestingly, the recruitment of 14-3-3ζ to the HDAC1 promoter was in strong correlation with the levels of ph/ac histone H3 (Figure 4B, panel v, lanes 1–4). In contrast, H3 phosphoacetylation and 14-3-3 recruitment were not observed at control genes such as β-actin (Figure 4B, panel v, lanes 5–8) or histone H4 (data not shown). As transcriptional activation can be accompanied by a reduction of nucleosome density within the proximal promoter region (Yuan et al, 2005), ChIP assays with C-terminal histone H3 antibodies showed that activation of the HDAC1 promoter resulted in only a slight reduction in histone occupancy (Figure 4B, panel iii; Supplementary Figure S4B). However, similar changes in nucleosome density upon anisomycin/TSA treatment were also observed for the actin control region. In accordance with the data from the mass spectrometry analysis, TSA treatment had no effect on the anisomycin-mediated H3S10 phosphorylation on the HDAC1 promoter (Figure 4C, panel iv). ChIP assays with H3K9me2 antibodies showed reduced dimethylation in response to anisomycin and in particular to anisomycin and TSA (Figure 4C, panel iii). However, dot blot analysis with different modified histone H3 peptides revealed that additional S10 phosphorylation reduced the binding affinity of the H3K9me2 antibodies (Supplementary Figure S4). Therefore, we cannot deduce a reduction in H3K9 dimethylation in response to stimulation by anisomycin. To circumvent this problem, we used in the following experiment antibodies directed against H3K9me2S10ph (Supplementary Figure S4A).
Figure 4.
Localization of 14-3-3 proteins to the HDAC1 promoter correlates with histone H3 phosphoacetylation. (A) Phosphorylation and acetylation of histone H3 synergize in HDAC1 gene expression. Resting 3T3 fibroblasts were either left untreated (rest) or stimulated with TSA, anisomycin (An) or both drugs simultaneously (A/T) for 6 h. HDAC1 mRNA expression was analyzed by quantitative real-time RT–PCR and normalized to β-actin. Data are depicted relative to the untreated control as average of two independent experiments (mean±s.d.). (B) Localization of 14-3-3ζ to the HDAC1 promoter correlates with phosphoacetylation of histone H3. Chromatin was prepared from resting and TSA-, anisomycin- or TSA/anisomycin-stimulated 3T3 fibroblasts and ChIP assays were performed with the indicated antibodies. Rabbit pre-immune serum was used as unspecific antibody control (un). Precipitated and input DNAs were analyzed by semi-quantitative PCR with primers specific for the HDAC1 promoter and the β-actin promoter. Intensities were quantified relative to input DNA (arbitrary units) and are indicated above the panels. (C) The H3K9me2 and H3S10ph epitopes are inversely present at the HDAC1 promoter. Induction of histone H3 phosphoacetylation via anisomycin (An) or combinatorial treatment (A/T) leads to reduced epitope availability for the H3K9me2 antibody whereas the H3S10ph antibody is only slightly affected. (D) Transcriptional super-induction of HDAC1 requires the nucleosomal response. Resting 3T3 fibroblasts were treated with anisomycin and TSA (A/T), or in addition pretreated with H89 (10 μM) 15 min before A/T treatment (A/T+H89). Expression analysis was performed as described for panel A. (E) The nucleosomal response is required for localizing 14-3-3ζ to the HDAC1 promoter and to generate the phosphomethyl dual mark. ChIP analysis with antibodies specific for modified histone H3, 14-3-3ζ and HP1γ was performed as described for panel B.
