Abstract
Imprinting control regions (ICRs) are known to repress genes by utilizing one of two mechanisms, CTCF-mediated insulation or the transcription of non-coding RNAs (ncRNAs). The KvDMR1 ICR contains both the promoter for the Kcnq1ot1 ncRNA and two CTCF-binding sites located within sequences exhibiting repressive activity in enhancer-blocking assays. Deletion of KvDMR1 results in ubiquitous biallelic expression of eight maternal-specific genes in distal chromosome 7. Here we report that while truncation of the Kcnq1ot1 transcript results in the loss of imprinted expression of these genes in the placenta, it does not affect imprinted expression of Cdkn1c in a subset of embryonic tissues despite universal loss of paternal-specific methylation at Cdkn1c. Consistent with tissue-specific loss of imprinted expression, growth deficiency of these mutant mice was less severe than that observed previously in mice with deletion of KvDMR1. This study demonstrates that the KvDMR1 locus can silence Cdkn1c by a mechanism independent of Kcnq1ot1 transcription, perhaps by CTCF-associated repression, making it the first example of an ICR capable of silencing the same gene by two distinct mechanisms.
Keywords: DNA methylation, genomic imprinting, growth restriction, Kcnq1ot1, non-coding RNA
Introduction
Imprinted genes are members of several gene families and encode gene products involved in a diversity of physiological processes, many of which are intimately involved in growth control and development (Tycko and Morison, 2002). The process of genomic imprinting involves the establishment of parent-specific epigenetic modifications in the germ line that result in monoallelic expression of genes in a parent-of-origin dependent manner (reviewed in Reik and Walter, 2001; Tycko and Efstratiadis, 2002). It is not completely understood how the two alleles of an imprinted gene are differentially expressed despite being present in the same nucleus. However, the genomic organization of imprinted genes, namely their tendency to occur in clusters, and the analysis of targeted deletions in mice suggest that relatively short DNA sequences, known as imprinting control regions (ICRs), operate in cis to impose monoallelic expression in imprinted domains (Spahn and Barlow, 2003; Verona et al, 2003). One of the most intensely studied ICRs is the H19/Igf2 DMD (differentially methylated domain); targeted deletion of this sequence in the mouse resulted in loss of imprinted expression of the Igf2 and H19 genes (Thorvaldsen et al, 1998). Paternal-specific expression of the Igf2 gene is due to the ability of this regulatory element to function as a chromatin insulator, which blocks enhancer–promoter interactions between the promoter and downstream enhancers. Importantly, it has been shown that the multifunctional transcriptional regulator CTCF binds to the H19/Igf2 DMD in a methylation-sensitive and parent-of-origin dependent manner, and that binding of CTCF is required for enhancer-blocking activity (Verona et al, 2003). Similar CTCF-associated mechanisms may also be functioning at the Rasgrf1 and DLK1/GTL2 loci (Rosa et al, 2005; Yoon et al, 2005).
Most imprinted domains contain at least one non-coding RNA (ncRNA) that exhibits reciprocal imprinted expression with respect to the protein coding genes, and is therefore implicated in their allele-specific silencing (O'Neill, 2005). At the Igf2r locus, the long (>100 kb) Air transcript is initiated from its promoter in an intronic differentially methylated region (DMR) termed Region 2 (Lyle et al, 2000). Targeted deletion of Region 2 results in biallelic expression of Igf2r, Slc22a2, and Slc22a3, three genes normally expressed only from the maternal alleles (Wutz et al, 1997). Evidence for the involvement of ncRNAs in imprinted expression of autosomal genes was first provided by truncating the Air transcript in the mouse, which resulted in derepression of these same genes on the paternal chromosome (Sleutels et al, 2002). The KvDMR1 imprinted domain (also called the Kcnq1 or IC2 domain) is similar to the Igf2r locus in that it has a maternally methylated intronic DMR, which contains the promoter for the long (>60 kb) paternally expressed ncRNA, Kcnq1ot1 (Smilinich et al, 1999). Following paternal transmission of a deletion of the KvDMR1 locus, eight genes normally expressed exclusively or preferentially from the maternal chromosome are expressed biallelically in all embryonic and extra-embryonic tissues (Fitzpatrick et al, 2002; Lewis et al, 2004; Mancini-DiNardo et al, 2006), and mutant mice are 22% smaller than wild-type (wt) littermates (Fitzpatrick et al, 2002; Salas et al, 2004). These independent deletions included both the entire differentially methylated CpG island as well as the promoter for the Kcnq1ot1 ncRNA. In a cell culture assay, truncation of Kcnq1ot1 in an episomal expression vector carrying flanking reporter genes demonstrated that Kcnq1ot1 can silence gene expression in a bidirectional manner (Thakur et al, 2004). More recently, truncation of Kcnq1ot1 in the mouse was reported to result in loss of imprinted expression of all genes tested in e11.5 placenta, as well as biallelic expression of Cdkn1c in e11.5 embryos and multiple neonatal and adult tissues (Mancini-DiNardo et al, 2006).
These studies have led to the notion that ICRs employ one of two distinct mechanisms, one of which utilizes enhancer blocking or ‘insulation' that relies on the protein CTCF and is represented by the Igf2/H19 DMD, and the other operating through ncRNAs and exemplified by the Igf2r/Region 2 domain and perhaps the KvDMR1 domain (Lewis and Reik, 2006; Mancini-DiNardo et al, 2006; Pauler and Barlow, 2006). However, in addition to the Kcnq1ot1 promoter, the KvDMR1 locus contains two CTCF-binding sites (Fitzpatrick et al, 2007) within sequences shown to have insulator or silencer activity in various enhancer-blocking assays (reviewed in Fitzpatrick et al, 2007). In the present study, a more extensive analysis of a Kcnq1ot1 truncation allele constructed in our laboratory suggests that KvDMR1 employs more than one silencing mechanism in vivo. Using semi-quantitative allelic expression analysis, we show that while truncation of the Kcnq1ot1 ncRNA results in the loss of imprinted expression of all KvDMR1-controlled genes in the placenta and in many embryonic lineages, the imprinted expression of Cdkn1c is maintained in several fetal and neonatal tissues. Although these Kcnq1ot1 truncation mutants exhibit significant growth deficiency in utero, postnatal growth restriction is considerably less severe than that observed in mice with a complete deletion of KvDMR1 (Fitzpatrick et al, 2002; Salas et al, 2004), consistent with tissue-specific retention of Cdkn1c-imprinted expression. These findings indicate that the truncation of Kcnq1ot is not equivalent to deletion of the entire KvDMR1 region, and suggest that this locus can also silence Cdkn1c by a second distinct mechanism that functions in an ncRNA-independent manner.
