Abstract
Sister chromatid cohesion is mediated by cohesin, but the process of cohesion establishment during S-phase is still enigmatic. In mammalian cells, cohesin binding to chromatin is dynamic in G1, but becomes stabilized during S-phase. Whether the regulation of cohesin stability is integral to the process of cohesion establishment is unknown. Here, we provide evidence that fission yeast cohesin also displays dynamic behavior. Cohesin association with G1 chromosomes requires continued activity of the cohesin loader Mis4/Ssl3, suggesting that repeated loading cycles maintain cohesin binding. Cohesin instability in G1 depends on wpl1, the fission yeast ortholog of mammalian Wapl, suggestive of a conserved mechanism that controls cohesin stability on chromosomes. wpl1 is nonessential, indicating that a change in wpl1-dependent cohesin dynamics is dispensable for cohesion establishment. Instead, we find that cohesin stability increases at the time of S-phase in a reaction that can be uncoupled from DNA replication. Hence, cohesin stabilization might be a pre-requisite for cohesion establishment rather than its consequence.
Keywords: chromosome segregation, cohesin, Mis4/Ssl3, Schizosaccharomyces pombe, Wapl
Introduction
In eukaryotes, newly replicated sister chromatids are linked together upon S-phase exit by a conserved protein complex known as cohesin (Haering et al, 2002). Cohesin consists of four core, conserved subunits called Scc1, Scc3, Smc1 and Smc3 in Saccharomyces cerevisiae (Rad21, Psc3, Psm1 and Psm3 in the fission yeast Schizosaccharomyces pombe). Smc1 and Smc3 are ATPases of the ABC family. They interact through a dimerization domain separated from their ATPase head by a long, flexible, coiled–coil segment. Scc1 bridges each ATPase head of the Smc1/3 heterodimer, thereby creating a tripartite ring and Scc3 binds to the ring through its interaction with Scc1 (Haering et al, 2002; Arumugam et al, 2003; Weitzer et al, 2003). How cohesin ensures cohesion is unknown, although experimental evidence suggests that the ring-shape structure may tether sister DNA strands by encircling them (Haering et al, 2002; Ivanov and Nasmyth, 2005, 2007).
The establishment of cohesion is a two-step process. Cohesin is first deposited on unreplicated chromatin in a reaction requiring ATP hydrolysis by the Smc heads and the cohesin-loading complex Scc2/Scc4 (Ciosk et al, 2000; Arumugam et al, 2003; Weitzer et al, 2003). The establishment of cohesion occurs during the ensuing S-phase, and several observations point to the idea that cohesion establishment is coupled with DNA replication. Mutations in factors related to DNA replication affect sister chromatid cohesion. These include the DNA polymerase alpha-binding protein Ctf4 (Miles and Formosa, 1992; Hanna et al, 2001), Chl1 helicase (Petronczki et al, 2004) and components of an alternative replication factor C complex (Mayer et al, 2001). The putative acetyl-transferase Eco1 (Ctf7) is associated with the replisome (Kenna and Skibbens, 2003; Lengronne et al, 2006; Moldovan et al, 2006) and mutations in this factor do not prevent cohesin loading in G1, but cohesion is not established during S-phase (Skibbens et al, 1999; Toth et al, 1999). In budding yeast, cohesion establishment during DNA replication can proceed without further cohesin recruitment and without need for cohesin to reengage an ATP hydrolysis motif that is critical for its initial chromatin binding in G1 (Lengronne et al, 2006). The replication fork might use prebound cohesin and convert it into functional cohesion, but the mechanism is still enigmatic, as no evidence has been reported so far showing a difference in cohesin structure before and after cohesion establishment (Weitzer et al, 2003). However, a recent study in mammalian cells revealed an interesting link between cohesin dynamics and S-phase progression. Live-cell imaging has shown that cohesin is not stably bound to chromatin during the G1 phase of the cell cycle. Chromatin-bound cohesin exchanges with the soluble nuclear pool and this reaction is controlled by the Wapl gene product (Gerlich et al, 2006; Kueng et al, 2006). Importantly, a fraction of cohesin becomes stably bound to chromatin while cells progress through S-phase. As cohesion is established at that time, this raised the possibility that the change in cohesin dynamics might be instrumental in the process of cohesion establishment. Stabilization of cohesin might be a manifestation of cohesion establishment or, alternatively, a stable cohesin association with chromatin might be a prerequisite for cohesion establishment. We address these questions using fission yeast as a model organism. If cohesin dynamics are important for the function of sister chromatid cohesion, then it should be conserved across evolution. Indeed, we present evidence that similar to mammalian cells, fission yeast cohesin displays dynamic behavior on G1 chromatin and this process requires a functional wpl1 gene. Strikingly, wpl1 is not essential for viability, indicating that a wpl1-dependent change in cohesin dynamics is not crucial for the establishment of cohesion. Furthermore, we show that cohesin association with chromatin is stabilized at the time of S-phase, but this stabilization can proceed independently of DNA replication. These observations indicate that stabilization of cohesin might be a pre-requisite for cohesion establishment rather than a consequence.
