Abstract
Kinetoplast DNA (kDNA), the trypanosome mitochondrial genome, is a giant network containing several thousand interlocked DNA rings. Within the mitochondrion, kDNA is condensed into a disk-shaped structure positioned near the flagellar basal body. The disk is linked to the basal body by a remarkable transmembrane filament system named the tripartite attachment complex (TAC). Following kDNA replication, the TAC mediates network segregation, pulling the progeny networks into the daughter cells by their linkage to the basal bodies. So far TAC has been characterized only morphologically with no known protein components. By screening an RNAi library, we discovered p166, a protein localizing between the kDNA and basal body in intact cells and in isolated flagellum–kDNA complexes. RNAi of p166 has only small effects on kDNA replication, but it causes profound defects in network segregation. For example, kDNA replication without segregation causes the networks to grow to enormous size. Thus, p166 is the first reported molecular component of the TAC, and its discovery will facilitate study of kDNA segregation machinery at the molecular level.
Keywords: genome segregation, kinetoplast, mitochondrial DNA, tripartite attachment complex, trypanosome
Introduction
Trypanosoma brucei causes African sleeping sickness. In addition to its importance as a pathogen, this parasite is noted for its unusual biology, with many characteristics differing from those in more familiar eukaryotes. One remarkable feature is its unusual mitochondrial DNA, known as kinetoplast DNA (kDNA) (for reviews, see Shlomai, 2004; Liu et al, 2005a). kDNA is a giant network of interlocked DNA rings, with a topology like that of medieval chain mail. The rings are of two types, including several thousand minicircles and a few dozen maxicircles. Maxicircles, like more conventional mitochondrial DNAs, encode rRNAs and subunits of respiratory complexes. Minicircles, however, are unconventional, encoding guide RNAs that are templates for editing maxicircle transcripts. Editing involves insertion or deletion of uridylate residues at specific sites in the transcript to generate an mRNA with an open reading frame. Since many guide RNAs are needed, a kDNA network contains numerous minicircle sequence classes, each encoding different guide RNAs (for review on editing, see Stuart et al, 2005).
A trypanosome has one kDNA network in its single mitochondrion. The kDNA is condensed into a disk-like structure positioned within the mitochondrial matrix near the flagellar basal bodies. The reason for this proximity is that the kDNA disk is connected to the basal bodies by a transmembrane filament system known as the tripartite attachment complex (TAC) (Robinson and Gull, 1991; Ogbadoyi et al, 2003; Gluenz et al, 2007). The TAC consists of unilateral filaments linking the kDNA disk to a specialized region of the mitochondrial membrane, and a set of exclusion zone filaments connecting the membrane to the basal bodies. The exclusion zone is named because it is free of cytoplasmic ribosomes, which presumably have been displaced by the filaments. Recent EM studies indicate that the unilateral filaments are subdivided into inner and outer segments (Gluenz et al, 2007). Lysis of trypanosomes with NP-40 or hypotonic buffer, followed by depolymerization of the sub-pellicular microtubules, releases the flagellum with its basal body still linked to the kDNA network, demonstrating the stability of the TAC (Robinson and Gull, 1991; Ogbadoyi et al, 2003). The TAC positions the kDNA in a specialized region of the mitochondrial matrix; in addition, the separation of the old and new basal bodies before cytokinesis mediates segregation of progeny kDNA networks following kinetoplast replication. So far there has been no identification of TAC's protein components.
The kDNA disk is also surrounded by its replication proteins, most of which are positioned in discrete locations. The current model for minicircle replication, reviewed in (Shlomai, 2004; Liu et al, 2005a), begins with the topoisomerase-mediated release of individual covalently closed minicircles from the network into the kinetoflagellar zone (KFZ), the space between the kDNA disk and the mitochondrial membrane near the flagellar basal body (Drew and Englund, 2001). The KFZ is traversed by the unilateral filaments. In the KFZ are proteins that initiate replication of the free minicircles, propagate the replication fork, and likely segregate the daughter minicircles (Li and Englund, 1997; Abu-Elneel et al, 2001; Klingbeil et al, 2002). These progeny are thought to migrate to the antipodal sites, two protein assemblies flanking the kDNA disk and positioned ∼180° apart. Within these sites are enzymes that remove replication primers and repair most but not all gaps in the minicircles (Engel and Ray, 1999; Saxowsky et al, 2003; Downey et al, 2005); they also contain a topoisomerase II that attaches newly replicated and still-gapped minicircles to the network periphery (Melendy et al, 1988; Wang and Englund, 2001). Minicircle replication results in the growth of the network until it finally reaches double size. Then the network splits and the two progeny segregate into the daughter cells during cytokinesis. As discussed above, segregation is mediated by separation of the basal bodies, each linked to a sister kDNA network by the TAC.
A major effort of our laboratory has been to identify and study new proteins involved in kDNA replication or maintenance. RNAi knockdown of such proteins often causes shrinkage and loss of kDNA. We therefore developed an RNAi library screen for cells that have undergone RNAi-mediated kDNA loss (Englund et al, 2005). Here we describe a protein, named p166, discovered using this approach. p166 localizes between the kDNA disk and the flagellar basal body both in cells and in isolated kDNA–flagellum complexes. RNAi indicates a crucial role in kDNA segregation rather than in replication. By these criteria, we conclude that p166 is the first example of a molecular component of the TAC.
Results
RNAi library screen reveals the gene for p166
Our RNAi library screen identified a gene encoding a 166-kDa protein, named p166, whose knockdown causes kDNA loss. p166 is encoded by a single-copy gene (accession number, XP_828411), and its predicted product has 1501 residues and a pI of 5.2. Homologous genes are present in related Trypanosoma and Leishmania species but not in other species. Based on software at http://elm.eu.org, p166 has a predicted N-terminal mitochondrial targeting signal and a predicted transmembrane domain (residues 1440–1462). Therefore, the protein's N-terminal region (including the targeting signal) is 1439 residues, its predicted transmembrane sequence is 23 residues, and its C-terminal region is 39 residues.
p166 localizes with the TAC
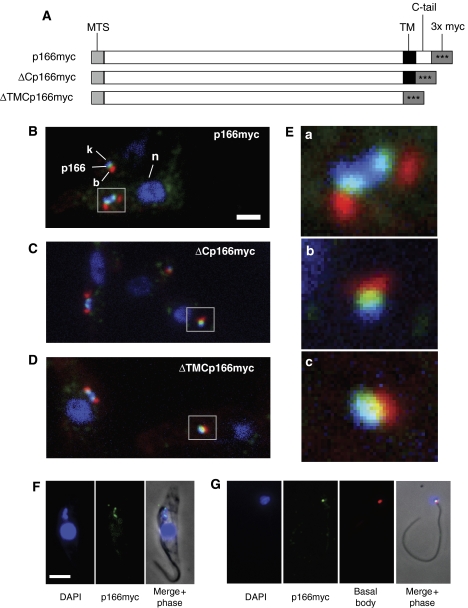
Figure 1A shows three constructs encoding p166 fusion proteins, each with three tandem myc epitopes (comprising a total of 42 amino-acid residues), at a different location at or near the C-terminus. In these studies, we assumed that residues 1440–1462 constitute a transmembrane domain. Using immunofluorescence (IF) with anti-myc antibody, we found that full-length p166myc (green) localizes between the kDNA (blue) and the flagellar basal body (red) (Figure 1B, with enlargement in Figure 1E, panel a; the enlarged image showed a dividing kinetoplast associated with basal bodies that had already separated). There was a similar pattern in virtually all cells examined from an asynchronous culture (including those with two kinetoplasts), indicating that this localization is independent of the cell cycle.