Results
Generation of Kcnq1ot1 truncation mice
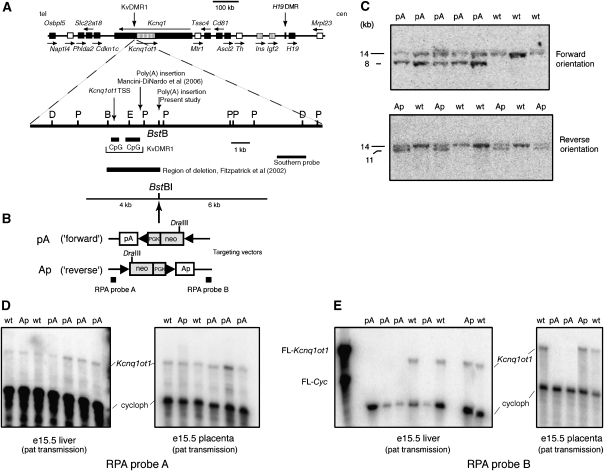
To determine whether the Kcnq1ot1 RNA or its transcription is required to silence genes regulated by the KvDMR1 ICR, a polyadenylation (poly(A)) cassette was inserted 2.6 kb downstream of the Kcnq1ot1 promoter by homologous recombination in ES cells. To exclude the possibility that an observed phenotype was due to physical disruption of the KvDMR1 locus itself, a similar construct with the poly(A) cassette in the reverse, non-functional orientation was inserted at the same location (Figure 1B). G418-resistant clones were screened by Southern hybridization using probes 3′ and 5′ of the targeted region (not shown). Positive ES clones were injected into C57BL/6J blastocysts to generate chimeric animals. Male chimeras derived from two independent positive ES cell clones with the poly(A) cassette in the forward orientation, and one derived from a positive ES cell clones with the poly(A) cassette in the reverse orientation, were then mated with C57BL/6J to establish germ-line mice. Mice were recovered with the correct targeting event for both the ‘forward' (pA) and ‘reverse' (Ap) orientations (Figure 1C). Mutant male mice were then crossed with Zp3-Cre (Lewandoski et al, 1997) females to promote the excision of the floxed neo-cassette. The analyses discussed below were carried out on both lines with the ‘forward' mutation with the same results.
Figure 1.
Physical map of the imprinted domain in distal chromosome 7, targeting strategy for insertion of poly(A) cassette in KvDMR1, and effect of insertion on Kcnq1ot1 expression. (A) Upper diagram: Genomic map showing the location of imprinted genes in mouse distal chromosome 7. Maternally expressed genes are indicated as black boxes, paternally expressed genes as stippled boxes, and non-imprinted (biallelic) genes as white boxes. Arrows above or below each gene indicate the direction of transcription. tel, telomere; cen, centromere. Lower diagram: An enlargement of the mouse KvDMR1 locus and flanking DNA showing the location of the two adjacent CpG islands and the position of the BstBI site into which the poly(A) cassette was inserted. B, BsrGI; D, DraIII; P, PstI, E, EagI. The positions of the Kcnq1ot1 transcription start site (TSS), as well as the insertion locations for poly(A) truncation cassettes use in this study and in the Mancini-Dinardo et al (2006) report, are indicated. (B) Schematic representation of the insertion vector showing the extent of the left-hand (6 kb) and right-hand (4 kb) homologous regions used for recombination. The ‘floxed' neomycin-resistance gene (neo) is driven by the Pgk promoter and contains an SV40 polyadenylation site. The SV40 polyadenylation site results in the introduction of an additional DraIII site, which facilitates genotyping the ES cell-derived mice by Southern hybridization. The rabbit β-globin poly(A) cassette was ligated to the neo-cassette and the resultant fragment inserted into the BstBI site in either forward (pA) (i.e., same orientation as Kcnq1ot1 transcript) or in reverse orientation (Ap). The extent of the deletion described by Fitzpatrick et al (2002) and the positions of the Southern and RPA probes are indicated. (C) Southern hybridization to genomic DNA digested with DraIII from fetuses derived from agouti colored chimeric males and C57BL/6J females. (D, E) Following excision of the neo-cassette by mating with female Zp3-cre mice, the effect of the poly(A) cassette on the expression of the Kcnq1ot1 ncRNA was determined by RPA using RNA from e15.5 conceptuses (liver and placenta) following paternal transmission of the poly(A) cassette (pA or Ap) and wt littermates. The positions of the cyclophilin and Kcnq1ot1 full-length probes (FLP) and protected probes are indicated.
Expression of Kcnq1ot1 ncRNA in mutant mice was first assessed by ribonuclease protection assays (RPA). Following paternal transmission, Kcnq1ot1 expression upstream of the insertions (RPA probe A) appeared unaltered in placenta and fetal liver from mutant conceptuses containing either the pA or Ap insertions (Figure 1D). More extensive analysis using quantitative RT–PCR confirmed that Kcnq1ot1 expression was not significantly affected in placenta and several fetal tissues by insertion of the pA cassette (Supplementary Figure S1). Southern hybridization using a methylation-sensitive restriction endonuclease showed that methylation at KvDMR1 was unchanged in mutant mice (not shown). Thus, targeted mutation of the locus did not affect the methylation and function of the Kcnq1ot1 promoter. In contrast, no expression of Kcnq1ot1 was detected in either e15.5 liver or placenta downstream of the truncation cassette (RPA probe B) when inserted in the pA orientation (Figure 1E); downstream expression was not changed when the cassette was inserted in the Ap orientation. As expected, maternal transmission of the poly(A) cassette had no effect on Kcnq1ot1 expression (not shown).