Results
The loading machinery is continuously required for cohesin binding to chromatin during the G1 phase
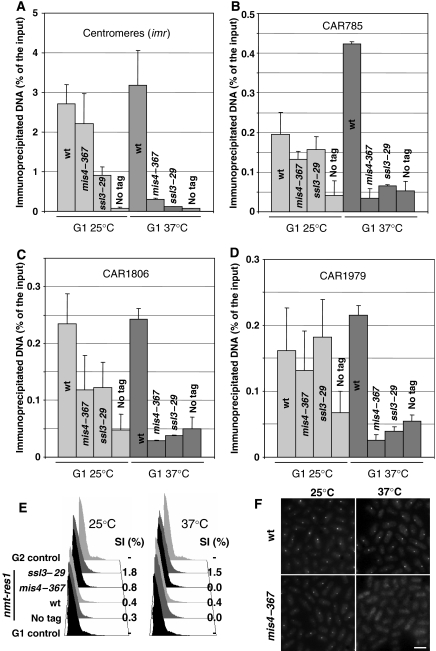
We recently reported (Bernard et al, 2006) the identification of Ssl3, which, together with Mis4 (Furuya et al, 1998), forms a complex analogous to the Scc2/Scc4 cohesin-loading machinery from budding yeast (Ciosk et al, 2000). In the course of this work, we were curious to see whether cohesin loading was an irreversible event. We reasoned that if cohesin were stably bound to chromatin once loaded, then it should remain bound after inactivation of the loading machinery. To test this, cells carrying the thermosensitive allele mis4–367 or ssl3–29 (Bernard et al, 2006) were arrested in late G1 by titrating out the Cdc10 transcription factor by overexpression of a C-terminal fragment of its binding partner Res1 (Ayte et al, 1995). Once arrested in G1 at permissive temperature, cells were shifted to 37°C while remaining arrested in G1. Cohesin association to chromatin was assessed by chromosome spreads and chromatin immunoprecipitation (ChIP) before and after the shift. As shown in Figure 1, the Rad21 cohesin subunit was detected on nuclear spreads of G1-arrested cells, both in wild-type (wt) and mutant background at the permissive temperature. By contrast, Rad21 detection dropped close to background levels after the inactivation of Mis4 or Ssl3. Centromeric heterochromatin is a major site of cohesin binding in fission yeast (Tomonaga et al, 2000; Bernard et al, 2001b; Nonaka et al, 2002). ChIP analyses showed that Rad21 dissociated from centromeres, when either Mis4 or Ssl3 was inactivated in G1 (Figure 2A). Similarly, Rad21 binding was abolished at three cohesin-associated regions (CARs) along chromosome 2 (Figure 2B–D). We conclude that the cohesin-loading complex is required to maintain chromosome association of cohesin during G1.
Figure 1.
The loading machinery is required for sustained cohesin binding to chromatin in G1. Cells bearing thermosensitive mutations in genes encoding the cohesin-loading complex mis4 and ssl3 were arrested in G1 at permissive temperature by overexpressing a C-terminal Res1 fragment, under the control of the inducible nmt1 promoter (Ayte et al, 1995). Cells were then shifted to 37°C for 2 h to inactivate cohesin loading. (A) Rad21-HA chromatin association was monitored on chromosome spreads by immunofluorescence at 25°C and after shift to 37°C. DNA was stained with 4′-6-diamidino-2-phenylindole (DAPI). Scale bar, 5 μm. (B) Quantification of Rad21-HA fluorescence intensity. A total of 50–100 nuclei were analyzed for each sample. The error bars show the confidence interval of the mean with α=0.05. (C) DNA content analysis and septation index (SI) show that cells remained arrested in G1 throughout the course of the experiment.
Figure 2.
Inactivation of the cohesin-loading complex in G1 causes loss of cohesin from centromeres and chromosome arms. Rad21-HA association with chromosomes was determined by ChIP before and after Mis4 or Ssl3 inactivation as in Figure 1. Rad21-HA enrichment was monitored at centromeres (A), and three cohesin-binding sites along chromosome 2 (B–D, the numbering refers to their coordinates in kb). The error bars show the s.d. calculated from at least two independent experiments. (E) DNA content analysis and septation index (SI) confirm the arrest in G1. (F) Rec8 accumulation at centromeres in G1 cells relies on functional Mis4. Strains bearing Rec8-GFP in a mis4–367 or wt background were arrested in G1 by nitrogen starvation at 25°C to induce Rec8-GFP accumulation at centromeres and then shifted to 37°C for 2 h. Scale bar, 5 μm.
Next, we asked whether the meiotic cohesin subunit Rec8 would behave similarly. In response to nitrogen starvation, fission yeast cells arrest in G1, in preparation for mating and meiosis. At this time, Rec8 is enriched at centromeres, which are clustered close to the spindle pole body (Bernard et al, 2001a) and can be visualized by a Rec8-GFP fusion protein as a fluorescent dot. As shown in Figure 2F, the Rec8-GFP dot disappeared when Mis4 was inactivated, whereas it persisted throughout the course of the experiment in wt background. From these experiments, we conclude that Mis4 and Ssl3 are continuously required for cohesin binding to G1 chromosomes. A possible explanation is that Mis4/Ssl3 is directly involved in anchoring cohesin to chromosomes in G1. This seems unlikely, however, as neither Mis4 nor Ssl3 was detected at centromeres in G1-arrested cells (Supplementary Figure S1), whereas sustained Rad21 binding at this locus requires Mis4/Ssl3. In line with this observation, distinct binding sites for cohesin and its loading machinery have been reported along whole chromosomes in budding yeast (Lengronne et al, 2004). A more likely explanation for our finding is that, like in human cells, fission yeast cohesin is bound to G1 chromosomes in a dynamic fashion and requires iterative reloading by Mis4/Ssl3 to maintain chromosome association. Cohesin may be intrinsically unstable in G1, or an antagonistic activity may exist that counteracts the loading reaction.