Figure 1.
Localization of p166. (A) Diagrams, not drawn to scale, of myc-tagged proteins expressed in genomic locus and used in localization. p166myc is the full-length protein and ΔCp166myc and ΔTMCp166myc are truncated as indicated. MTS, mitochondrial targeting signal; TM, predicted transmembrane domain. 3 × myc, three copies of myc epitope. (B–D) Localization of p166myc, ΔCp166myc, and ΔTMCp166myc, respectively. Fluorescence micrographs used anti-myc antibody to recognize p166 (green), anti-tyrosinated tubulin antibody for basal body (red), and DAPI to stain kDNA and nucleus (blue). Boxes are regions enlarged in (E). Scale bar, 2 μm. (E) Enlargements showing association of kDNA, p166, and basal body. a, p166myc; b, ΔCp166myc; c, ΔTMCp166myc. (F) Localization of p166myc in isolated cytoskeleton by fluorescence microscopy. Anti-myc antibody detected p166myc (green) and DAPI stained the kDNA and the nucleus (blue). (G) Isolated flagellum stained as in panel F, except that the basal body was also detected by anti-tyrosinated tubulin (Tyr–Tu). Scale bar in panels F and G, 5 μm. Fluorescence images were modified uniformly for contrast and brightness with Adobe Photoshop.
We next examined ΔCp166myc (lacking the 39 residue C-terminal tail) and found that its localization closely resembled that of the full-length protein (Figure 1C, with enlargement in Figure 1E, panel b). Surprisingly, ΔTMCp166myc, lacking the putative transmembrane domain as well as the C-terminal tail (totaling 62 residues), did not redistribute in the mitochondria. Instead, as shown in Figure 1D (with enlargement in Figure 1E, panel c), it had a localization similar to those in Figures 1B and C. This latter observation raised the possibility that the protein not only traverses the mitochondrial membrane but also is held in place by some other interaction, possibly with a cytoskeletal structure.
To assess whether the myc tag might affect p166's function and/or localization, we myc tagged the products of both alleles. After using PCR to show that each of the two wild-type alleles had been replaced by a sequence encoding p166myc (Supplementary Figure S1, Panel A), we then demonstrated that the myc-tagged protein was actually expressed (see western blot in Supplementary Figure S1, Panel B) and that the double myc tag had no effect on growth (Supplementary Figure S1, Panel C). We also detected no change in the kDNA, as visualized by DAPI staining (data not shown). Finally, we demonstrated that p166myc, in cells producing only the tagged version of the protein, also localizes between the kDNA disk and the basal body (Supplementary Figure S1, Panel D). These data provide strong evidence that the myc tag does not interfere with this p166's function or localization.
p166 stably associates with the TAC
The TAC system remains intact following cell lysis with NP-40 (Robinson and Gull, 1991). Therefore, we used IF to examine cytoskeletons (Figure 1F) from cells extracted with NP-40 and MgCl2 (a treatment that removes membranes but does not disrupt sub-pellicular microtubules). Despite this rather strong treatment, we found that p166myc localizes between the kDNA and flagellar base (Figure 1F) as it does in intact cells. In this cell, the kDNA and the adjacent basal bodies had undergone segregation and the p166myc remains associated with both.
We next tested whether p166myc remains associated with the isolated flagella even after depolymerization of the sub-pellicular microtubules. In fact, we observed this association whether the flagellum is linked to a single kDNA network (Figure 1G) or, if derived from a cell that has finished kDNA replication and initiated segregation, to two networks (not shown). As in intact cells, p166 is positioned between the basal body and the kDNA. Although we cannot rule out the possibility that some p166myc was lost or partially degraded during isolation of these flagellum–kDNA complexes, an examination of 34 complexes revealed that all contain p166myc. Therefore, these experiments provided strong evidence that p166 is a stable component of the TAC filament system.
Localization of p166myc by immunoelectron microscopy
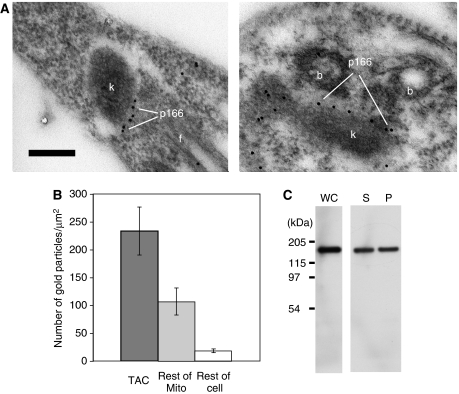
To localize p166myc at the EM level, we used gold-labeled anti-myc antibodies (Figure 2A). Both images show gold particles aligned between the kDNA disk and the basal body but closer to the kDNA. This localization provided further evidence that p166 is a component of TAC. To assess the specificity of the immunolocalization, we examined 18 EM images and counted the gold particles per μm2 in the TAC region, the remainder of the mitochondrial matrix, and in the rest of the cell. As shown in Figure 2B, the density of gold particles is highest in the TAC region and slightly less than half as high in the remainder of the mitochondrial matrix. Taking into account the relative surface areas, about 2/3 of the particles in the mitochondrion are in TAC. This ratio may be inaccurate because the images were chosen to include the kinetoplast/basal bodies, and parts of the tubular mitochondrion may not appear in these sections. Of the particles associated with TAC, 97% appeared to be in the region of the unilateral filaments or mitochondrial membrane, and virtually none were in the region of the exclusion zone filaments. The remaining particles in the mitochondrial matrix did not appear associated with the mitochondrial membrane. Figure 2C shows an experiment that may address the nature of p166myc in the mitochondrial matrix. Using SDS–PAGE and western blotting to measure the fraction of p166myc that is soluble in 1% Triton X-100, we found that of the material present in the whole cell (lane WC), about half is soluble (lane S), and about half is in the pellet (lane P). We speculate on the significance of this finding in the Discussion.