Expression of the Kcnq1ot1 ncRNA is necessary for imprinted expression of most genes under the control of the KvDMR1 ICR
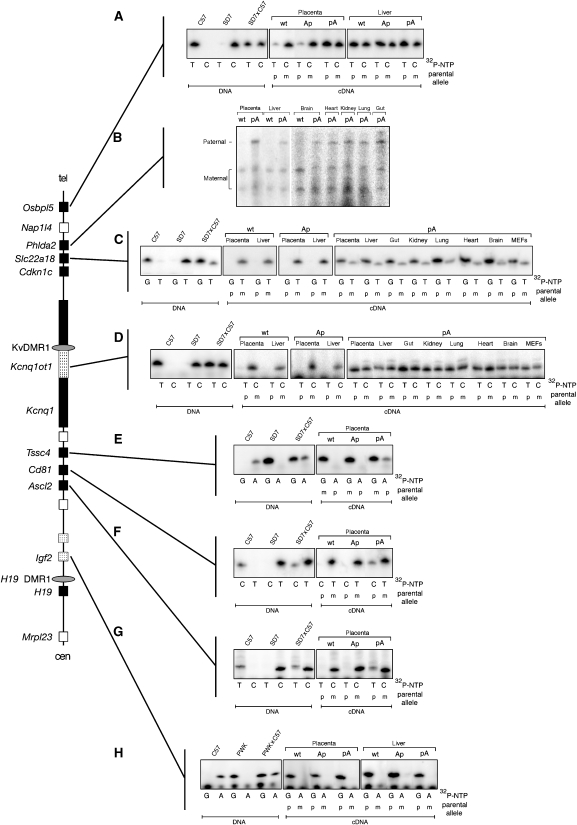
The effect of the truncated Kcnq1ot1 RNA on imprinted expression of the eight genes known to be under the control of KvDMR1 was assessed by allele-specific expression assays. With the exception of Phlda2, this was initially carried out using single-nucleotide primer extension (SNuPE) assays (Shen et al, 1998) utilizing SNPs between 129SvJae and SD7 mice. In these semi-quantitative assays, gene expression is considered biallelic if the ratio of radionucleotide incorporation in cDNA is similar to that in heterozygous (SD7 × C57) DNA, which contains 50% of each allele. Several of the genes in this domain (Osbpl5, Tssc4, Cd81, Ascl2) are imprinted only in the placenta (Engemann et al, 2000). Consistent with earlier findings, expression of Osbpl5 exhibited strong maternal allele expression bias in wt e15.5 placenta but not in e15.5 liver (Figure 2A). However, Osbpl5 expression was biallelic in e15.5 placenta following paternal inheritance of the pA cassette (Figure 2A). Importantly, preferential expression of the maternal allele was maintained in placenta from conceptuses carrying a paternally inherited Ap construct. Together with the Kcnq1ot1 expression results discussed above, these data indicate that loss of imprinting of Osbpl5 observed in placenta with the pA cassette is due to truncation of the Kcnq1ot1 transcript and not simply disruption of the KvDMR1 locus. Biallelic expression was also observed for Tssc4, Cd81, and Ascl2 in e15.5 placenta following paternal transmission of the pA truncation cassette but not with the Ap cassette (Figure 2E–G). Note that for these three genes, the radionucleotide corresponding to paternal (C57BL/6J) allele is not incorporated as efficiently as the nucleotide corresponding to the maternal (SD7) allele (see SD7 × C57 DNA lanes). This explains why in the pA cDNA lanes the paternal allele is less intense than might be expected for biallelic expression.
Figure 2.
Paternal transmission of the Kcnq1ot1 truncation results in the loss of imprinted expression of maternal-specific genes in cis. Allele-specific SNuPE (A, C–G) or RPA (B) assays were used to analyze the expression of seven maternally expressed genes in placenta and various fetal tissues from e15.5 day conceptuses derived from crosses of female SD7 mice and male mice heterozygous for the poly(A) cassettes. Although the mutated mice in this study are of a mixed genetic background of 129SvJae and C57BL/6J, these mouse strains are closely related and share many of the same polymorphisms. Thus, for simplicity these mice are designated as C57BL/6J. (A) An expressed polymorphism in Osbpl5 was amplified by RT–PCR and analyzed by SNuPE (Materials and methods). Analysis of DNA showed that the C57BL/6J (C57) and M. spretus (SD7) alleles incorporates only radiolabeled TTP or dCTP, respectively, whereas DNA from F1 offspring of a wt SD7 × C57 cross incorporated both in roughly equal proportion. (B) Allele-specific RPA for Phdla2. Preferential or exclusive expression of the maternal allele (lower two protected bands) is seen in RNA from placenta and fetal liver and brain. Placenta and fetal tissues obtained from pA-mutant conceptuses also expressed the paternal (C57) allele, indicating loss of imprinted expression. (C) SNuPE analysis of Slc22a18. In wt and Ap-mutant placenta and fetal liver, imprinted expression is observed as indicated by exclusive incorporation of TTP into cDNA. In contrast, placental and embryonic tissues from pA mutants showed biallelic expression. (D) SNuPE analysis of Kcnq1 shows maternal-specific expression (i.e., incorporation of dCTP only) in placenta and fetal liver from wt and Ap conceptuses. In all tissues tested from pA conceptuses, Kcnq1 becomes biallelic. (E–G) Tssc4, Cd81, and Ascl2 have imprinted expression only in placenta (Paulsen et al, 2000). In each case, loss of imprinted expression is observed following paternal transmission of the pA insertion. (H) Paternal-specific expression of Igf2 is seen in wt and both Ap and pA mutations.
The four other genes (Phlda2, Slc22a18, Cdkn1c, Kcnq1) under the control of KvDMR1 are imprinted in the embryo proper as well as in the placenta (Hatada and Mukai, 1995; Qian et al, 1997; Cooper et al, 1998; Gould and Pfeifer, 1998). We exploited a 6-bp deletion in the Mus spretus allele of SD7 mice to develop an allele-specific RPA for Phlda2. Figure 2B shows that the maternal-specific expression of Phlda2 in wt placenta and embryonic tissues became biallelic following paternal inheritance of the pA cassette. Similar to Phlda2, both Slc22a18 (radionucleotide corresponding to the paternal allele (G) is incorporated much more efficiently in this case) and Kcnq1 were biallelically expressed in mutant tissues (Figure 2C and D). As expected from our previous analysis of a KvDMR1 deletion mouse (Fitzpatrick et al, 2002), truncation of Kcnq1ot1 had no effect on the imprinted expression of Igf2 (Figure 2H), consistent with the notion that it is regulated by a different ICR.