Our in vitro studies show that 14-3-3ζ is a high-affinity detector protein for ph/ac histone H3. To confirm that 14-3-3ζ localization to the HDAC1 promoter is indeed dependent on ph/ac histone H3, we used the kinase inhibitor H89, a potent suppressor of the nucleosomal response (Thomson et al, 1999). Pretreatment of cells with H89 abolished anisomycin/TSA induced H3 phosphoacetylation at the HDAC1 promoter (Figure 4E, panel iii; Supplementary Figure S4B) and significantly reduced transcriptional activation (Figure 4D). Importantly, the recruitment of 14-3-3ζ to the promoter was strongly impaired upon inhibition of the nucleosomal response (Figure 4E, panel iv). Phosphomethylation of histone H3, which was increased upon anisomycin/TSA treatment, was also sensitive to H89 (Figure 4E, panel v). In agreement with data from the mass spectrometry analysis, this finding indicates that at responsive loci the K9 methylated histone H3 is converted into a multiple modified form. HP1 proteins recognize and bind methylated K9 at histone H3 and were previously shown to be involved in the repression of euchromatic genes (Nielsen et al, 2001; Ogawa et al, 2002; Hediger and Gasser, 2006). Therefore, we analyzed the effect of H3 phosphorylation on the binding of HP1 in a euchromatic environment. ChIP assays revealed that HP1γ was located at the HDAC1 promoter in resting cells, but was released upon induction of phosphoacetylation and phosphomethylation (Figure 4E, panel vi, lanes 1 and 2). HP1 dissociation was blocked by H89, indicating that the nucleosomal response is important not only for the recruitment of 14-3-3ζ but also for the release of HP1γ (Figure 4E, panel vi, lane 3). We conclude that activation of the HDAC1 gene is associated with differential localization of two histone H3-binding modules HP1 and 14-3-3. These show opposing binding affinities for combinatorial modifications of histone H3. The transcriptional ‘off-state' is characterized by H3K9me2 and HP1γ binding, while activation is linked to phosphoacetylation/phosphomethylation, recruitment of 14-3-3ζ and concomitant HP1γ displacement.
14-3-3ζ Is required for transcriptional induction of the HDAC1 gene
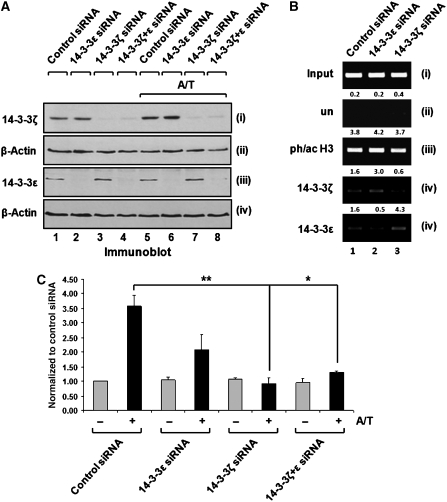
Given that 14-3-3ζ is recruited to the HDAC1 promoter region in a phosphoacetylation-dependent manner, we wanted to determine whether this recruitment has an impact on transcriptional induction. To address this question, we used siRNA-mediated depletion of 14-3-3ɛ and ζ proteins in HeLa cells, as pilot experiments indicated that 14-3-3 knockdown was most efficient in this cell system. In addition, several phosphoacetylation target genes that were previously identified in 3T3 mouse fibroblasts were confirmed to be activated by anisomycin and TSA in this system (data not shown). Protein and mRNA levels of 14-3-3ɛ and ζ were efficiently reduced by siRNA-mediated gene silencing, while the levels of both isoforms were unaffected by an unspecific control siRNA (Figure 5A, panels i and iii, lanes 1–4; Supplementary Figure S5A). Anisomycin and TSA treatment did not affect the efficiency or specificity of the knock down (Figure 5A, panels i and iii, lanes 5–8; Supplementary Figure S5A).
Figure 5.