Figure 2.
Localization of p166 by EM. (A) Immunogold EMs, using anti-myc antibody, showing localization of p166myc. k, kinetoplast; b, basal body; f, flagellum. Scale bar, 250 nm. (B) Bar graph showing distribution of gold particles. Bars show mean±standard deviation. (C) Western blot showing the solubility of p166 in Triton X-100. Cells (1 × 107) were centrifuged and the pellet was treated at 4°C with either 100 μl 1 × SDS–PAGE loading buffer (containing 1% SDS) or with 50 μl of 1% Triton X-100. In both cases, the solution contained protease inhibitor (Pierce, catalog no. 78410, used at 2.5 times the recommended concentration). In the treatment with Triton X-100, the solution was incubated (20 min, 4°C) and then centrifuged (16 000 g, 20 min, 4°C). The supernatant was removed and the pellet was resuspended in 50 μl of 1% Triton (containing protease inhibitors). SDS–PAGE loading buffer (50 μl, 2 × concentration) was added to the supernatant and pellet. A 20-μl volume of each sample was loaded on an SDS–PAGE gel. After electrophoresis, the p166myc in the cell lysate (WC), supernatant (S), and pellet (P) were detected on a western blot probed with anti-myc antibody described in the legend of Supplementary Figure S1.
RNAi of p166 reduces growth rate
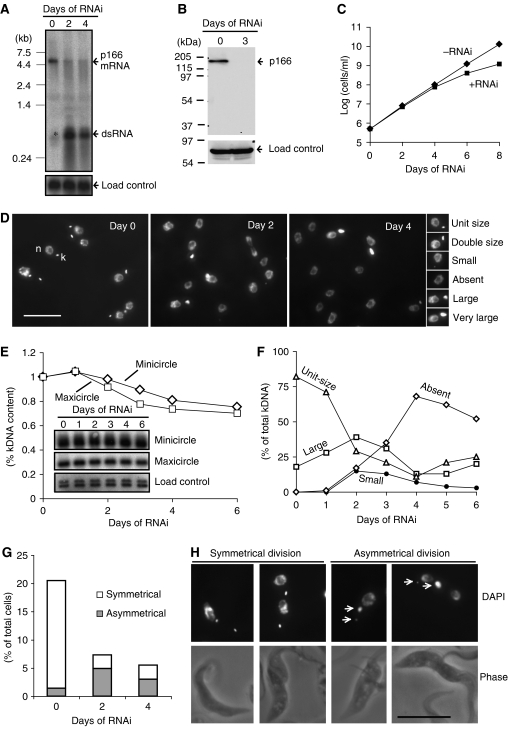
The discovery of p166 in an RNAi library screen relied on the fact that its knockdown caused shrinking or loss of kDNA. We therefore performed more comprehensive RNAi experiments, using the inducible pZJM vector, to identify the effects of p166 knockdown. In these experiments, the pZJM insert (encoding p166 residues 442–651) differed from the one sequenced from the cell line cloned from the RNAi library (encoding residues 334–511). A northern blot in Figure 3A showed that RNAi for 2 or 4 days caused robust production of dsRNA (∼600 bp, expected from the size of the pZJM insert); phosphorimaging of the blot showed that the level of p166 mRNA (∼5 kb) was reduced by 86% at day 2 and by 89% at day 4. There was a low level of dsRNA produced at day 0 (band marked by * in Figure 3A) presumably due to the inability of the tetracycline repressor to fully inhibit transcription of the pZJM insert. A western blot (Figure 3B), probed with antibody against a recombinant fragment of p166, showed that RNAi for 3 days caused a nearly complete loss of p166 protein.
Figure 3.
Effects of RNAi on p166. (A) Northern blot showing effect of RNAi on p166 mRNA level. The blot was probed with a 32P-labeled fragment used as the insert in the pZJM RNAi vector and then reprobed with trypanosome hexose transporter (THT1, NCBI accession no. AAA92489) as a load control. * Indicates dsRNA expressed before tetracycline induction. (B) Effect of RNAi on the level of p166 protein. Western blot was probed with anti-p166 and then with an antibody recognizing mitochondrial HSP70 as the load control. (C) Effect of RNAi on cell growth. Values of cells/ml on y-axis are the measured value multiplied by the dilution factor. (D) Effect of RNAi on kinetoplast structure. Cells were DAPI-stained to visualize the nucleus (n) and kinetoplast (k). Examples of each kinetoplast category are shown on the right. Scale bar, 10 μm. (E) Effect of RNAi on kDNA loss. A Southern blot of total DNA digested with HindIII and XbaI was probed for minicircles, maxicircles, and THT1 (load control) (Wang and Englund, 2001). Inset figures are segments of autoradiographs showing 1-kb linearized minicircle fragment, 1.4-kb maxicircle fragment, and load control. Band intensities were quantitated by phosphorimaging, corrected for the load control, and plotted. (F) Kinetics of RNAi-mediated change in kinetoplast structure. Cells (∼150) like those in panel D were categorized. The ‘large' category includes double-size, large, and very large (as defined in panel D). (G) Effect of RNAi on the production of cells with two kinetoplasts. Bars show fraction of cells with kinetoplasts dividing symmetrically and asymmetrically. (H) Images of cells with kinetoplasts dividing symmetrically and asymmetrically. Scale bar, 10 μm.
We next evaluated the effect of RNAi on cell growth. RNAi cells grew normally for 4 days and then continued at a lower rate, indicating that p166 is required for maximum growth rate (Figure 3C). The doubling time of the uninduced cells in Figure 3C (13.5 h) is about 10% higher than that of untransfected 29–13 cells (12 h), a difference possibly due to the low level of dsRNA produced in uninduced cells (Figure 3A, day 0). It may seem surprising that normal growth of RNAi cells persists through day 4, a time when 70% of cells lack kDNA (see next section and Figure 3F). Although procyclic trypanosomes require kDNA gene products (e.g., respiratory complex subunits), these proteins must be in excess and stable enough to sustain growth for a few days after kDNA loss.