The Cdkn1c gene can be silenced by a mechanism independent of Kcnq1ot1 transcription
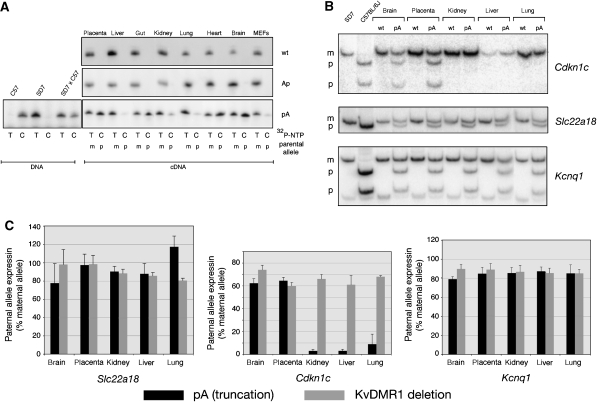
Following paternal inheritance of the pA truncation cassette, imprinted expression of Cdkn1c was lost in the placenta and several embryonic tissues including heart, brain, and gut. In contrast, maternal-specific expression of Cdkn1c was, in large part, maintained in other embryonic tissues and cells including liver, kidney, lung and embryonic fibroblasts (MEFs) (Figure 3A). Tissue-specific loss of imprinted expression at Cdkn1c was confirmed by a second independent assay, which takes advantage of restriction fragment length polymorphisms (RFLP) in RT–PCR products generated from SD7 and C57 alleles. Hot-stop PCR (Uejima et al, 2000) was performed to circumvent the effect of incomplete digestion due to heteroduplex PCR products. Consistent with the SNuPE results, imprinted expression of Cdkn1c was lost in the truncation mutant in the placenta and fetal brain, but retained in fetal kidney, liver, and lung (Figure 3B). To examine this finding more closely, RFLP-RT–PCR analysis of Slc22a18, Cdkn1c, and Kcnq1 was repeated in triplicate for the same five tissues from two pA-mutant conceptuses and two wt littermates, and the results quantitated using the PhosphorImager (Figure 3B and C). Because we found that the paternal C57 allele of these genes is consistently expressed at a reduced level compared with the maternal SD7 allele, tissues from a KvDMR1 deletion fetus (considered as complete loss of imprinted expression) were also analyzed for comparison. For Slc22a18 and Kcnq1, imprinted expression was lost in the pA mutant to roughly the same extent as in the KvDMR1 deletion mutant (Figure 3C, left and right panels). In contrast, paternal allele expression of Cdkn1c was equivalent to the maternal allele only in placenta and brain. In e15.5 kidney, liver, and lung, expression from the paternal allele of Cdkn1c represented only 3–9% of the maternal allele (Figure 3C, center panel). This low-level of expression from the paternal allele could be a consequence of the loss of paternal methylation at Cdkn1c observed in poly(A) truncation mutants (see below and Discussion).
Figure 3.
Insertion of poly(A) cassette affects imprinted expression of Cdkn1c in only a subset of embryonic tissues. (A) SNuPE analysis of Cdkn1c in e15.5 placenta and embryonic tissues from e15.5 day conceptuses derived from crosses of female SD7 mice and male mice heterozygous for the poly(A) cassettes. Exclusive expression of the maternal allele (T) is observed in all tissues examined from wt and Ap-mutant conceptuses. Imprinted expression is lost (indicated by incorporation dCTP into cDNA) in placenta, gut, heart, and brain from pA conceptuses while primarily maternal-specific expression is retained in liver, kidney, lung, and MEFs. (B) RFLP-RT–PCR analysis of Cdkn1c, Slc22a18, and Kcnq1 in multiple tissues from pA truncation mutants and wt littermates. Hot-stop RT–PCR products were digested by an allele-specific restriction enzyme (Supplementary Table S1) and separated by 10% PAGE. (C) Three independent RFLP-RT–PCR assays were performed for each of five tissues from two pA truncation-mutant embryos, two wt embryos, and one KvDMR1 deletion embryo for Slc22a18, Cdkn1c, and Kcnq1. Maternal and paternal bands were quantitated using the PhosphorImager. The histograms show the expression (average of 3–6 determinations plus STDEV) of the ‘relaxed' paternal alleles in the two mutants relative to that of the maternal allele. Expression of paternal alleles in wt tissues was undetectable.
Tissue-specific maintenance of imprinted expression of Cdkn1c in truncation mutants is not due to the retention of paternal-specific methylation
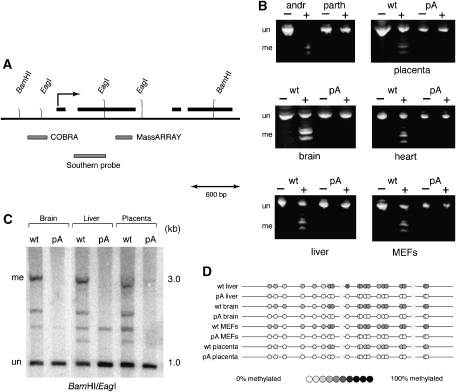
Differential methylation of the paternal allele of Cdkn1c occurs between e6.5 and e8.5 and is dependent on the KvDMR1 locus (Bhogal et al, 2004). Thus, a simple explanation for the tissue-specific retention of Cdkn1c imprinting in truncation mutants is concomitant maintenance of paternal-specific methylation. We assessed the methylation status of the Cdkn1c promoter by combined bisulfite restriction analysis (COBRA) (Xiong and Laird, 1997). Primers defining a 296-bp amplicon (Figure 4A) spanning nine BstUI sites (CGCG) in the native sequence were used to amplify bisulfite-treated DNA from the eight tissues used in the expression analyses as well as from control androgenetic (100% methylated) and parthenogenetic (0% methylated) MEFs. Methylation protects the BstUI sites from conversion during bisulfite treatment, allowing the subsequent PCR products to be cleaved by the restriction enzyme. As expected, DNA from androgenetic and parthenogenetic MEFs was completely digested or completely resistant to cleavage, respectively (Figure 4B, upper left hand panel). For each of the other tissues, DNA from wt littermates was digested to approximately 50% as expected for a differentially methylated locus (Figure 4B). However, PCR products from the truncation mutant were resistant to digestion, indicating that all or most of the BstUI sites were lost during bisulfite conversion due to the absence of methylation. Thus, in the pA truncation mutant, all tissues showed loss of methylation (LOM) at the Cdkn1c promoter irrespective of expression status of the paternal allele. Methylation within the body of the Cdkn1c gene was assessed using both Southern hybridization (Figure 4C) and MassARRAY analysis (Ehrich et al, 2005; Figure 4D, see Materials and methods). Once again LOM was observed in all tissues tested from Kcnq1ot1 truncation mice, including liver, and MEFs, which maintain imprinted expression at Cdkn1c. These results clearly demonstrate that retention of imprinting at Cdkn1c in specific embryonic tissues in Kcnq1ot1 truncation mice is not due to the maintenance of paternal methylation.
Figure 4.