Fission yeast Wapl controls cohesin binding to chromatin in G1
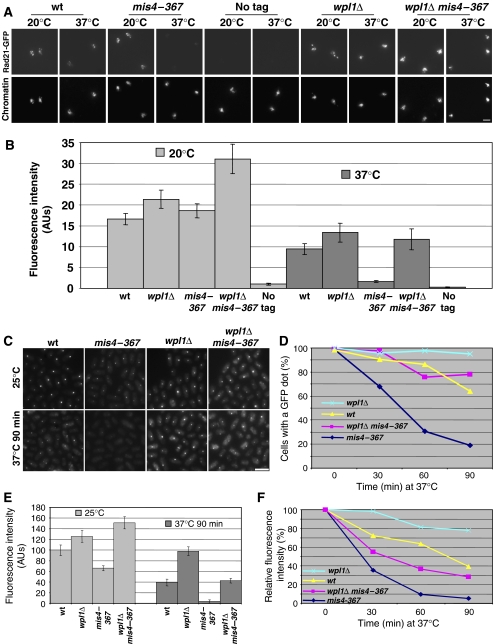
During the course of this study, two papers reported a role for Wapl in promoting cohesin removal from chromosomes during early mitosis in higher eukaryotes (Gandhi et al, 2006; Kueng et al, 2006). Intriguingly, Wapl depletion also increased the residence time of cohesin on chromatin in interphase. Given that Wapl is conserved in fission yeast, we asked whether it might control cohesin binding to G1 chromosomes. First, the wpl1 gene was deleted and it proved nonessential for growth at all temperatures tested (Decottignies et al, 2003; Supplementary Figure S2). Next, we examined whether the absence of Wapl would affect cohesin behavior in G1 cells. As shown in Figure 3, the steady-state amount of chromatin-bound Rad21 was slightly increased in wpl1Δ cells as compared with wt, both at 20 and 37°C. Similarly, Rec8-GFP signal intensity was increased in wpl1Δ cells versus wt (Figure 3C and E), consistent with the idea that Wapl negatively regulates cohesin binding to chromatin. When the loading machinery was now inactivated in G1-arrested wpl1Δ cells, a substantial amount of Rad21 remained chromatin bound. Similarly, Rec8 foci stayed readily visible after mis4–367 wpl1Δ cells were shifted to the restrictive temperature (Figure 3C–F). Even though wpl1 deletion clearly stabilized cohesin binding, chromatin-bound cohesin appeared partially reduced after Mis4 inactivation. We conclude that wpl1 deletion suppressed removal of a substantial fraction of cohesin from chromosomes during G1. We also noticed that the amount of chromatin-bound cohesin appeared increased in mis4–367 wpl1Δ cells at the permissive temperature when compared with wpl1Δ cells (Figure 3B and E). The reason for this is currently unclear.
Figure 3.
Fission yeast Wapl regulates cohesin dynamics in G1. (A) Cells were arrested in G1 at 20°C by Res1 C-terminal overexpression and then shifted to 37°C for 2 h. Rad21-GFP association with chromatin was monitored on chromosome spreads by immunofluorescence using anti-GFP antibodies. Scale bar, 5 μm. (B) Rad21-GFP fluorescence intensity was measured for 50–100 nuclei per sample. The error bars show the confidence interval of the mean with α=0.05. (C) Strains bearing rec8-GFP were arrested in G1 by nitrogen starvation at 25°C to induce Rec8-GFP accumulation at centromeres and then shifted to 37°C for 90 min. Samples were taken every 30 min and examined for Rec8-GFP fluorescence. Scale bar, 5 μm. (D) Proportion of cells with a dot of Rec8-GFP after inactivation of Mis4. More than 150 cells were examined for each sample. (E) Quantification of Rec8-GFP fluorescence before and 90 min after the temperature shift. More than 150 cells were analyzed for each sample. The error bars show the confidence interval of the mean with α=0.05. (F) Relative Rec8-GFP fluorescence intensity after shift to 37°C. Rec8-GFP fluorescence for each strain is normalized to its value at the time of temperature shift.
In summary, we found using two different assays that cohesin binding to G1 chromatin is unstable, as sustained cohesin binding requires the continuous activity of the loading machinery, and that cohesin instability depends at least in part on Wapl function. These observations suggest that, as in mammals, cohesin's binding to chromatin is dynamic during the G1 phase of the fission yeast cell cycle, the steady-state amount being tuned by the balance between opposing activities, a loading activity provided by the Mis4/Ssl3 complex and a destabilizing, or unloading, activity provided by Wapl.
Cohesin binding to chromosomes is stabilized during S-phase independently of DNA replication
Experiments in budding and fission yeast have shown that the cohesin-loading factors are dispensable for viability in G2, when cohesion has been established (Ciosk et al, 2000; Bernard et al, 2006). In fission yeast, inactivation of the loading machinery at that time has little effect on cohesin binding to centromeres as assayed by ChIP (Bernard et al, 2006), and by nuclear spreads about 60% of chromatin-bound Rad21 can be detected after Mis4 inactivation (Supplementary Figure S3). In mammalian cells, about one-third of nuclear cohesin becomes stably bound to chromatin in G2 (Gerlich et al, 2006). As the binding of cohesin to chromosomes appears labile in G1, but stabilized in G2, we asked how cohesin becomes stable during the intervening S-phase. Stabilization might be the consequence of the establishment of sister chromatid cohesion during DNA replication (Uhlmann and Nasmyth, 1998), or of cell-cycle regulation in S-phase, independently of cohesion establishment. Cells bearing the mis4–367 or ssl3–29 mutations were arrested in early S-phase using hydroxyurea (HU) at permissive temperature and shifted to 37°C for 2 h, while still arrested. Cohesin binding to chromatin was again monitored before and after the shift by chromosome spreading and Rad21-HA immunofluorescence. As shown in Figure 4, Rad21 levels only slightly decreased after Mis4 or Ssl3 inactivation, in contrast to the almost complete loss of cohesin observed in G1-arrested cells (Figure 4A and B; compare with Figure 1). This implies that Rad21 became more stably bound to chromosomes in S-phase cells.