p166 RNAi inhibits kDNA segregation
To study the effects of RNAi on kDNA, we prepared a Southern blot of total DNA digested with HindIII and XbaI and probed for minicircles and maxicircles. Minicircle levels declined about 25% by day 6 and maxicircles fell slightly faster (Figure 3E). We also assessed kDNA loss by microscopy of DAPI-stained cells (Figure 3D). Small images on the right in Figure 3D are examples of kinetoplast size categories. Cells not induced for RNAi (day 0, left panel) contained a nucleus and a kinetoplast (some of the latter, which are undergoing or have finished replication, appeared elongated or double-size). After 2 days (middle panel) or 4 days of RNAi (right panel), there were increasing numbers of cells lacking a kinetoplast as well as some with kinetoplasts larger than the usual double-size replicative forms. We next determined the kinetics of the RNAi-mediated change in kinetoplast structure by daily examining ∼150 DAPI-stained cells (Figure 3F). At day 0, before RNAi, all cells had both a nucleus and a kinetoplast. Most kinetoplasts were unit-size but others were larger. Cells with larger kinetoplasts, undergoing replication, constituted about 20% of the total, a percentage characteristic of an unsynchronized wild-type culture (Ferguson et al, 1994; Liu et al, 2005b). Following induction of RNAi, cells with unit-size kinetoplasts gradually disappeared, reaching a minimum at day 4; then they increased in number, probably due to outgrowth of cells that had recovered from RNAi. Following RNAi induction, there was an increase in cells with large kinetoplasts, reaching a peak at day 2, and then declining to a minimum at day 4 (large kinetoplasts include double-size, large, and very large categories; see right-hand panel in Figure 3D). The fact that few cells with small-size kinetoplasts accumulate during the 6 days of RNAi (only about 15% at days 2 and 3) suggests that the kinetoplasts in most cells do not gradually shrink in size as might be expected from a replication defect (Wang and Englund, 2001). Instead, the accumulation of cells with either very large kinetoplasts or none at all raises the possibility that p166 RNAi causes a defect in kinetoplast segregation.
As a direct measure of segregation efficiency, we determined the effect of RNAi on the production of cells with two kinetoplasts (2K) (Figure 3G; see Figure 3H for examples of DAPI-stained cells with two kinetoplasts). At day 0 (before RNAi induction), 21% of the cells were 2K. This value dropped to about 7% after 2 days of RNAi and 5% at day 4, consistent with a segregation defect (Figure 3G). Also indicating a defect in segregation, RNAi sharply raised the fraction of cells with asymmetrically dividing kinetoplasts (see examples in Figure 3H); of the 2K cells, 5% at day 0 (before RNAi induction) divided asymmetrically, and the percentage rises to 67% on day 2 and 55% at day 4. Since wild-type kinetoplasts almost never divide asymmetrically, the 5% observed at day 0 could be due to the low level of p166-specific dsRNA detected at day 0 (Figure 3A).
RNAi of p166 produces giant kDNA networks
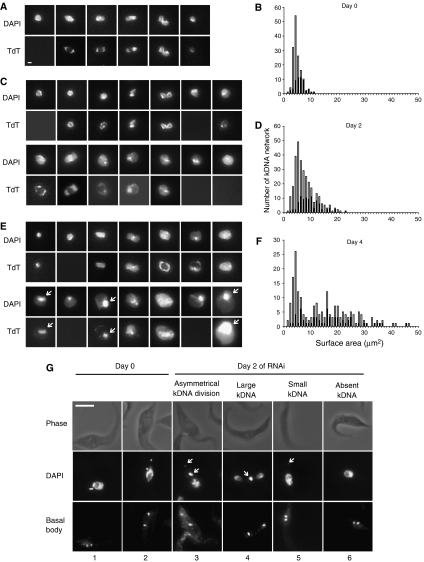
We next investigated whether the large kinetoplasts (Figure 3D) were due to a single larger-than-normal kDNA network or to two or more normal-size networks that were closely packed within the mitochondrial matrix. For this purpose, we isolated networks from cells after 0, 2, or 4 days of RNAi, labeled the minicircle gaps with a fluorescent dNTP using terminal deoxynucleotidyl transferase (TdT), stained them with DAPI, and captured images by fluorescence microscopy. See Figures 4A, C, E for examples of DAPI-stained networks in the upper row and TdT labeling in the lower row; the TdT labeling will be discussed later in this paper. We then measured surface areas of the DAPI-stained networks and the results are in panels B, D, and F. The bar graph for uninduced control cells (Figure 4B, day 0) had a large major peak of unit-size networks averaging 4.5 μm2 with a shoulder of slightly larger networks ranging up to approximately double-size. The networks in the shoulder are larger because they are undergoing replication. We then measured surface areas of networks isolated from cells that had undergone RNAi for 2 days (Figures 4C and D) or 4 days (Figures 4E and F). At day 2 there is a peak of unit-size networks, but in this case the large networks (roughly half of the total) included not only double-size but also triple- and even quadruple-size. At day 4, some of the networks were even larger, up to about 10 times unit-size. In no case did we observe networks that were significantly smaller than unit-size, although some could have been preferentially lost because network isolation involves a centrifugation step.
Figure 4.
Effect of RNAi on kDNA networks (A–F) and on the flagellar basal body (G). Isolated networks, prepared as described (Pérez-Morga and Englund, 1993a), were DAPI-stained (0.2 μg/ml), and TdT-labeled (Liu et al, 2005b). Surface areas of networks were measured using IPLab software (version 3.9). (A) Networks from cells uninduced for RNAi (day 0) showing DAPI staining and TdT labeling. Scale bar, 1 μm. (B) Bar graph showing surface areas of networks from cells uninduced for RNAi (day 0). Black portion of bars represents TdT-positive networks. (C, D) Same as panels A and B, except that RNAi was for 2 days. (E, F) Same as panels A and B, except that RNAi was for 4 days. For unclear reasons, possibly due to differences in spreading, the average surface area of unit-size kDNA networks, ∼4.5 μm2, was about 1/3 of that previously reported (Liu et al, 2005b). (G) Effect of RNAi (2 days) on association of kDNA with the basal body. All cells shown have two basal bodies, which were identified by antibodies to tyrosinated-tubulin. Scale bar, 5 μm.
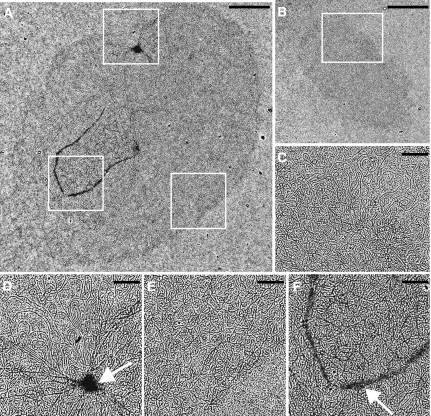
We next used EM to examine networks from control cells and from cells after 2 days of RNAi (Figure 5). A giant network from RNAi cells is shown in Figure 5A, with enlargements, corresponding to the white boxes in Figure 5A, presented in Figures 5D–F. For comparison, a wild-type network (either a pre-replication or early-replicating form) is shown in Figure 5B (with an enlargement in Figure 5C). The surface area of the RNAi network (Figure 5A) is about six times larger than that of the control network (Figure 5B). In general, the RNAi networks appeared elliptically shaped and had a density of minicircles comparable to that of control networks. Condensed areas (marked by arrow in Figure 5D) were sometimes found in RNAi networks but almost never in control networks. Markings like the horseshoe shape on the RNAi network (see arrow in enlargement in Figure 5F) were rare on either RNAi or control networks.