Truncation of Kcnq1ot1 results in the loss of paternal methylation at Cdkn1c regardless of expression. (A) Genomic map of the mouse Cdkn1c locus, indicating the positions of the regions assayed for methylation by COBRA and MassARRAY, and the position of the probe used in the Southern analysis. Exons are shown as black boxes. The transcription start site is indicated by the broken arrow. (B) The promoter region was analyzed by COBRA (Xiong and Laird, 1997) using primers that amplify a 296-bp fragment that contains nine sites for BstUI in the unmodified sequence (Bhogal et al, 2004). Bisulfite-treated DNA from the eight tissues used in the expression analyses as well as from control androgenetic (100% methylated) and parthenogenetic (0% methylated) MEFs was amplified and digested with BstUI (+) or incubated in a mock digest (−). The smaller bands result from the digestion of methylation-protected BstUI sites. (C) DNAs from wt and pA brain, liver, and placenta were digested with BamHI and the methylation-sensitive endonuclease EagI. Following electrophoresis and transfer to nylon, hybridization was performed using a probe within the gene body of Cdkn1c. The uncut (i.e., methylated) band at 3 kb is virtually absent in all three tissues from the pA mutant. (D) Bisulfite-treated DNA from wt and pA brain, liver, MEFs, and placenta were amplified using primers spanning part of exon 2 and analyzed by MassARRAY. In each case, quantitation of mass spectra indicated approximately 50% methylation of DNA from wt tissues but no methylation in tissues from pA mutants. me, methylated; un, unmethylated.
Tissue-specific maintenance of Cdkn1c imprinted expression is associated with a less severe growth phenotype compared with ubiquitous biallelic expression
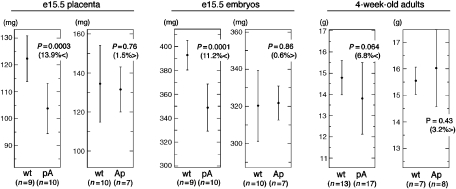
Following paternal transmission of the poly(A) insertion in the forward orientation (pA), e15.5 placentas and embryos were 14 and 11%, respectively, smaller than their wt littermates (Figure 5). This is comparable to the growth retardation observed in KvDMR1 deletion mutants (Fitzpatrick et al, 2002; Salas et al, 2004). The analysis of several 4-week-old litters from crosses between wt females and mutant males indicated that the number of wt and Kcnq1ot1 truncation offspring were approximately equal demonstrating that there was no pre- or postnatal lethality associated with this mutation. At this point in development however, mutant mice were only 7% smaller than wt littermates, a difference that reached only marginal significance (Figure 5). Thus, growth deficiency in adult mice was markedly reduced compared with KvDMR1 deletion mice, which were more than 20% smaller than wt counterparts at this stage (Fitzpatrick et al, 2002; Salas et al, 2004). This observation suggests that while resulting in a similar phenotype in e15.5 placenta and embryo, these two mutations are not equivalent in later development. The less severe growth restriction is presumably due to the retention of Cdkn1c-imprinted expression in a subset of tissues as described above.
Figure 5.
Growth deficiency in mice with a paternally inherited truncation of Kcnq1ot1 is less severe than in mice with a complete deletion of KvDMR1. Diagrams show the mean weights and standard deviation of offspring at different times during development from males carrying a heterozygous pA or Ap mutation crossed to wt females. Statistical significance using Student's t-test is indicated as a ‘P' value. Differences in weight are indicated in parentheses.
Discussion
Two mechanisms have been described by which ICRs function; one, exemplified by the Igf2/H19 locus, utilizes a methylation-sensitive CTCF-regulated enhancer blocker or insulator, while the other mechanism, for which the Igf2r/Air locus is the prototype, involves the transcription of an ncRNA (Lewis and Reik, 2006; Pauler and Barlow, 2006). Truncations of Kcnq1ot1 in an episomal expression vector in cell culture (Thakur et al, 2004), and in the mouse (Mancini-DiNardo et al, 2006), have previously implicated the Kcnq1ot1 ncRNA or its transcription in gene silencing, placing the KvDMR1 domain in the same category as the Igf2r/Air locus (Mancini-DiNardo et al, 2006; Pauler and Barlow, 2006). Consistent with earlier studies, our results demonstrate that the Kcnq1ot1 ncRNA, or its transcription, plays a major role in silencing genes under the control of the KvDMR1 ICR. The exact mechanism by which transcription of Kcnq1ot1 results in gene silencing is unknown but likely involves recruitment of repressive chromatin modifications, possibly associated with localization to a suppressive subnuclear compartment (Mager et al, 2003; Lewis et al, 2004; Umlauf et al, 2004; Yang and Kuroda, 2007). Although not mutually exclusive, another possibility is that silencing of the paternal domain is brought about by Kcnq1ot1 transcription through, and activation of, a domain-wide repressor, or inactivation of a domain-wide activator (Pauler and Barlow, 2006). Regardless of the exact mechanism of Kcnq1ot1 transcription-dependent gene silencing, the results presented here clearly demonstrate that a second mechanism is capable of establishing and maintaining the imprinted expression of Cdkn1c in some tissues. Analysis of both allele-specific expression and growth phenotype indicate that truncation of Kcnq1ot1 is not equivalent to deletion of the entire KvDMR1 locus as previously implied (Mancini-DiNardo et al, 2006; Pauler and Barlow, 2006). In the present truncation mutant, several embryonic tissues retain maternal-specific expression of Cdkn1c, which results in a less severe growth deficiency at 4 weeks of age (7% smaller) compared with the complete deletion of KvDMR1 (20–22% smaller) (Fitzpatrick et al, 2002; Salas et al, 2004). It should be reiterated that these molecular and growth phenotypes were observed in mice derived from two independently targeted ES clones.
In a previous report, Mancini-DiNardo et al (2006) also using RFLP-RT–PCR showed that, in mice with a truncated Kcnq1ot1 gene, Cdkn1c lost imprinted expression in all tissues tested. Why this earlier account failed to observe maintenance of Cdkn1c imprinting in certain tissues is not clear. In the earlier study, retention of imprinted expression in fetal tissues may have been masked by the analysis of whole embryos rather than individual tissues (present study). However, this explanation cannot account for the disparities in results seen at later times in development. Unlike most imprinted genes in this domain, Cdkn1c imprinted expression is not lost in neonates. Mancini-DiNardo et al (2006) reported biallelic expression of Cdkn1c in all neonatal tissues tested (brain, heart, liver, lung, spleen, and kidney) from their truncation mutant. In contrast, we found that imprinted expression of Cdkn1c in neonatal mice carrying the paternal pA mutation is maintained in the same tissue-specific manner as observed in e15.5 embryonic tissues (Supplementary Figure S2). The lack of tissue-specific retention of Cdkn1c imprinted expression in the mice generated by Mancini-Dinardo and co-workers may be a consequence of inserting the poly(A) cassette closer (1.5 kb) to the Kcnq1ot1 promoter than was inserted in the present study (2.6 kb). Insertion closer to the promoter may have disrupted other sequences in KvDMR1 important for Cdkn1c silencing in certain tissues (e.g., silencer/insulator). Alternatively, it is formally possible that the 2.6 kb of Kcnq1ot1 transcript or transcription in our truncation mutant is sufficient to silence the paternal Cdkn1c allele in some tissues, while the shorter 1.5-kb allele is not. However, considering that Kcnq1ot1 is 60–70 kb long, this explanation seems unlikely. Finally, it is also possible that differences in the stability or processing of the truncated Kcnq1ot1 ncRNA from the two truncation alleles could explain the discrepancies between our results and those of Mancini-DiNardo and co-workers. In this regard it is noteworthy that quantitative RT–PCR analysis of six embryonic tissues showed no significant differences in the steady state level of Kcnq1ot1 in mutant tissues that lose imprinted expression (brain, placenta, heart) compared with those that retain imprinted expression (kidney, lung, liver) (Supplementary Figure S1).