Figure 4.
Rad21 remains chromatin bound when cohesin loading is inactivated in S-phase. Cells bearing rad21-HA and the thermosensitive mutations mis4–367 or ssl3–29 were arrested in early S-phase by hydroxyurea (HU) treatment at 25°C and then shifted to 37°C for 2 h in the presence of HU. (A) Rad21-HA association with chromatin was monitored on chromosome spreads by immunofluorescence before and after the shift to 37°C. Scale bar, 5 μm. (B) Rad21-HA fluorescence was measured in 50–100 nuclei. The error bars show the confidence interval of the mean with α=0.05. (C) DNA content analysis and septation index (SI) show that cells remained arrested with close to 1C DNA content throughout the experiment.
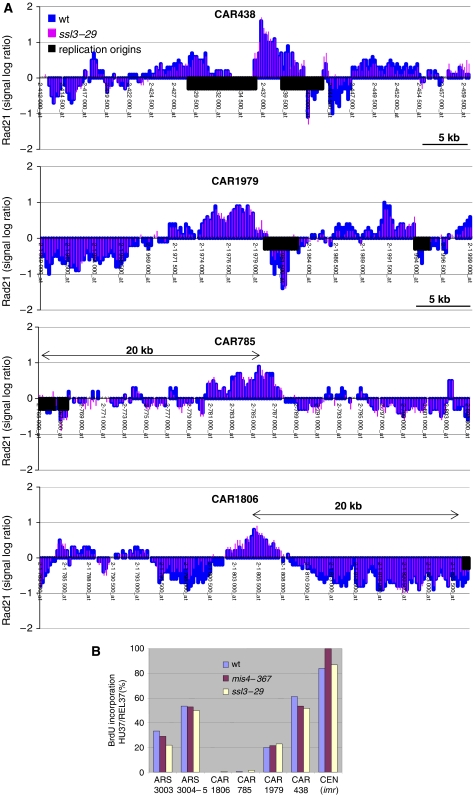
We next wanted to analyze whether cohesin association was equally stabilized at all its association sites along chromosome arms. Rad21 binding was therefore analyzed on a chromosome-wide scale by ChIP followed by hybridization to an oligonucleotide tiling array covering chromosomes 2 and 3. We compared the Rad21-binding pattern in HU-arrested wt versus ssl3–29 cells after the shift to the restrictive temperature. Four 50 kb regions from chromosome 2 are shown in Figure 5, and the complete chromosome 2 in Supplementary Figure S4. This showed that cohesin peaks remained indistinguishable in their relative height and positions whether or not Ssl3 was inactivated. We conclude that, unlike in G1, the loading machinery is dispensable for the stable binding of cohesin to chromosomes in S-phase cells. An interesting explanation for cohesin stabilization in S-phase would link it to the process of cohesion establishment. During replication fork passage through cohesin-binding sites, cohesin's binding to DNA might be stabilized by the process of cohesion establishment between the newly synthesized sister chromatids. This seemed unlikely, however, because very little DNA is replicated in HU-arrested cells. Replication is initiated, but DNA synthesis rapidly ceases due to depletion of deoxynucleotides. As a consequence, regions of only 4–5 kb to both sides of early firing replication origins are replicated (Patel et al, 2006). It remained possible that cohesin-binding sites might be preferentially located close to early replication origins and hence might have been replicated. To investigate this possibility, we marked replication origins known to initiate replication in HU-treated cells on the Rad21 map (Heichinger et al, 2006). About half of the cohesin-binding sites on chromosome 2 were located within 10 kb of an active origin, a conservative estimate for the maximum region that might have been replicated in HU-treated cells. Another half is clearly located too far from an origin to be replicated. As all cohesin peaks remain similarly strong after Ssl3 inactivation, this suggests that cohesin becomes stabilized on chromosomes in S-phase regardless of the replication status.
Figure 5.
Rad21 stabilization occurs independent of DNA replication. (A) Wt and ssl3–29 cells bearing rad21-HA were arrested in early S-phase by hydroxyurea (HU) treatment at 25°C and then shifted to 37°C for 3 h in the presence of HU. Rad21-HA binding to chromosomes was assessed by ChIP and hybridization of the chromatin immunoprecipitate to oligonucleotide tiling arrays covering chromosomes 2 and 3. The data sets from the two strains were merged to facilitate comparison. Intergenic regions containing replication origins known to fire in HU (Heichinger et al, 2006) are depicted as black boxes. Four regions of chromosome 2 are shown as an example. (B) Determination of the replication status of selected loci in the HU arrest. Cells were arrested in G1 by nitrogen starvation and released into the cell cycle at 25°C in the presence of HU and BrdU. After 2 h at 37°C (HU37), HU was washed out and cells were allowed to resume replication for 1 h in the presence of BrdU (REL37). Replicated DNA was immunoprecipitated using anti-BrdU antibodies and quantified by real-time PCR. The extent of DNA replication in the HU arrest at 37°C is given by the ratio HU37/REL37.