Figure 5.
EM of isolated kDNA networks. (A) Network from cell after 2 days of RNAi. (B) Network from control cell (strain 29–13), which could be a pre-replication form or in the early stages of replication. (C) Higher magnification of the network in panel B, corresponding to the region within the white box. (D–F) Higher magnification images of the network shown in panel A, corresponding to the regions within the white boxes. The long strands associated with the network, prominent in panels C and D, are segments of maxicircles. White arrows in panels D and F are discussed in the text. Scale bar for panels A and B, 2 μm. Scale bar for panels C–F, 0.5 μm.
RNAi of p166 does not inhibit the duplication or segregation of basal bodies
Because kDNA is linked to the flagellar basal body via the TAC, we used IF detection of tyrosinated-tubulin (a component of the basal body; Sherwin et al, 1987) to measure the effect of p166 RNAi on basal bodies. Before RNAi (day 0), 39% of cells had two basal bodies (each is presumably associated with a probasal body) and at days 2 and 4 of RNAi, and this percentage was 40 and 31%, respectively. These percentages agree well with published values (Pradel et al, 2006). These relatively small changes, especially at day 2, indicate that p166 RNAi has little or no effect on basal bodies. Figure 4G shows images of cells with two basal bodies. In uninduced cells (day 0) the basal bodies, as expected, associate with the kinetoplasts whether they are in the early stages (cell 1) or late stages (cell 2) of segregation (Robinson and Gull, 1991). In RNAi cells (day 2) the basal bodies appear to segregate normally and associate with the kDNA even if the two kinetoplasts had formed by an asymmetric division (cell 3). If there was a single large (cell 4) or small (cell 5) kinetoplast, it was associated with one of the two basal bodies. The basal bodies also segregate normally in cells with no kinetoplast (cell 6).
p166 RNAi has only limited effects on kDNA replication
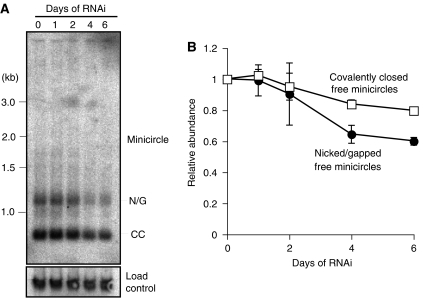
As described in the Introduction, kDNA synthesis involves release of covalently closed minicircles from the network, replication of the free minicircles, and reattachment of nicked/gapped progeny to the network periphery adjacent to the antipodal sites. These free minicircles species are resolved by electrophoresis if the gel buffer contains ethidium bromide. RNAi of kDNA replication enzymes can cause loss of free minicircles, a change in ratio of the species, or appearance of new minicircle species. Figure 6 shows that p166 RNAi, over 6 days, has relatively small effects on free minicircles, with only a 20% decrease in covalently closed free minicircles and a 40% decline in nicked/gapped forms.
Figure 6.
Effect of RNAi on free minicircle replication intermediates. Total DNA was fractionated on an agarose gel with TBE buffer containing 1 μg/ml ethidium bromide (Wang and Englund, 2001). (A) Autoradiograph of gel probed for minicircles and trypanosome hexose transporter (THT1) as loading control. N/G, nicked and gapped free minicircles that are the products of replication; CC, covalently closed free minicircles that are the substrates for replication. (B) Quantitation of free minicircles by phosphorimaging with values corrected for loading control. Values are the mean (±standard deviation) of two experiments, one of which is shown in panel A.
Replication can also be evaluated at the network level by TdT labeling of newly replicated gapped minicircles. Figure 4A (bottom row) shows the well-characterized progression of network replication in cells not undergoing RNAi (Hoeijmakers and Weijers, 1980; Ferguson et al, 1994; Robinson and Gull, 1994; Guilbride and Englund, 1998; Liu and Englund, 2007). This pathway starts with unit-size TdT-negative networks, and as it proceeds, the network elongates and the polar TdT-labeled zones expand. Finally, the uniformly labeled network becomes dumbbell-shaped and segregates into TdT-positive progeny. Gap repair quickly converts this network to TdT-negative and completes the cycle. Consistent with this interpretation, the ratio of TdT-labeled networks (black segment of the bars in Figure 4B) to total networks (total bar) increases as the networks get larger.
In day 0 cells, 40% of the networks are TdT-positive and this percentage decreases to about 30% at days 2 and 4. Inspection of the TdT labeling patterns at day 2 (Figure 4C) reveals only a small fraction of networks with polar labeling. Some networks label in a peripheral ring as previously observed following RNAi of several replication proteins (Liu and Englund, 2007). We saw the same pattern at day 4 (Figure 4E), with many of the largest networks being TdT-positive. Labeling is often uniform except for a prominent bright zone detectable in some networks by DAPI and TdT labeling (see arrows on Figure 4E). The bright spots could be clusters of maxicircles as observed previously by DAPI staining (Ferguson et al, 1994), or they could be regions of high minicircle density.
Discussion
By screening an RNAi library, we discovered p166, a protein with sequence characteristics typical of a mitochondrial membrane protein. IF localization of p166myc showed it to be positioned between the kDNA disk and the flagellar basal body. Removal of the 39-residue C-terminal tail or the tail plus the predicted transmembrane domain did not change the localization. Assuming that p166 actually traverses the membrane, the latter observation could be explained if the protein was anchored in the membrane and also by interaction with other proteins, possibly cytoskeletal. p166's predicted leucine zipper motif (residues 1074–1098) and other coiled-coil motifs (residues 458–484, 1167–1203, and 1247–1276; identified by software at http://elm.eu.org) could be involved in these interactions. Extraction of cells with NP-40 (Figure 1F) or NP-40 plus EDTA (Figure 1G) does not alter the localization of p166 between the kDNA and the flagellar basal body. In either case the NP-40 is likely to disrupt the mitochondrial membrane, providing further evidence that the transmembrane domain is not the only means by which the protein is held in place. Of course an alternative explanation is that this hydrophobic sequence does not in fact serve as a transmembrane domain. If it is a transmembrane domain, we do not know which mitochondrial membrane it traverses or the protein's orientation in the membrane.
ImmunoEM (Figures 2A and B) confirmed the IF results and provided evidence that p166myc resides within the region of the unilateral filaments or the TAC-associated mitochondrial membrane. However, about 1/3 of the gold particles appeared to be in the mitochondrial matrix outside the TAC region and not associated with membranes. We also found using SDS–PAGE and western blotting that about one half of the p166myc is soluble in 1% Triton X-100 and the other half is insoluble (Figure 2C). We speculate that the soluble fraction corresponds to the mitochondrial p166myc that is not associated with the TAC, and that it is a precursor not yet assembled into the TAC filament system.