Analysis of methylation at Cdkn1c indicates that the Kcnq1ot1 ncRNA or its transcription regulates the establishment of the somatic DMR present at this gene. Surprisingly paternal-specific methylation at Cdkn1c does not appear to play a major role in maintaining imprinted expression in truncation mutants, since methylation was absent in all tissues regardless of Cdkn1c expression. Unlike in the mouse, the human CDKN1C locus is not differentially methylated and exhibits low-level expression from the paternal allele (Chung et al, 1996; Hatada et al, 1996). In contrast, the paternal allele of Cdkn1c in wt mice is completely repressed (Hatada and Mukai, 1995) and it was suggested that paternal-specific methylation at the mouse Cdkn1c locus is responsible for its more robust silencing (Chung et al, 1996). Thus, while the traces of paternal Cdkn1c expression in e15.5 liver, kidney, and lung may reflect a small proportion of cells within the heterogeneous primary tissues that in fact lose imprinting, an alternative explanation is that LOM at Cdkn1c in truncation mutants results in the slight relaxation of imprinted expression observed in these tissues. This model supports the notion that, in the mouse, paternal-specific methylation at this gene serves to potentiate the silencing effects elicited by Kcnq1ot1 transcription or other components of the KvDMR1 locus.
Cdkn1c encodes p57Kip2, a cyclin-dependent kinase inhibitor known to regulate cellular proliferation and differentiation (Yan et al, 1997; Zhang et al, 1997). Twofold expression of Cdkn1c in transgenic mice leads to offspring that display a growth deficiency phenotype similar to KvDMR1 deletion mice (Andrews et al, 2007). Furthermore, growth restriction in mice with a paternally inherited deletion of KvDMR1 is rescued by a maternal deletion of Cdkn1c, and the absence of Cdkn1c expression results in significantly larger organs from 4-week-old mice than those from wt littermates (J-Y Shin, MJ Higgins, unpublished data). These findings indicate that Cdkn1c is the major determinant of embryonic growth in this imprinted domain. Not surprisingly then, abnormal overexpression of Cdkn1c is likely responsible for at least some cases of Silver–Russell syndrome (SRS), a heterogeneous disorder characterized by severe intrauterine and postnatal growth retardation (Schonherr et al, 2007). On the other hand, aberrant silencing of Cdkn1c is associated with the overgrowth condition Beckwith–Wiedemann syndrome (BWS) in cases that exhibit LOM at the maternal KvDMR1 ICR and consequent maternal expression of KCNQ1OT1 (Diaz-Meyer et al, 2003). Thus, the importance of Cdkn1c/CDKN1C expression levels in embryonic growth may be the driving force responsible for the evolution of a second silencing mechanism specifically targeted to this locus. Whether this second silencing mechanism serves a backup role to the Kcnq1ot1 RNA-dependent mechanism or is normally used to establish imprinted expression of Cdkn1c in some tissues is unknown; however, it is apparent that this mechanism is sufficient to set up and maintain maternal-specific expression in certain cell types in the absence of full-length Kcnq1ot1 transcription. If the alternative silencing mechanism is more than a backup system and is instead necessary for the establishment of CDKN1C in certain tissues, one would predict that in BWS patients with LOM at KvDMR1, this second mechanism, as well as maternal expression of KCNQ1OT1, would be activated by the epimutation. In this context it is interesting that a detailed analysis of LOM at KvDMR1 in BWS patients has shown that methylation is lost across the entirety of the CpG island including the KCNQ1OT1 promoter regions and the region known to exhibit silencer activity in enhancer-blocking assays (Beatty et al, 2006).
The identity of the second silencing mechanism associated with KvDMR1 is uncertain but must involve sequence elements located within the KvDMR1 deletion (Fitzpatrick et al, 2002; Mancini-DiNardo et al, 2006). Several studies have shown that in addition to providing the promoter for Kcnq1ot1, sequences within this deleted region exhibit repressive activity (silencer or insulator) in enhancer-blocking assays (reviewed in Fitzpatrick et al, 2007), and we have recently shown that the multifunctional protein CTCF binds in a paternal-specific manner to the minimal repressive element in this locus (Fitzpatrick et al, 2007). A possible model by which a repressive element could regulate imprinted expression of Cdkn1c in some cell types and not in others proposes that the element is active in some cell types (e.g., hepatocytes, MEFs) but inactive in others (e.g., brain) (see Figure 6). Inactivity of the repressive element in cells where imprinted expression of Cdkn1c is lost in the truncation mutant, could be due to the lack of binding of CTCF or some tissue-specific cofactor in these cells. If a chromatin insulator mechanism is responsible for the retention of Cdkn1c imprinting in some cell types, KvDMR1 will represent the first example of an ICR that utilizes both of the previously documented silencing mechanisms (Lewis and Reik, 2006; Pauler and Barlow, 2006). Previous studies have indicated that imprinted expression is regulated differently in extra-embryonic tissues and in the embryo proper (Lewis et al, 2004; Lin et al, 2007). Our present findings demonstrate that mechanistic differences in imprinted gene silencing also exist among various embryonic lineages.
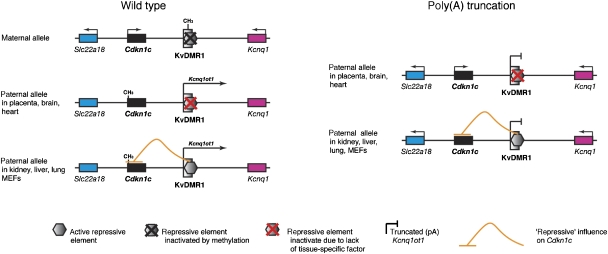
Figure 6.
Model for tissue-specific retention of Cdkn1c imprinted expression. A repressive element (insulator or silencer likely regulated by CTCF) that acts specifically on Cdkn1c is active in only a subset of tissues (e.g., kidney, liver, lung). This repressive activity is either a backup silencing system to the Kcnq1ot1 ncRNA mechanism, or is involved in setting up the initial silencing of Cdkn1c and maintaining it thereafter. In either case, the lack of transcription through the Kcnq1ot1 locus has no affect on Cdkn1c-imprinted expression in these tissues. Note that paternal-specific methylation at Cdkn1c is lost in pA truncation-mutant tissues regardless of expression.