To confirm this assumption, we analyzed the replication status of four CARs (Figure 5A), two located close to predicted origins (CAR1979 and CAR438) and two located far from an origin (CAR785 and CAR1806). Cells engineered to take up and incorporate 5-bromo-2-deoxyuridine (BrdU) during DNA synthesis (Patel et al, 2006) were arrested in G1 by nitrogen starvation and released at 25°C into rich medium containing HU and BrdU. Once arrested in S-phase, cells were shifted to 37°C for 2 h, after which HU was washed out to allow completion of replication at 37°C in the presence of BrdU. DNA was extracted just before release from HU (HU37) and after release and completion of S-phase (REL37). BrdU incorporation was monitored by immunoprecipitation with anti-BrdU antibody, followed by quantification of immunoprecipitated DNA by real-time PCR. For each site examined, the extent of DNA replication in HU-treated cells at 37°C was given by the ratio HU37/REL37 (Figure 5B). As even strong fission yeast replication origins do not fire in every cell cycle, we did not expect complete replication even of origin proximal loci (Patel et al, 2006). For instance, two early firing origins, ars3003 and ars3004-5, were shown by DNA combing to fire in ∼30 and ∼70% of cells, respectively (Patel et al, 2006). Our assay showed those two sites to be replicated to ∼30 and ∼50%, respectively, thus validating this approach. The two CARs close to an origin showed replication to ∼20 and ∼50%, respectively. Centromeres are known as early replicated regions (Kim et al, 2003), and indeed showed nearly complete replication in our analysis. By contrast, no incorporation of BrdU could be detected in the HU arrest at CAR785 and CAR1806, consistent with their location far from known origins. The pattern of BrdU incorporation was similar in wt, mis4–367 and ssl3–29 cells, showing that inactivation of the cohesin-loading complex did not affect the pattern of origin firing. Therefore, we conclude that, unlike in G1, Rad21 remains bound to chromatin after inactivation of the loading machinery in HU-arrested cells, and that this stabilization occurs independently of DNA replication.
The contribution of Eso1 to cohesin stabilization during S-phase
Two recent studies in budding yeast have demonstrated that DNA double-strand breaks initiate reinforcement of genome-wide cohesion (Strom et al, 2007; Unal et al, 2007). This process can take place outside S-phase, does not require DNA synthesis, but is dependent on a functional Eco1 (Ctf7) gene. A similar mechanism may operate in HU-arrested cells to stabilize cohesin binding to chromosomes on a genome-wide scale. To investigate this possibility, we asked whether the ECO1 homolog eso1 was required for stable cohesin binding to chromosomes in HU-arrested cells by using the thermosensitive allele eso1-H17 (Tanaka et al, 2000). We took advantage of the fact that eso1-H17 becomes efficiently inactivated at 32°C, whereas mis4–367 still supports cell growth at this temperature (Supplementary Figure S5). We therefore arrested mis4–367 and mis4–367 eso1-H17 double mutant cells by HU treatment at 32°C. Thus mis4–367 eso1-H17 cells entered the HU arrest without Eso1 function. Rad21 was bound to chromosomes under these conditions, albeit at somewhat reduced levels when compared with wt cells. This could be because of partial loss-of-function of Mis4 at this higher permissive temperature (Figure 6A). Now, Mis4 was completely inactivated by shift to 37°C, to assess the stability of cohesin on chromosomes. Rad21 remained stably bound to chromatin independently of the presence of a functional eso1 gene. We conclude that Eso1 is not required for stabilization of cohesin binding in HU-arrested cells.
Figure 6.
The contribution of Eso1 to cohesin stabilization on chromosomes. (A) Eso1 is not required for sustained Rad21 binding to chromatin in HU-arrested cells. HU was added to cycling cells at 25°C and the culture was immediately shifted to 32°C to induce the early S-phase arrest at 32°C, a restrictive temperature for eso1-H17. Cells were then shifted to 37°C for 2 h to inactivate mis4–367 in the presence of HU. Rad21-HA association with chromatin was monitored on chromosome spreads by immunofluorescence before and after the shift to 37°C. Rad21-HA fluorescence was measured in 50–100 nuclei. The error bars show the confidence interval of the mean with α=0.05. DNA content analysis and septation index (%) show that cells remained arrested in early S-phase throughout the experiment. (B) Cohesin stability on chromosomes is compromised after passage through an unperturbed S-phase without Eso1. Cells were arrested in G1 by nitrogen starvation (25°C−N) and released to pass through S-phase at 32°C, a restrictive temperature for eso1-H17. At 3.75 h after release, when cells had completed S-phase (32°C+N), the temperature was raised to 37°C for 1.5 h to inactivate Mis4 and probe the stability of cohesin on chromosomes in G2 (37°C+N). Samples were analyzed before and after temperature shift to 37°C as in (A). Scale bar, 5 μm. The quantification is based on two independent experiments with the error bars indicating the s.d. of the means. The fluorescence for each sample is normalized to the value of the mis4–367 sample before the 37°C shift.
Our finding that cohesin residence on chromosomes is stabilized in HU-treated cells independently of DNA replication, and of Eso1, does not exclude the possibility that an additional or alternative pathway acts to regulate cohesin dynamics during unperturbed S-phase progression. Such a pathway has been proposed based on FRAP experiments in mammalian cells, showing that the residence time of cohesin on chromatin increases as cells progress through S-phase (Gerlich et al, 2006). To see whether this holds true in fission yeast, we asked whether Rad21 binding to chromatin is affected when cohesion establishment is compromised by Eso1 inactivation during normal S-phase. Cells bearing mis4–367 and eso1-H17 were arrested in G1 by nitrogen starvation, and then released into the cell cycle at 32°C to inactivate Eso1, whereas Mis4 remained at least partially active. After DNA replication, the temperature was raised to 37°C to inactivate Mis4 and probe the stability of cohesin on chromosomes in G2 (Figure 6B). In an eso1+ background, the amount of chromatin-bound Rad21 remained nearly constant after inactivation of Mis4 in early G2, consistent with the notion that sustained Rad21 binding to chromatin in G2 no longer requires the loading machinery. However, in the eso1-H17 background, the amount of chromatin-bound Rad21 significantly dropped during the incubation time at 37°C. The decrease in Rad21 levels to approximately half of the level in the eso1+ control may be an underestimate of Eso1's contribution to cohesin stabilization, as in our synchronization procedure only about two-thirds of cells underwent S-phase in the absence of Eso1 function. We conclude that altering Eso1 function compromises stabilization of cohesin binding to chromatin during undisturbed S-phase.