RNAi of p166 has a major effect on kDNA segregation, and the strongest evidence is that when 70% of cells at day 4 had no detectable kDNA (Figure 3F) but the total minicircles has declined only ∼20% (Figure 3E). The reason for this difference is that the remaining networks had grown in size (Figure 4F) due to their continued replication and inability to segregate. As would be expected from a segregation defect, RNAi also sharply reduced the number of cells with two kinetoplasts, and it increases the frequency of kinetoplasts dividing asymmetrically (Figure 3G). Surface area measurements of DAPI-stained networks reveal considerable growth at days 2 and 4 of RNAi, with some networks about 10 times that of wild type (Figure 4A–F). EM of RNAi networks revealed little difference from control networks except for size (Figure 5).
RNAi of p166 only modestly affected kDNA replication. Knockdown produces few small kinetoplasts (Figure 3F) that often form when RNAi blocks replication (Wang and Englund, 2001), and the small networks we detected could have formed from asymmetric division (Figures 3G and H). The fact that networks grow in size throughout 4 days of RNAi (Figures 4B, D, F) provides evidence that the replication machinery must be functional. The large networks that are TdT-negative show that gap repair can still follow replication. However, replication may not operate at top efficiency when the network is several fold larger than normal. This reduced efficiency could explain the small decline in free minicircle replication intermediates (Figure 6). Also, the loss of only 2% of the total minicircles at day 2 and 20% at day 4 (when segregation defects are already robust at day 2) provides more evidence that the effect of RNAi on replication is relatively small and unlikely to be a primary effect (Figure 3E).
The moving apart of flagellar basal bodies, each linked to a sister kDNA network via the TAC, is thought to provide the force mediating kinetoplast segregation (Robinson and Gull, 1991). In analyzing cells with two basal bodies (Figure 4G), we observed that they segregate normally despite the status of the kinetoplast (either asymmetrically dividing, large, small, or absent). Inspection of 16 cells containing two nuclei and one kinetoplast revealed that 15 of them had the kinetoplast associated with the old basal body (the most anterior; see example in Figure 4G, cell number 4). Similarly, analysis of eight cells undergoing asymmetric division of the kinetoplast showed that all had the larger progeny associated with the old basal body (see example in Figure 4G, cell number 3). Thus, as RNAi gradually depletes p166, the linkage of the kDNA to the basal body becomes progressively weaker, causing the unilateral filaments to loosen their grip on the kDNA. Because of the declining level of p166, this loosening predominantly affects the kinetoplast associated with the new basal body; initially this loosening causes asymmetric division, but ultimately when the kinetoplast completely disconnects from the new basal body, kinetoplast segregation cannot occur at all. The daughter cell with the new basal body completely loses its kDNA.
There are other mechanisms that regulate kinetoplast segregation. One involves UMSBP (universal minicircle sequence-binding protein), a minicircle replication origin-binding protein that triggers replication of free minicircles. Recent RNAi studies show that it is also involved in kDNA segregation, but since RNAi also affects nuclear segregation and basal body separation (Milman et al, 2007), the mechanism must be different from the TAC-related segregation mechanism described in this paper.
The TAC is a remarkable transmembrane filament system linking the kDNA disk to the flagellar basal body that mediates kinetoplast segregation (Robinson and Gull, 1991; Ogbadoyi et al, 2003). Are there analogous systems in other organisms? As recently reviewed (Chen and Butow, 2005), there is developing evidence that in Saccharomyces cerevisiae the mitochondrial nucleoids, composed of a few mitochondrial DNA molecules and multiple proteins, are positioned close to the mitochondrial membrane because they are bound to a protein assembly that traverses both membranes. This assembly is thought to mediate segregation of the mitochondrial genome because it is linked to the actin cytoskeleton.
Based on the intracellular location of p166 and the striking effect of its knockdown on kDNA segregation, we conclude that p166 is the first molecular component of TAC to be identified. Upcoming studies should reveal whether p166 is in fact an integral membrane protein in a mitochondrial membrane and what its orientation may be. Identification of its interaction partners could lead to identification of other TAC components. Finally, the discovery of p166 demonstrates the power of the RNAi library. Given the absence of a counterpart in unrelated species, and the lack of a suitable assay for its isolation, p166 would have been difficult to discover by any other means.
Materials and methods
Trypanosomes and RNAi
For expression of myc-tagged p166, we used procyclic T. brucei 927 strain (from Dr Elisabetta Ullu). RNAi experiments utilized the procyclic T. brucei 29-13 strain (from Dr George Cross) that harbors genes for T7 RNA polymerase and the tetracycline repressor (Wirtz et al, 1999). Both strains were cultured at 27°C in SDM-79 medium containing 10% fetal bovine serum (Brun and Shonenberger, 1979). Media were supplemented with appropriate selection agents (hygromycin, 50 μg/ml; G418, 15 μg/ml; phleomycin, 2.5 μg/ml; blasticidin, 10 μg/ml). For RNAi, T. brucei 29-13 was transfected with the pZJM RNAi vector (Wang et al, 2000) whose insert was a PCR-amplified fragment of the p166 gene (encoding amino-acid residues 442–651). After drug selection, the transfected cells were cloned by limiting dilution. RNAi was induced by tetracycline (1 μg/ml).
RNAi library screening
We screened the library to identify kDNA replication genes (see Morris et al, 2004 for library methodology). The library, described previously (Morris et al, 2002), consisted of T. brucei 29-13 cells transfected with the pZJM vector, each containing a random genomic fragment (average size 660 bp). Briefly, to screen for kDNA loss, a lethal phenotype, our strategy (previously described in Motyka et al, 2004, 2006) has been to clone cell lines from the library, induce RNAi in each clone, and assay for kDNA loss. We then used nested PCR and sequencing to identify the pZJM insert from an uninduced replica of the relevant cell line. In our screen, we analyzed 1400 cell lines that had been cloned by limiting dilution. We induced RNAi in each cell line, evaluated kDNA loss after 7 days by staining live cells with Hoechst 33354 dye (5 μg/ml), and then examined each cell line by fluorescence microscopy. Of three cell lines with small or absent kinetoplasts, one had part of the p166 gene as the pZJM insert.