14-3-3ζ Is required for transcriptional activation of the HDAC1 gene by anisomycin and TSA. (A) Isoform-specific depletion of 14-3-3 proteins by RNA interference. HeLa cells were transfected with siRNAs against 14-3-3ɛ or ζ and either left untreated or stimulated with anisomycin and TSA (A/T). Whole-cell lysates were prepared and analyzed for 14-3-3 protein levels by immunoblotting. Equal loading was confirmed with a β-actin antibody. (B) 14-3-3 Depletion has no impact on the phosphoacetylation status of histone H3. HeLa cells, depleted of 14-3-3ɛ or ζ isoforms, and control cells were stimulated with TSA and anisomycin (sAn) for 1 h. ChIP assays were performed with the indicated antibodies. (C) Depletion of 14-3-3ζ interferes with transcriptional activation of the HDAC1 gene. HeLa cells were transfected with siRNAs as described for panel A. HDAC1 expression levels were determined by real-time RT–PCR, normalized to GAPDH levels and are depicted as fold increase compared with untreated samples, which were transfected with control siRNA. Values represent three independent experiments (mean±s.d.). Depletion of 14-3-3ζ interferes with the induction of HDAC1 by anisomycin and TSA treatment (lane 6, **P=0.002 and lane 8, *P=0.01, t-test), whereas 14-3-3ɛ depletion caused a more moderate reduction (lane 4, P=0.07, t-test).
Next, we examined the impact of 14-3-3 depletion on H3 phosphoacetylation. Since 14-3-3 proteins are important components of MAP kinase pathways (Xing et al, 2000), we first analyzed whether the signal transduction cascades mediating H3 phosphoacetylation remain intact in cells depleted for particular isoforms. No significant change in bulk histone phosphoacetylation was observed in 14-3-3 knockdown cells (Supplementary Figure S5B), indicating that the p38 MAP kinase pathway remains fully functional. Furthermore, ChIP experiments of 14-3-3 knockdown cells confirmed that phosphoacetylation at the HDAC1 promoter was not impaired upon loss of either of the two 14-3-3 proteins (Figure 5B, panel iii).
Finally, we examined the effect of 14-3-3 knockdown on the induction of the HDAC1 gene, which can be induced by combinatorial treatment with anisomycin and TSA in proliferating HeLa cells (Figure 5C, lanes 1 and 2). Depletion of 14-3-3ɛ had a negative impact on HDAC1 activation (Figure 5C, lanes 3 and 4). However, this effect was not statistically significant (P=0.07) and therefore a direct role for 14-3-3ɛ in HDAC1 expression cannot be reliably inferred. However, loss of 14-3-3ζ strongly interfered with transcriptional induction, suggesting an activating role of 14-3-3ζ at the HDAC1 promoter (Figure 5C, lanes 5 and 6). The same effect was observed for the 14-3-3ɛ and ζ double knockdown (Figure 5C, lanes 7 and 8).
Altogether, these data indicate that 14-3-3 proteins recognize ph/ac histone H3 and are required for the induction of the phosphoacetylation target gene HDAC1.
Discussion
Additional PTMs modulate the phosphorylation-dependent interaction of histone H3 with 14-3-3
In this report we investigated the function of 14-3-3ɛ/ζ as detector proteins for phosphoacetylated histone H3 and their roles in transcriptional regulation. The first important finding of this study is the modulation of the phosphorylation-dependent interaction between 14-3-3 and the histone H3 tail. Although the binding affinities of 14-3-3 proteins were correlated with the amino-acid sequence, flanking the phosphorylated residue (Yaffe et al, 1997; Uchida et al, 2006), this is to our knowledge the first report demonstrating that this interaction can be affected by PTMs. Phosphorylation of histone H3 is clearly required to mediate interaction of histone H3 with 14-3-3. In contrast, acetylation as single modification does not result in significant interaction. However, in the context of H3S10 phosphorylation, additional acetylation causes a significant shift in the equilibrium toward the bound state (Figures 2 and 3). This effect was observed in a more pronounced manner for S10 phosphorylation rather than for S28 (Figure 1C, and data not shown), which may be addressed to the initial higher affinity of 14-3-3 for the S28ph peptide (Macdonald et al, 2005; Figure 3). In this scenario, lysine acetylation could function as an ‘auxiliary modification' that supports the relatively weak interaction of 14-3-3 with H3S10ph, whereas the significantly stronger interaction with H3S28ph is less reliant on additional modifications. In addition, the dual modification would allow for an additional level of regulation. By relying on two modifications for complete stimulation, a more fine-tuned response could be achieved. Especially since the machineries laying down the two marks are different and respond to distinct signaling pathways. Although a previous study indicated no significant effect of dual H3K9/K14 acetylation on this interaction (Macdonald et al, 2005), our data set clearly indicates a modulation of the 14-3-3/histone H3 interaction by single acetylation. One explanation could be that the positive effect of single acetylation observed in our study is neutralized by the dual acetylation mark. It might be that a single acetylation helps stabilizing the interaction by generating additional contacts within the phosphoacetyl peptide and also with 14-3-3. These findings were also corroborated by in vivo experiments. MAP kinase activation resulted in low levels of ph/ac histone H3 and minor recruitment of 14-3-3ζ to the HDAC1 promoter region; additional TSA treatment increased local phosphoacetylation and 14-3-3 recruitment (Figure 4B). Given that the deacetylase inhibitor does not affect the abundance of the H3S10ph modification per se (Hauser et al, 2002; Supplementary Figure S3), these data suggest that the phosphoacetylated form is also preferred in native chromatin.