Discussion
Our results show that cohesin association with chromatin in G1 requires the continuous activity of the cohesin-loading machinery, and that the instability of cohesin on chromosomes in G1 relies on a functional wpl1 gene. This suggests that in fission yeast, as in higher eukaryotes, cohesin binding to chromatin is dynamic and is controlled by a conserved mechanism. Hence, cohesin dynamics in G1 might be universal among eukaryotes, suggesting an important but yet unresolved biological function. Wapl directly interacts with cohesin, and thus may alter the intrinsic stability of the complex (Gandhi et al, 2006; Kueng et al, 2006). Alternatively, Wapl might modulate cohesin's association with chromatin by facilitating an as yet uncharacterized unloading reaction. The wpl1 gene, although crucial for cohesin dynamics in G1, is not essential for proliferation in fission yeast, and wpl1Δ strains grow indistinguishably from wt. Wapl-mediated control of cohesin stability in G1, therefore, must be dispensable for the essential process of cohesion establishment during S-phase. Cohesin dynamics in G1 may facilitate sister chromatid cohesion-independent functions of cohesin at this cell-cycle stage.
The establishment of sister chromatid cohesion usually occurs during S-phase and data from mammalian cells have shown that cohesin stabilization correlates with S-phase progression (Gerlich et al, 2006). The question is therefore whether cohesin stabilization is integral to the process of cohesion establishment during DNA replication. We found that a substantial amount of Rad21 was not stably bound to chromatin in G2 when Eso1 function was compromised during the preceding S-phase. As Eso1 is required for the establishment of cohesion, this result suggests that stable cohesin binding during G2 might be attributable, at least in part, to the process of cohesion establishment.
This observation leaves undecided whether stable cohesin binding to chromosomes is a consequence of cohesion establishment or a prerequisite. Stable cohesin binding might be required before the passage of the replication fork for cohesion establishment to occur. Alternatively, stable cohesin binding might be a consequence of cohesion establishment. These two possibilities are not mutually exclusive. A change in the mode of cohesin binding might facilitate the establishment of cohesion, and in addition, the process of cohesion establishment might further stabilize cohesin binding to chromosomes. Our data do not allow us to differentiate between these possibilities. However, using HU-arrested cells, we show that the stabilization of cohesin binding to chromatin and DNA replication can be clearly uncoupled. Cohesin dissociation from chromosomes is downregulated at a stage when little DNA is replicated, and importantly, we show that cohesin is stabilized at unreplicated as well as at replicated genomic regions. Stabilization of cohesin under these conditions does not correlate with DNA replication, but instead appears to be regulated in a replication-independent, cell-cycle-dependent manner. We therefore suggest that cohesin stabilization can occur independently of DNA replication.
How do the observations in HU-treated cells relate to the process of cohesin stabilization that occurs in a normal S phase? We cannot exclude that, in response to HU, a pathway is activated that is not normally acting during undisturbed S-phase progression. At the same time, even during an unperturbed S phase, RPA-bound single-stranded DNA triggers a dose-dependent activation of the intra-S checkpoint that leads to the inhibition of late origins (Miao et al, 2003; Marheineke and Hyrien, 2004; Sorensen et al, 2004; Shechter and Gautier, 2005). It has therefore been suggested that HU-induced S-phase arrest is an amplification of the normal, low level of checkpoint activation (Shechter and Gautier, 2005). In this scenario, HU-mediated fork stalling may exacerbate a cohesin stabilization reaction that normally occurs at a lower level or only in the vicinity of the replication fork. In an unperturbed S-phase, cohesin stabilization and DNA replication may thus be temporally coupled but mechanistically distinct.
One attractive possibility that is consistent with our observations would be that cohesin stabilization might precede DNA replication. In this scenario, stabilization of cohesin would be a prerequisite for cohesion establishment. In other words, stably bound cohesin might be the substrate for cohesion establishment. Clearly, elucidation of the mechanism by which Eso1 and additional factors downregulate cohesin dynamics in S-phase will be an important challenge for the future, with the key prediction that interference with this process should alter cohesion establishment.
Materials and methods
Media, strains and molecular genetics
Media were prepared as described previously (Moreno et al, 1991). Complete YES medium was used unless otherwise stated. Synthetic medium is EMM2. Synthetic medium lacking a nitrogen source (EMM2-N) was used to arrest cells in G1 by nitrogen starvation. YPD medium (2% tryptone, 1% yeast-extract, 2% glucose, supplemented with adenine 150 mg/l, uracil 100 mg/l, histidine 100 mg/l and leucine 100 mg/l) was used to release cells into the cell cycle after nitrogen starvation induced G1 arrest. Strains used in this study are listed in Supplementary Table S1. The wpl1 gene (SPBC428.17c) was disrupted by replacement of the entire open reading frame with the kanR cassette using a PCR-based module method (Bahler et al, 1998). The nmt-res1Cter strain was constructed as follows. The 3′end of res1 (corresponding to amino acids 399–637) was amplified by PCR and cloned into pREP2 (Maundrell, 1993) to generate pREP2res1. The plasmid was linearized at the StuI site within ura4 and integrated at the ura4 locus of an ura4–294 recipient strain, selecting for Ura+ transformants. The strains engineered to incorporate BrdU were derived from strain YFS240 (Patel et al, 2006). Strains carrying the tagged alleles of rad21 and rec8 were described previously (Watanabe and Nurse, 1999; Bernard et al, 2001b; Yokobayashi et al, 2003).