Epitope tagging
One or both alleles of p166 were myc-tagged by transfecting trypanosomes with PCR products encoding three tandem copies of the myc epitope and either a G418 or a blasticidin selectable marker. To ensure homologous recombination at the desired site, the PCR product also contained at either end 80 bp of p166 gene sequences that flank the desired integration site. For using G418 as the selectable marker, downstream of the 80 nucleotides were the sequences 5′-GGTACCGGGCCCCCCCTCGAG and 5′-CGGCCGCTCTAGAACTAGTG, complementary to the left and right arms of the pMOTag (containing the G418 resistance gene), which served as template for PCR (Oberholzer et al, 2006). For using blasticidin as the selectable marker, downstream of the 80 nucleotides were sequences 5′-AGCTAGCGGCCGCTCTGAGC and 5′-TTAGCCCTCCCACACATAAC, complementary to the arms of the PCR template pXSMycbla (a plasmid containing the blasticidin-resistance gene provided by Dr Beiyu Liu). The PCR products were Qiagen column-purified and transfected into log-phase procyclic strain 927 cells (5 × 106 to 107 cells/ml). Transfectants were selected for 14 days in medium containing G418 or blasticidin. Using this approach, a single allele of p166 was myc-tagged (with the G418 marker) at either of three locations at or near the C-terminus. The resulting proteins are shown in Figure 1A. To tag both alleles of full-length p166 with the myc epitope, the first allele was tagged using G418 as selectable marker and then the second allele was tagged by the same method with blasticidin selection.
Antibody generation
Antibody to p166 was generated in rat using as antigen a soluble recombinant fragment (residues 284–505, expressed in Escherichia coli using pET28 (Novagen)). The immunization, performed at Cocalico Biologicals (Reamstown, PA), followed a standard protocol. Serum was collected after three boosts. This antibody works well for western blot, but not for IF under conditions we have tested.
Immunofluorescence microscopy of intact cells
Cells expressing p166myc, ΔCp166myc, or ΔTMCp166myc (Figure 1A) were grown to 5 × 106 cells/ml, washed twice in PBS, and adhered to poly-L-lysine-coated eight-well slides (6 mm well diameter). The cells were fixed in Mirsky's solution (National Diagnostics), rinsed twice in PBS, and permeabilized in methanol (overnight, −20°C). They were then treated with rabbit anti-myc antibody (Santa Cruz Biotechnology) diluted 1:100 in PBS containing 1% BSA. Immunodetection of basal bodies utilized a rat monoclonal antibody (YL 1/2) against yeast tyrosinated-tubulin diluted 1:100 in the same diluent (Chemicon) (Kilmartin et al, 1982). Secondary antibodies were AlexaFluor-488-conjugated goat anti-rabbit antibody (diluted 1:200) and AlexaFluor-594-conjugated goat anti-rat antibody (diluted 1:200), respectively (both antibodies from Molecular Probes). The cells were then DAPI-stained (0.5 μg/ml, 1 min). Slides were examined on an Axioskop microscope (Carl Zeiss Inc.) and images were captured with a CCD camera (Retiga™Exi QImaging Corp.) using IPLab software (Scanalytics Inc.).
Immunofluorescence microscopy of cytoskeletons and flagella
All operations were performed at room temperature. To prepare cytoskeletons, mid-log-phase cells expressing full-length p166myc (10 ml) were harvested, washed in PBS, resuspended in 500 μl of PBS, and spotted on poly-L-lysine-coated slides (50 μl per well on a 10-well slide). Cells were then extracted in NP-40 (0.25% in 100 mM PIPES, 1 mM MgCl2, pH 7.4, 4 min) and washed again (PBS, twice, 5 min each). Resulting cytoskeletons were fixed (3% paraformaldehyde in PBS, 30 min), neutralized (100 mM glycine, three times, 5 min each), and blocked (0.1% BSA in PBS, twice, 10 min each). Cytoskeletons were then incubated in rabbit polyclonal anti-myc antibody (diluted 1:50 in 0.1% BSA in PBS, 1 h), washed (PBS, twice, 10 min each), and incubated with FITC-conjugated goat anti-rabbit antibody (Sigma, diluted 1:100 in PBS, 1 h). Cells were washed (PBS, twice, 5 min each), DAPI-stained (10 μg/ml), and mounted for fluorescence microscopy in SlowFade Lite (Molecular Probes).
We used a new procedure to isolate flagellum–kDNA complexes. Whereas the original method used Ca++ to depolymerize sub-pellicular microtubules (Robinson and Gull, 1991), the new procedure uses EDTA. To isolate these complexes, mid-log phase cells (10 ml culture) expressing p166myc was treated 15 min with EDTA (final concentration, 10 mM). Cells were harvested (1000 g, 10 min) and washed three times in 1 ml PBS containing 10 mM EDTA (centrifuging 1000 g, 10 min, room temperature). Between washes cells were incubated 10 min at room temperature. Cells were resuspended in 500 μl PBS containing10 mM EDTA (room temperature) and spotted onto a 10-well poly-L-lysine-coated slide. Cells were extracted for 15 min in PBS containing 10 mM EDTA, 0.5% NP-40, and protease inhibitors (Calbiochem set III (catalog no. 539134) diluted 1:1000, 30 min, 4°C). Slides were kept on an ice-cold block during depolymerization of the sub-pellicular microtubules. The extent of depolymerization was checked every 15 min by phase-contrast microscopy and was complete in 45–60 min. Flagella were then washed with PBS containing 10 mM EDTA and a 1:1000 dilution of set III protease inhibitors (50 μl per well, three times, 15 min each, room temperature).
In preparing flagella for IF, all operations were performed at room temperature. Flagella were fixed (3% paraformaldehyde in PBS, 30 min), neutralized (100 mM glycine, three times, 5 min each), blocked (0.1% BSA in PBS, two times, 10 min each), then incubated 1 h in rabbit polyclonal anti-myc antibody (Sigma C-3956, diluted 1:50 in 0.1% BSA in PBS), washed (PBS, two times, 10 min each), and incubated with FITC-conjugated goat anti-rabbit antibody (Sigma, diluted 1:100 in PBS, 1 h). The flagella were then washed (PBS, three times, 10 min) and then incubated with antibody YL1/2 (Chemicon MAB 1864) that recognizes the basal body (diluted 1:450 in PBS, pH 7.2, containing 1% BSA, 60 min). Flagella were then washed (PBS, pH 7.2, three times, 10 min) and probed with chicken anti-rat IgG (Molecular Probes, Alexa Fluor® 594, A-21471), diluted 1:100 PBS, pH 7.2, for 60 min. Then the flagella were washed (PBS, two times, 5 min each), DAPI-stained (10 μg/ml), and mounted for fluorescence microscopy in SlowFade gold S36936 (Invitrogen).