Since several 14-3-3-interacting proteins such as HMGN1 or p53 are targeted by multiple different PTMs, this finding is of particular interest regarding 14-3-3 biology. We hypothesize that additional PTMs could also modulate 14-3-3 binding to other proteins. Since the amino-acid composition of histone H3 surrounding S10 does not match one of the high-affinity 14-3-3-binding motifs, it is possible that the postulated modulation of binding may only be relevant for a specific subset of 14-3-3-associated proteins with initial low-affinity binding.
Combinatorial histone modifications and detector proteins
While in mitosis the majority of histone H3 is decorated by the S10ph mark, activation of the MAP kinase pathway in resting cells leads to the phosphorylation of a minute fraction of H3 molecules. Our mass spectrometry analysis reveals the presence of additional lysine methylation and acetylation besides HS10ph in particular upon combinatorial treatment of cells (Table II). We did not observe an H3K9acS10ph form in anisomycin-stimulated cells, although previous research clearly demonstrated the generation of this species under these conditions. One possible reason may be the extremely low abundance of this species that could be picked up by high-affinity antibodies but not by bulk analysis using mass spectrometry. The potential modulation of protein–histone interactions by different PTM combinations was formulated as the histone code hypothesis (Cheung et al, 2000a; Strahl and Allis, 2000), postulating that PTMs can cooperate in directing the accessibility of chromatin. In agreement with this hypothesis, 14-3-3 binding to H3S10ph is enhanced by additional active marks (K14ac or K9ac). Interestingly, HP1γ as a detector of the repressive H3K9me2 mark shows a reciprocal affinity: phosphorylation or phosphoacetylation of histone H3 triggered the displacement of HP1 from mitotic chromatin (Mateescu et al, 2004; Fischle et al, 2005; Hirota et al, 2005). It is important to mention that 14-3-3ɛ and ζ are globally excluded from mitotic chromosomes (Macdonald et al, 2005, and data not shown), which may be attributed to the generation of a ‘14-3-3 sink' during mitosis (Margolis et al, 2006). Thus it appears that 14-3-3 proteins are specific detectors for phosphorylated histone H3 during interphase.
14-3-3 Proteins are required for transcriptional induction of the HDAC1 gene
Based on the diametrical binding affinities of 14-3-3 for ph/ac histone H3 and HP1γ for H3K9me2, it is tempting to speculate that the two factors epitomize different transcriptional states in the regulation of particular target genes. This concept is supported by a recent report, which demonstrated HP1γ displacement during transcriptional initiation concomitant with H3S10ph during progesterone receptor-mediated gene activation (Vicent et al, 2006).