Cell-cycle arrests
G1 arrest by Res1-Cter overexpression. Cells bearing the nmt-res1Cter construct were grown to late log phase in EMM2 containing 20 μM thiamine to repress expression from the nmt promoter. Cells were harvested, washed three times in EMM2 without thiamine and inoculated into fresh EMM2 without thiamine at a density of 2 × 105 cells/ml. Cell proliferation ceased after seven doublings. The time to achieve seven doublings (hence the arrest) can be controlled by modulating the temperature (20 and 25°C were used). Once arrested, cells were shifted to 36.6°C for 2 h. Cell-cycle arrest before and after the temperature shift was monitored by measuring DNA content by flow cytometry and the frequency of cells undergoing cytokinesis (septation index) by calcofluor staining of the septa.
Rec8-GFP induction by nitrogen starvation. Heterothallic cells bearing rec8-GFP were grown to saturation in YES medium, washed three times in EMM2-N and inoculated into EMM2-N at a density of 1 × 107 cells/ml at 25°C to arrest them in G1. After 22–24 h, all cells displayed a Rec8-GFP dot of fluorescence.
S-phase arrest by hydroxyurea treatment. Cells were grown in YES medium to mid-log phase at 25°C at which time HU was added to 20 mM. Cells were further incubated for 4.5 h at the same temperature to induce S-phase arrest. Cells were collected, the HU-containing medium was renewed and cells were incubated for 2 h at 36.6°C. Cell-cycle arrest before and after the temperature shift was monitored by measuring DNA content by flow cytometry and the septation index.
Nuclear spreads
Nuclear spreads were performed as described previously (Bahler et al, 1993) with some modifications. Lysing enzymes (Sigma) were used to digest the cell wall (5–10 mg/ml in 1.2 M sorbitol, 50 mM sodium citrate, 50 mM disodiumhydrogen phosphate, pH 5.6) at 30°C. After the digestion of the cell wall, spheroplasts were deposited onto 1 ml of a sucrose cushion (15% sucrose, 1.2 M sorbitol, 10 mM Tris–HCl, pH 7.5) and spun down at 2000 r.p.m. for 4 min at 4°C. The pellet was washed once in ice-cold Sorb/Tris (1.2 M sorbitol 10 mM Tris–HCl, pH 7.5) and once in Sorb/MES (0.1 M MES hydrate (2-(N-morpholino) ethane sulfonic acid), 1 mM EDTA, 0.5 mM MgCl2, 1 M sorbitol, pH 6.4). Spreading and immunofluorescence carried out were as described previously (Bahler et al, 1993) using monoclonal anti-HA (12CA5) or polyclonal anti-GFP antibodies (A11122, Molecular Probes).
Chromatin immunoprecipitation
ChIP was performed as described previously (Bernard et al, 2001b). The immunoprecipitated DNA was quantified by real-time PCR using a MX3000P cycler (Stratagene) and Absolute qPCR Sybr Green Mix (ABGene). A 10-fold dilution series of genomic DNA was used to calibrate the quantification. DNA was quantified in the input and immunoprecipitated samples and the ratio was calculated (%IP). Immunoprecipitations were performed in duplicate and PCR reactions were carried out at least twice for each sample. The mean and s.d. were calculated. A list of oligonucleotide primers used in this study is available upon request.
Determination of Rad21-binding sites on a genome-wide scale by ChIP-chip was conducted in strains carrying rad21-HA in an ssl3+ or ssl3–29 background. Cells were arrested for 8.5 h at the permissive temperature (20°C) in early S-phase using HU (20 mM final). To inactivate Ssl3, the cultures were shifted to 36°C for 3 h with a fresh batch of HU. ChIP and microarray analysis were carried out essentially as described previously (Katou et al, 2003). In short, 2 × 109 cells were fixed with 1% formaldehyde for half-an-hour at room temperature. Cell extracts were prepared using a multibeads shocker (Yasui Kikai). After sonication (Sanyo Soniprep150) genomic DNA fragments of 400–800 bp were retrieved as input for ChIP. We used anti-HA mouse monoclonal 16B12 antibody (Babco) in conjunction with protein A magnetic dynabeads (Dynal). Eluted immunoprecipitates were incubated overnight at 65°C to reverse the crosslinking. The genomic DNA was purified and amplified by random PCR. After labeling with biotin, the DNA cocktail was hybridized to an Affymetrix high-density oligonucleotide microarray covering fission yeast chromosomes 2–3 (S_pombea520106F, P/N 520106). The experiment was repeated twice with essentially identical results. The microarray data in this report have been deposited with the Gene Expression Omnibus, accession number GSE8450.