Materials and methods
Generation of Kcnq1ot1 truncation mice
A selection cassette containing neomycin driven by a PGK promoter and flanked by loxP sites was ligated to a 1.2-kb fragment containing the rabbit β-globin polyadenylation signal (Sleutels et al, 2002). This fragment was inserted into an 11-kb clone corresponding to nucleotides 527 668–538 169 of sequence NT_039453 at the BstBI site (position 532173) in both orientations. Constructs were electroporated into J1 ES cells and following selection on G418 and expansion of colonies, genotyping was by Southern hybridization. Male chimeras were mated to C57BL/6J females to establish germ-line transmission. Tail-clip DNA from these litters was analyzed by Southern hybridization as described above for ES cells (see Figure 1). Male mice carrying the poly(A) cassette were crossed with Zp3-Cre (Lewandoski et al, 1997) females to promote the excision of the floxed neo-cassette. Experiments using mice were approved by the RPCI Institute Animal Care and Use Committee (IACUC).
Quantitative RT–PCR
Expression levels of Kcnq1ot1 were determined using real-time quantitative RT–PCR. RNA extracted from five tissues (see Supplementary Figure S1) from two wt and two pA truncation mutants were digested with RNase-free DNase and purified using reagents from the RNeasy Mini kit (Qiagen). cDNA was prepared using the RT2 First-Strand kit (SuperArray), diluted threefold with water and 1 μl used in a 25 μl PCR using the RT2 Master Mix containing SYBR® Green (SuperArray) following the manufacturer's protocol. Primers were designed by SuperArray; the target sequence for Kcnq1ot1 was approximately position 46 970–47 020 of sequence NT_039453.2. Internal control primers amplified Ldhal6b (cat. no. PPM29380A) or Rpl13a (cat. no. PPM03694E). Real-time PCR was performed using an ABI 7900HT instrument and the raw data analyzed by SDS v2.3 software (ABI). Expression levels were determined using the ΔΔCt method.
Analysis of allelic expression in truncation mice
Following excision of the neo-cassette, mice derived from two independently targeted ES lines, and carrying the poly(A) cassette in either the forward or reverse orientation, were mated with SD7 mice (C57BL/6J congenic for distal chromosome 7 of M. spretus) or PWK (for Igf2 analysis only) in reciprocal crosses. For analyses, conceptuses were recovered at day e15.5, except for Kcnq1 where tissues were obtained at e14.5, since this gene loses imprinted expression later in development (Gould and Pfeifer, 1998). RNA was extracted from various tissues using TRIzol reagent (Invitrogen). With the exception of Phlda2, allelic expression was first assessed by SNuPE (Shen et al, 1998), utilizing previously identified SNPs. For semi-quantitative analysis, we carried out RFLP-RT–PCR using Hot-stop PCR (Uejima et al, 2000). Tissues from two wt embryos, two pA truncation embryos, and one KvDMR1 deletion embryo were tested. For each tissue, three independent assays were carried out. Standard RT–PCR was carried out in a 20-μl reaction for 25 cycles and 2 μl checked on a 1.5% agarose gel. A 10-μl volume of the PCR was then subjected to one additional cycle of PCR in a final volume of 20 μl containing 2 μl 32P-TTP (1000 Ci/mmol). The PCR was then digested with the appropriate restriction enzyme for 2 h and the products electrophoresed through the 10% TBE polyacrylamide gel. Gels were analyzed by a PhosphorImager using ImageQuant software. The sequences of primers used for RT–PCR and SNuPE, as well as the restriction enzyme used for each gene, are contained in Supplementary Table 1. Phlda2 expression was detected by an allele-specific RNase protection assays (below).
Ribonuclease protection assays
RPA probe A (upstream of poly(A) cassette insertion) and RPA probe B (downstream of insertion) contained nucleotides 142 034–142 535 and nucleotides 144 718–45 568, respectively, from sequence AJ271885. For Phlda2, the probe corresponded to nucleotides 59–671 from sequence NM_009434. Radioactive RNA probes were synthesized using the MaxiScript T7/T6 kit (Ambion) and 32P-UTP (800Ci/mmol). RPA was performed using the RPAIII kit (Ambion), with hybridization at 45°C (Kcnq1ot1 plus cyclophillin) or 42oC (Phlda2).
Methylation analysis
Methylation of KvDMR1 was determined by Southern hybridization as described (Smilinich et al, 1999). Methylation at the promoter of Cdkn1c was assessed by COBRA as described (Xiong and Laird, 1997). Bisulfite-treated DNA (EZ DNA Methylation kit, ZYMO Research) was PCR-amplified using primers designed to amplify both methylated and unmethylated DNA (Supplementary Table S1). PCR was carried out in 10 μl using 1 μl bisulfite-treated DNA, 200 nmole of each primer, 1.25 mM of each dNTP, 0.2 U of HotStar Taq polymerase (Qiagen), and 1 μl 10 × PCR buffer. The reaction mix was preactivated for 15 min at 94°C and then subjected to 45 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 1 min. Final extension was at 72°C for 3 min. PCR products were purified from a 1% agarose gel using the Qiagen Gel Extraction kit (Qiagen), digested by BstUI restriction enzyme, separated on a 10% polyacrylamide gel, and visualized using SYBR gold (Molecular Probes). Methylation of the Cdkn1c gene body was determined by digesting genomic DNA with BamHI plus EagI and Southern hybridization with a probe corresponding to nucleotides 145–540 of sequence NM_009876. Methylation within exon 2 using bisulfite-treated DNA was assayed using MassARRAY Quantitative Methylation Analysis (MA-QMA) using the MassARRAY Compact system (Sequenom) (Ehrich et al, 2005). This system utilizes mass spectrometry (MS) for the detection and quantitative analysis of DNA methylation using Homogeneous MassCLEAVE (hMC) base-specific cleavage and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS. Methylation calls are performed by the Quantitative Methylation software (Sequenom).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure Legends
Supplementary Table S1
Acknowledgments
We are grateful to Rosemary Elliott for PWK mice, to Wolf Reik for providing the SD7 mice and for critically reading the manuscript, and to Colin Stewart for providing androgenetic and parthenogenetic cell lines. We also thank Denise Barlow for the poly(A) truncation cassette and Andras Nagy for the PGK-neo cassette. This work utilized core facilities supported in part by Roswell Park Cancer Institute's NCI-funded Cancer Center Support Grant CA16056. This work was supported by a grant from the Roswell Park Alliance Foundation and NCI/NIH Grant 2RO1 CA089426 to MJH. The authors declare that they have no competing financial interests or other conflicts of interest.