We observed H3K9me2 and localization of HP1γ at the promoter of the transcriptional silent HDAC1 gene (Figure 4C and E). Stimulation of H3 phosphoacetylation resulted in the recruitment of 14-3-3ζ (Figure 4B and E), which correlated with the displacement of the transcriptional repressor HP1γ (Figure 4E). Inhibition of the nucleosomal response resulted in reduced transcriptional responsiveness accompanied by HP1γ retention and absence of 14-3-3 (Figure 4E). Therefore, one particular function of histone phosphorylation in this context may be replacement of the transcriptional repressive module. We cannot completely rule out that lack of 14-3-3 recruitment and persistence of HP1γ at the HDAC1 promoter may be caused by reduced transcriptional responsiveness of the promoter upon kinase inhibition. However, the adaptations of the chromatin embedding the HDAC1 promoter region occur much earlier than transcriptional initiation (Hauser et al, 2002). Together with the contrary binding affinities of HP1 and 14-3-3 for phosphomethylated/acetylated histone H3 and the appearance or absence of these marks in the different treatment conditions, we consider a direct effect via the reduction of H3S10 phosphorylation more likely than an indirect via reduced transcriptional responsiveness.
Our ChIP experiments suggest that the H3K9me2 mark at the HDAC1 promoter is converted to K9me2S10ph rather than being actively removed (Figure 4E). This scenario is also supported by mass spectrometry results where corresponding double and triple modified states of histone H3 have been identified (Table II). These observations expand the binary switching model (Fischle et al, 2003) from a mitosis specific mechanism to interphase transcription and implicate a particular function of histone H3 phosphorylation in this process.
The 14-3-3 RNAi experiments demonstrate a requirement for 14-3-3 proteins in the transcriptional induction of HDAC1 (Figure 5C). In agreement with earlier studies on the function of 14-3-3 proteins in MAP kinase cascades (Xing et al, 2000), a reduction did not impair H3 phosphoacetylation. However, depletion of 14-3-3ζ almost completely abolished the transcriptional induction of HDAC1. Thus, 14-3-3 proteins are crucial factors for mediating the switch from transcriptional repressive to active chromatin in vivo, although they appear not essential for establishing histone phosphoacetylation per se.
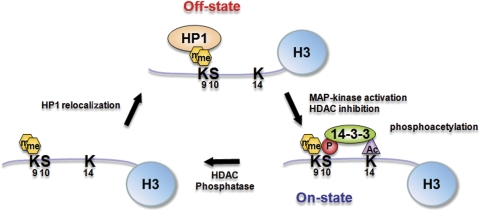
Based on our analysis, we suggest the model shown in Figure 6 for induction of the HDAC1 gene. The transcriptional silent state encountered in resting cells is epitomized by the presence of H3K9me2 bound by HP1γ and the absence of ph/ac histone H3. Stimulation of histone H3 phosphoacetylation leads to HP1γ displacement (Figure 4B and E; E Simboeck and C Seiser, unpublished observations) and consolidated binding of 14-3-3, which can now trigger the switch to transcriptional competent chromatin environment in the presence of H3K9me2. Binding of 14-3-3, in turn, might stabilize the ph/ac mark on histone H3. We also hypothesize that the activity of phosphatases and deacetylases may be required for the conversion of me2/ph and me2/ph/ac histone H3 into a K9me2 form, which could trigger subsequent relocalization of HP1 to the promoter region and transcriptional ‘shut-down' of the target.
Figure 6.
Model for the role of histone H3 modifications in the regulation of HP1 and 14-3-3 binding during transcriptional induction of the HDAC1 gene. In the transcriptional silent state, the HDAC1 promoter region is occupied by H3K9me2-modified nucleosomes and HP1γ. Activation of MAP kinase signaling via anisomycin and inhibition of HDAC activity via TSA leads to the formation of triple modified histone H3, stable binding of 14-3-3 and transcriptional induction of the HDAC1 gene. Removal of the phosphorylation and acetylation marks via phosphatases and deacetylases, respectively, can regenerate K9 dimethylated histone H3 and allow the re-association of HP1 proteins.
To examine a more general role of the dual H3 modification in gene activation, we have recently identified a set of novel phosphoacetylation target genes in mouse fibroblasts (E Simboeck, C Hauser and C Seiser, unpublished observations). Importantly, all tested target genes showed recruitment of 14-3-3 to their promoters and a requirement for 14-3-3ζ for full gene activation. These results suggest that 14-3-3 proteins have a more general role in the regulation of genes targeted by histone H3 phosphoacetylation.