Immunoelectron microscopy
Mid-log-phase cells expressing p166myc (50 ml culture) were harvested and resuspended in 3% paraformaldehyde in PBS (25 ml, 30 min, 4°C). Cells were then washed, dehydrated, and embedded in Lowicryl HM20 mono-step (Electron Microscopy Sciences). All embedding and polymerization procedures in this section were conducted at −25°C. Sections were cut, neutralized (100 mM glycine, 10 min), blocked (1% BSA in PBS, 10 min), and probed with rabbit polyclonal anti-myc tag (Sigma C-3956, diluted 1:20 in 0.1% BSA in PBS, 4°C, overnight). After washing, sections were probed (10 nm gold-conjugated protein A (Aurion), diluted 1:30 in blocking buffer, 2 h). After a second wash, sections were treated with 1% glutaraldehyde to fix the gold particles to the grid, stained with 5% uranyl acetate in 1% acetic acid for 30 min, and visualized on a Philips CM10 electron microscope.
To count gold particles, 18 EM images were scanned and processed using Adobe Photoshop. Areas of the whole cell, the TAC, and the rest of the mitochondrion were measured using Photoshop. Gold particles in each area were counted.
Electron microscopy of kDNA
kDNA networks isolated from cells after 2 days of RNAi or control cells (strain 29–13) were spread on grids using the formamide method (Pérez-Morga and Englund, 1993b) and rotary shadowed with Pt–Pd using a Denton Vacuum evaporator (DV502-A). Networks were photographed using a Hitachi 7600 transmission electron microscope and a DVC 1412M-FW digital camera with AMT Image Capture Engine Software (5.4.2.307). Images were enhanced by uniformly adjusting brightness and contrast using Adobe Photoshop.
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank members of our laboratory for encouragement and discussion, and we especially thank Gokben Yildirir for technical help. We thank Rob Jensen for valuable discussion and Michael Delannoy and Michael McCaffrey for help with EM. NIH (grant AI058613) provided financial support.
References
- Abu-Elneel K, Robinson DR, Drew ME, Englund PT, Shlomai J (2001) Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J Cell Biol 153: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Shonenberger M (1979) Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop 36: 289–292 [PubMed] [Google Scholar]
- Chen XJ, Butow RA (2005) The organization and inheritance of the mitochondrial genome. Nat Rev Genet 6: 815–825 [DOI] [PubMed] [Google Scholar]
- Downey N, Hines JC, Sinha KM, Ray DS (2005) Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot Cell 4: 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew ME, Englund PT (2001) Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J Cell Biol 153: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel ML, Ray DS (1999) The kinetoplast structure-specific endonuclease I is related to the 5′ exo/endonuclease domain of bacterial DNA polymerase I and colocalizes with the kinetoplast topoisomerase II and DNA polymerase beta during replication. Proc Natl Acad Sci USA 96: 8455–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund PT, Agbo EE, Lindsay ME, Liu B, Liu Y, Motyka SA, Yildirir G, Zhao Z (2005) RNAi libraries and kinetoplast DNA. Biochem Soc Trans 33 (Part 6): 1409–1412 [DOI] [PubMed] [Google Scholar]
- Ferguson ML, Torri AF, Perez-Morga D, Ward DC, Englund PT (1994) Kinetoplast DNA replication: mechanistic differences between Trypanosoma brucei and Crithidia fasciculata. J Cell Biol 126: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E, Shaw MK, Gull K (2007) Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol Microbiol 64: 1529–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbride DL, Englund PT (1998) The replication mechanism of kinetoplast DNA networks in several trypanosomatid species. J Cell Sci 111: 675–679 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JHJ, Weijers PJ (1980) The segregation of kinetoplast DNA networks in Trypanosoma brucei. Plasmid 4: 97–116 [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C (1982) Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol 93: 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil MM, Motyka SA, Englund PT (2002) Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol Cell 10: 175–186 [DOI] [PubMed] [Google Scholar]
- Li C, Englund PT (1997) A mitochondrial DNA primase from the trypanosomatid Crithidia fasciculata. J Biol Chem 272: 20787–20792 [DOI] [PubMed] [Google Scholar]
- Liu B, Liu Y, Motyka SA, Agbo EEC, Englund PT (2005a) Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol 21: 363–369 [DOI] [PubMed] [Google Scholar]
- Liu Y, Englund PT (2007) The rotational dynamics of kinetoplast DNA replication. Mol Microbiol 64: 676–690 [DOI] [PubMed] [Google Scholar]
- Liu Y, Motyka SA, Englund PT (2005b) Effects of RNA interference of Trypanosoma brucei structure-specific endonuclease-I on kinetoplast DNA replication. J Biol Chem 280: 35513–35520 [DOI] [PubMed] [Google Scholar]
- Melendy T, Sheline C, Ray DS (1988) Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell 55: 1083–1088 [DOI] [PubMed] [Google Scholar]
- Milman N, Motyka SA, Englund PT, Robinson D, Shlomai J (2007) Mitochondrial origin binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Drew ME, Englund PT (2002) Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J 21: 4429–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Motyka SA, Drew ME, Englund PT (2004) An RNAi-based genomic library for forward genetics in the African trypanosome. In Gene Silencing by RNA Interference: Technology and Application, Sohail M (ed), pp 241–257. Boca Raton: CRC Press [Google Scholar]
- Motyka SA, Drew ME, Yildirir G, Englund PT (2006) Overexpression of a cytochrome b5 reductase-like protein causes kinetoplast DNA loss in Trypanosoma brucei. J Chem 281: 18499–18506 [DOI] [PubMed] [Google Scholar]
- Motyka SA, Zhao Z, Gull K, Englund PT (2004) Integration of pZJM library plasmids into unexpected locations in the Trypanosoma brucei genome. Mol Biochem Parasitol 134: 163–167 [DOI] [PubMed] [Google Scholar]
- Oberholzer M, Morand S, Kunz S, Seebeck T (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol 145: 117–120 [DOI] [PubMed] [Google Scholar]
- Ogbadoyi EO, Robinson DR, Gull K (2003) A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell 14: 1769–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Morga D, Englund PT (1993a) The attachment of minicircles to kinetoplast DNA networks during replication. Cell 74: 703–711 [DOI] [PubMed] [Google Scholar]
- Pérez-Morga DL, Englund PT (1993b) Microtechnique for electron microscopy of DNA. Nucleic Acids Res 21: 1328–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel LC, Bonhivers M, Landrein N, Robinson DR (2006) NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J Cell Sci 119 (Part 9): 1852–1863 [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352: 731–733 [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K (1994) The configuration of DNA replication sites within the Trypanosoma brucei kinetoplast. J Cell Biol 126: 641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxowsky TT, Choudhary G, Klingbeil MM, Englund PT (2003) Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J Biol Chem 278: 49095–49101 [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K (1987) Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol 104: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J (2004) The structure and replication of kinetoplast DNA. Curr Mol Med 4: 623–647 [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK (2005) Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30: 97–105 [DOI] [PubMed] [Google Scholar]
- Wang Z, Englund PT (2001) RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J 20: 4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 275: 40174–40179 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1