Measurement of DNA replication by BrdU incorporation
The method was derived from two previous studies (Cimbora et al, 2000; Patel et al, 2006). Cells were grown to late log phase (107 cells/ml) in EMM2 at 25°C and then transferred to EMM2-N at 25°C at the same density for 16 h to induce cell-cycle arrest in G1. Cells were collected by centrifugation and transferred to YPD medium containing 20 mM HU and 500 nM BrdU at a density of 2 × 107 cells/ml. Samples were incubated for 7 h at 25°C to allow cells to recover from the G1 arrest and enter S-phase. Cells were collected by centrifugation, the medium was renewed and samples were incubated at 37°C for 2 h (HU37). Cells were collected by centrifugation, washed once in YPD medium containing 500 nM BrdU but without HU and further incubated in the same medium for 1 h at 37°C to allow cells to complete replication. Samples were collected before the addition of HU and BrdU (G1 control), after the 2 h incubation time at 37°C (HU37) and after the release (REL37). Cell-cycle arrest and release were confirmed by flow cytometry. DNA was extracted from cell samples and sonicated to approximately 1000 bp fragments. DNA was denatured by heating to 95°C and rapid cooling on ice. Immunoprecipitations were carried out with 10 ng DNA in 250 μl IP buffer (0.14 M NaCl, 0.05% Triton X-100, 10 mM sodium phosphate, pH 7), containing 10 μg sonicated, denatured salmon sperm DNA and 0.5 μg anti-BrdU antibody (Becton). After 45 min at 26°C, 10 μg of anti-mouse antibodies were added and incubation was continued for 45 min at the same temperature. One-tenth of the reaction volume was taken at that time (total fraction). The remaining (IP fraction) was centrifuged at 14 000 r.p.m. for 5 min at 4°C to collect immune complexes. The pellet was dispersed in 750 μl ice-cold IP buffer and spun again as above. The total and IP fractions were digested overnight with proteinase K, extracted with phenol and phenol/chloroform. DNA was precipitated with ethanol and dissolved in 40 μl of water. Quantification of immunoprecipitated DNA was performed by real-time PCR as for ChIP experiments. DNA was quantified in the total and IP samples, and the ratio was calculated (%IP). PCR reactions were repeated at least twice. Unspecific background was estimated from the %IP of the G1 samples. Finally, the extent of BrdU incorporation for a genomic locus was given by the formula 100 × (%IP HU37−%IP G1)/(%IP REL37−%IP G1).
Microscopy
Fluorescence images were acquired with a Leica DMRD microscope equipped with a cooled CCD camera. Signals were quantified using Metamorph software. For nuclear spreads, signal intensity was measured in a square surface containing the spread nucleus. Background signal was measured by moving the square surface in an adjacent region devoid of nuclei. The background value was subtracted for each nucleus. The signal was quantified for at least fifty nuclei for each sample. The mean and the confidence interval of the mean were calculated with α=0.05.
To observe Rec8-GFP, cells were resuspended in water containing 0.4 μg/ml Hoechst 33342 (Molecular Probes) and mounted on poly-L-lysine-coated coverslips in Vectashield medium. Eight z-sections (0.4 μm increment) were acquired and converted to single two-dimensional images by maximum intensity projection. The Rec8-GFP signal was quantified by placing a square surface (4 × 4 pixels) onto the Rec8-GFP dot of fluorescence. Background was estimated by placing the square surface outside the nuclear region, and the value was subtracted. The signal was quantified for at least 150 nuclei per sample. The mean and the confidence interval of the mean were calculated with α=0.05.
Measurement of DNA content by flow cytometry
Flow cytometry was performed after propidium iodine staining of ethanol-fixed cells, as described previously (Moreno et al, 1991). The cell-cycle mutant strains cdc10–129 and cdc25–22 arrested at the restrictive temperature were used as G1 and G2 DNA content controls, respectively.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Acknowledgments
We thank Nick Rhind for providing the BrdU incorporating strain, Hiroto Okayama for the eso1 mutant strain and Mark Groudine for the BrdU immunoprecipitation protocol. This work was supported by the Centre National de la Recherche Scientifique, l'Université Victor Segalen Bordeaux 2, and grants from l'Association pour la Recherche sur le Cancer and l'Agence Nationale de la Recherche (BLAN06-2_135754). SV was supported by a fellowship from l'Association pour la Recherche sur le Cancer and l'Agence Nationale de la Recherche. JD was supported by a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
References
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol 13: 1941–1953 [DOI] [PubMed] [Google Scholar]
- Ayte J, Leis JF, Herrera A, Tang E, Yang H, DeCaprio JA (1995) The Schizosaccharomyces pombe MBF complex requires heterodimerization for entry into S phase. Mol Cell Biol 15: 2589–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wyler T, Loidl J, Kohli J (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol 121: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP (2006) A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol 16: 875–881 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Javerzat JP (2001a) Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol 3: 522–526 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001b) Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Cimbora DM, Schubeler D, Reik A, Hamilton J, Francastel C, Epner EM, Groudine M (2000) Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol Cell Biol 20: 5581–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Decottignies A, Sanchez-Perez I, Nurse P (2003) Schizosaccharomyces pombe essential genes: a pilot study. Genome Res 13: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M (1998) Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev 12: 3408–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J (2006) Live-cell imaging reveals a stable cohesin–chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Molecular Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P (2006) Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849–860 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K (2007) A physical assay for sister chromatid cohesion in vitro. Mol Cell 27: 300–310 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kenna MA, Skibbens RV (2003) Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol Cell Biol 23: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA (2003) Early-replicating heterochromatin. Genes Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Marheineke K, Hyrien O (2004) Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem 279: 28071–28081 [DOI] [PubMed] [Google Scholar]
- Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P (2001) Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7: 959–970 [DOI] [PubMed] [Google Scholar]
- Miao H, Seiler JA, Burhans WC (2003) Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J Biol Chem 278: 4295–4304 [DOI] [PubMed] [Google Scholar]
- Miles J, Formosa T (1992) Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol 12: 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N (2006) DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, Davis RW, Watanabe Y, Nasmyth K (2004) Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci 117: 3547–3559 [DOI] [PubMed] [Google Scholar]
- Shechter D, Gautier J (2005) ATM and ATR check in on origins: a dynamic model for origin selection and activation. Cell Cycle 4: 235–238 [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Lukas J, Bartek J (2004) ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle 3: 941–945 [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C (2007) Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol 20: 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev 14: 2757–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K (1998) Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D (2007) DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P (1999) Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y (2003) Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol 23: 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1