Materials and methods
Plasmid construction and site-directed mutagenesis
Expression vectors for N-terminal GST-tagged mouse 14-3-3ɛ and 14-3-3ζ were produced by conventional PCR cloning of cDNA into the pGEX-5X2 vector (Amersham Biosciences). For T7 promoter-driven in vitro transcription/translation reactions, 14-3-3ɛ and 14-3-3ζ cDNAs were cloned into the pCINeo vector (Promega). Histone H3 mutant constructs were generated in the pET-3d vector using the QuickChange site-directed mutagenesis kit II (Stratagene). All constructs were verified by DNA sequencing. Specific sequence data are available upon request.
Cell culture, transfection and reagents
Swiss 3T3 mouse fibroblasts and HeLa human cervix carcinoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS). 3T3 cultures were rendered quiescent by incubation in DMEM containing 0.2% (v/v) FCS for 72 h. The siRNAs specific for human 14-3-3ɛ (sc-29588) and 14-3-3ζ (sc-29583), and unspecific control siRNA (sc-37007), were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cells were transfected with siRNAs at a final concentration of 24 nM using Lipofectamine 2000 (Invitrogen) and grown for additional 72 h. Control transfection experiments were carried out using an unspecific control siRNA at a final concentration of 48 nM.
The following drugs were used in this study: TSA (165 nM; Wako Pure Chemical Industries), anisomycin (180 nM; Sigma) and H89 (10 μM; Alexis Biochemicals).
RNA isolation and real-time RT–PCR analysis
Total RNA was isolated with TRIZOL reagent (Invitrogen) as specified by the manufacturer. For cDNA, 1 μg of total RNA was reverse transcribed with iScript cDNA synthesis kit (BioRad). Real-time RT–PCR reactions were performed with 0.5 μl of the RT reaction by iCycler iQ system (BioRad) using SYBER green (Molecular probes) for labeling. Primer sequences are available upon request.
Western blot analysis and antibodies
Histone preparation and western blot analyses were performed as previously described (Hauser et al, 2002), or using the Odyssey® infrared imaging system (LI-COR Biosciences). The following antibodies were used in this study: modification-specific histone antibodies from Upstate Biotechnology (Lake Placid, NY, USA), C-terminal histone H3 antibody from Abcam; 14-3-3ɛ (T16) and 14-3-3ζ (C16) were purchased from Santa Cruz Biotechnology and affinity-purified via recombinant GST–14-3-3ɛ and ζ proteins, respectively.
GST pull-down assay
Recombinant GST-tagged proteins were expressed in and purified from the Escherichia coli strain BL21 RIL and GST pull-down experiments were performed as described previously (Doetzlhofer et al, 1999) and in Supplementary data.
Preparation of nuclear extracts
Extracts were prepared as described at http://www.celldeath.de/apometh/emsa.html, except that all buffers were supplemented with Complete-Protease inhibitor cocktail (Roche), phosphatase inhibitors (PPI): 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, 50 mM sodium fluoride, 20 mM sodium pyrophosphate and 10 mM sodium butyrate.
In vitro peptide pull-down assay
For a detailed description, see Supplementary data. Histone H3 peptides were purchased from Peptide Specialty Laboratories GmbH (Heidelberg, Germany). Peptides were covalently coupled to agarose beads and 10 μl of peptide slurry were incubated with 2.5 μl IVT, 35S-methionine-labeled 14-3-3 proteins (TNT rabbit reticulocyte lysate system; Promega) in 50 μl total reaction volume.
Fluorescence polarization binding measurements
Measurements were performed as described previously (Jacobs et al, 2004) and in Supplementary data.
Chromatin immunoprecipitation assays
Preparation of soluble chromatin and chromatin immunoprecipitation assays were carried out as described previously (Hauser et al, 2002).