References
- Andrews SC, Wood MD, Tunster SJ, Barton SC, Surani MA, John RM (2007) Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty L, Weksberg R, Sadowski PD (2006) Detailed analysis of the methylation patterns of the KvDMR1 imprinting control region of human chromosome 11. Genomics 87: 46–56 [DOI] [PubMed] [Google Scholar]
- Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis TL (2004) Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics 84: 961–970 [DOI] [PubMed] [Google Scholar]
- Chung WY, Yuan L, Feng L, Hensle T, Tycko B (1996) Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms' tumors. Hum Mol Genet 5: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Cooper PR, Smilinich NJ, Day CD, Nowak NJ, Reid LH, Pearsall RS, Reece M, Prawitt D, Landers J, Housman DE, Winterpacht A, Zabel BU, Pelletier J, Weissman BE, Shows TB, Higgins MJ (1998) Divergently transcribed overlapping genes expressed in liver and kidney and located in the 11p15.5 imprinted domain. Genomics 49: 38–51 [DOI] [PubMed] [Google Scholar]
- Diaz-Meyer N, Day CD, Khatod K, Maher ER, Cooper W, Reik W, Junien C, Graham G, Algar E, Der Kaloustian VM, Higgins MJ (2003) Silencing of CDKN1C (p57(KIP2)) is associated with hypomethylation at KvDMR1 in Beckwith–Wiedemann syndrome. J Med Genet 40: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D (2005) Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 102: 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J (2000) Sequence and functional comparison in the Beckwith–Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum Mol Genet 9: 2691–2706 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Pugacheva EM, Shin J-Y, Abdullayev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ (2007) Allele-specific binding of CTCF binds to the multipartite imprinting control region KvDMR1. Mol Cell Biol 27: 2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431 [DOI] [PubMed] [Google Scholar]
- Gould TD, Pfeifer K (1998) Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet 7: 483–487 [DOI] [PubMed] [Google Scholar]
- Hatada I, Inazawa J, Abe T, Nakayama M, Kaneko Y, Jinno Y, Niikawa N, Ohashi H, Fukushima Y, Iida K, Yutani C, Takahashi S, Chiba Y, Ohishi S, Mukai T (1996) Genomic imprinting of human p57KIP2 and its reduced expression in Wilms' tumors. Hum Mol Genet 5: 783–788 [DOI] [PubMed] [Google Scholar]
- Hatada I, Mukai T (1995) Genomic imprinting of p57-KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat Genet 11: 204–206 [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR (1997) Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 7: 148–151 [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W (2004) Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Lewis A, Reik W (2006) How imprinting centres work. Cytogenet Genome Res 113: 81–89 [DOI] [PubMed] [Google Scholar]
- Lin SP, Coan P, da Rocha ST, Seitz H, Cavaille J, Teng PW, Takada S, Ferguson-Smith AC (2007) Differential regulation of imprinting in the murine embryo and placenta by the Dlk1–Dio3 imprinting control region. Development 134: 417–426 [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP (2000) The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 25: 19–21 [DOI] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T (2003) Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet 33: 502–507 [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MJ (2005) The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum Mol Genet 14 (Spec No 1): R113–R120 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Barlow DP (2006) Imprinting mechanisms—it only takes two. Genes Dev 20: 1203–1206 [DOI] [PubMed] [Google Scholar]
- Paulsen M, El-Maarri O, Engemann S, Strodicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J (2000) Sequence conservation and variability of imprinting in the Beckwith–Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet 9: 1829–1841 [DOI] [PubMed] [Google Scholar]
- Qian N, Frank D, O'Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B (1997) The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum Mol Genet 6: 2021–2029 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J (2001) Genomic imprinting: parental influence on the genome. Nature 2: 21–32 [DOI] [PubMed] [Google Scholar]
- Rosa AL, Wu YQ, Kwabi-Addo B, Coveler KJ, Reid Sutton V, Shaffer LG (2005) Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res 13: 809–818 [DOI] [PubMed] [Google Scholar]
- Salas M, John R, Saxena A, Barton S, Frank D, Fitzpatrick G, Higgins MJ, Tycko B (2004) Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev 121: 1199–1210 [DOI] [PubMed] [Google Scholar]
- Schonherr N, Meyer E, Roos A, Schmidt A, Wollmann HA, Eggermann T (2007) The centromeric 11p15 imprinting centre is also involved in Silver–Russell syndrome. J Med Genet 44: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SI, Gao C, Singer-Sam J (1998) Use of a reverse transcriptase–polymerase chain reaction assay to analyze allele-specific expression in individual hippocampal neurons. Mol Genet Metab 63: 96–102 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith–Wiedemann syndrome. Proc Natl Acad Sci USA 96: 8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn L, Barlow DP (2003) An ICE pattern crystallizes. Nat Genet 35: 11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Gondor A, Grange T, Ohlsson R, Kanduri C (2004) An antisense RNA regulates the bidirectional silencing property of the kcnq1 imprinting control region. Mol Cell Biol 24: 7855–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and igf2. Genes Dev 12: 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B, Efstratiadis A (2002) Genomic imprinting: piece of cake. Nature 417: 913–914 [DOI] [PubMed] [Google Scholar]
- Tycko B, Morison IM (2002) Physiological functions of imprinted genes. J Cell Physiol 192: 245–258 [DOI] [PubMed] [Google Scholar]
- Uejima H, Lee MP, Cui H, Feinberg AP (2000) Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet 25: 375–376 [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R (2004) Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Verona RI, Mann MR, Bartolomei MS (2003) Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol 19: 237–259 [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka O, Schweifer N, Schellanders K, Wagner E, Barlow D (1997) Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389: 745–749 [DOI] [PubMed] [Google Scholar]
- Xiong Z, Laird PW (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25: 2532–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Frisen J, Lee M-H, Massague J, Barbacid M (1997) Ablation of the CDK inhibitor p57KIP2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev 11: 973–983 [DOI] [PubMed] [Google Scholar]
- Yang PK, Kuroda MI (2007) Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell 128: 777–786 [DOI] [PubMed] [Google Scholar]
- Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, Piel JC, Loukinov DI, Lobanenkov VV, Soloway PD (2005) Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol Cell Biol 25: 11184–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Liegeois N, Wong C, Finegold M, Hou H, Thompson J, Silverman A, Harper J, DePinho R, Elledge S (1997) Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role for in Beckwith–Wiedemann syndrome. Nature 387: 151–158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure Legends
Supplementary Table S1