PCR analysis of immunoprecipitated DNA
All PCRs were performed on a Biometra D3 thermocycler, using Eppendorf PCR Master Mix. Primer sequences are available upon request. The linear range for each primer pair was determined empirically using different amounts of input DNA. PCRs with increasing amounts of genomic DNA were carried out along with the PCRs of the immunoprecipitated DNA. PCR products were resolved on 2% agarose–TAE gels.
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Bernd Schuettengruber, Reinhard Brunmeir, Peter Steinlein, Tim Clausen, Franck Dequiedt, Andrea Pichler, Stefan Schuechner, Oliver Pusch and Leonie Ringrose for helpful discussions. We are grateful to Thomas Jenuwein and Christian Muchardt for histone antibodies and Markus Hengstschlaeger for plasmids. This work was supported by the Austrian Science Fund (FWF P18746-B12) and the GEN-AU project ‘Epigenetic Plasticity of the Mammalian Genome' (Austrian Federal Ministry for Education, Science and Culture). Stefan Winter is a fellow of the Vienna Biocenter PhD program (Austrian Science Fund).
References
- Barratt MJ, Hazzalin CA, Cano E, Mahadevan LC (1994) Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc Natl Acad Sci USA 91: 4781–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C (1997) Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol 17: 5033–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P (2000a) Signaling to chromatin through histone modifications. Cell 103: 263–271 [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD (2000b) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Mahadevan LC (2003) MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett 546: 51–58 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Rose S, Barratt MJ, Mahadevan LC (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J 19: 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T (2002) Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12: 2090–2097 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C (1999) Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 19: 5504–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC (2005) MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci 118: 2247–2259 [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD (2005) Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003) Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Hauser C, Schuettengruber B, Bartl S, Lagger G, Seiser C (2002) Activation of the mouse histone deacetylase 1 gene by cooperative histone phosphorylation and acetylation. Mol Cell Biol 22: 7820–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Gasser SM (2006) Heterochromatin protein 1: don't judge the book by its cover!. Curr Opin Genet Dev 16: 143–150 [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM (2005) Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Fischle W, Khorasanizadeh S (2004) Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol 376: 131–148 [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J (2006) Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res 14: 393–404 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124 [DOI] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC (2005) Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell 20: 199–211 [DOI] [PubMed] [Google Scholar]
- Mahadevan LC, Clayton AL, Hazzalin CA, Thomson S (2004) Phosphorylation and acetylation of histone H3 at inducible genes: two controversies revisited. Novartis Found Symp 259: 102–111, discussion 111–104, 163–109 [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ (1991) Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65: 775–783 [DOI] [PubMed] [Google Scholar]
- Margolis SS, Perry JA, Forester CM, Nutt LK, Guo Y, Jardim MJ, Thomenius MJ, Freel CD, Darbandi R, Ahn JH, Arroyo JD, Wang XF, Shenolikar S, Nairn AC, Dunphy WG, Hahn WC, Virshup DM, Kornbluth S (2006) Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell 127: 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, England P, Halgand F, Yaniv M, Muchardt C (2004) Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep 5: 490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ (2006) The relationship between histone H3 phosphorylation and acetylation throughout the mammalian cell cycle. Biochem Cell Biol 84: 640–657 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Simboeck E, Khier H, Seiser C (2003) Autoregulation of mouse histone deacetylase 1 expression. Mol Cell Biol 23: 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS (2003) MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J 22: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Strelkov IS, Davie JR (2002) Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res 62: 75–78 [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC (1999) The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J 18: 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Mahadevan LC (2001) Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell 8: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Uchida S, Kubo A, Kizu R, Nakagama H, Matsunaga T, Ishizaka Y, Yamashita K (2006) Amino acids C-terminal to the 14-3-3 binding motif in CDC25B affect the efficiency of 14-3-3 binding. J Biochem (Tokyo) 139: 761–769 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M (2006) Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 24: 367–381 [DOI] [PubMed] [Google Scholar]
- Xing H, Zhang S, Weinheimer C, Kovacs A, Muslin AJ (2000) 14-3-3 Proteins block apoptosis and differentially regulate MAPK cascades. EMBO J 19: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91: 961–971 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures