Abstract
Guanine-rich nucleic acid sequences can adopt noncanonical four-stranded secondary structures called guanine (G)-quadruplexes1. Bioinformatics analysis suggests that G-quadruplex motifs are prevalent in genomes2, which raises the need to elucidate their function. There is now evidence for the existence of DNA G-quadruplexes at telomeres with associated biological function3. A recent hypothesis supports the notion that gene promoter elements contain DNA G-quadruplex motifs that control gene expression at the transcriptional level4. We discovered a highly conserved, thermodynamically stable RNA G-quadruplex in the 5′ untranslated region (UTR) of the gene transcript of the human NRAS proto-oncogene. Using a cell-free translation system coupled to a reporter gene assay, we have demonstrated that this NRAS RNA G-quadruplex modulates translation. This is the first example of translational repression by an RNA G-quadruplex. Bioinformatics analysis has revealed 2,922 other 5′ UTR RNA G-quadruplex elements in the human genome. We propose that RNA G-quadruplexes in the 5′ UTR modulate gene expression at the translational level.
The existence of RNA G-quadruplexes in vivo is more inevitable than the existence of DNA G-quadruplexes, given that (i) the former are generally more thermodynamically stable in the folded form than their DNA counterparts5, and (ii) RNA is single-stranded, which implies that quadruplex formation does not have to compete with hybridization to a complementary strand. In this study we have focused on the 5′ UTRs of mRNA, which are known to be involved in translational regulation, particularly for growth factors, transcription factors and oncoproteins6. The neuroblastoma RAS viral oncogene homolog (NRAS)-encoded protein p21 mediates both signal transduction across the plasma membrane and the intracellular signaling pathways responsible for cell proliferation and differentiation7. Activating mutations in the coding region of NRAS are responsible for increased cell proliferation8. The suppression of oncogenic NRAS by small interfering RNA causes apoptosis of tumor cells9, which suggests that inhibiting the expression of oncogenic NRAS is a potential therapeutic strategy. Using a computational search algorithm we developed for locating quadruplex sequence motifs2, we identified a putative G-quadruplex forming sequence in the 5′ UTR of the human NRAS proto-oncogene mRNA. This 254-nucleotide-long NRAS 5′ UTR10 contains the NRAS RNA G-quadruplex (NRQ) motif (5′-GGGAGGGGCGGGUCUGGG-3′), which is located 14 nucleotides downstream of the 5′ cap and 222 nucleotides upstream of the translation start site (Supplementary Fig. 1 online). This motif is highly conserved, in both its sequence and its position relative to the translation start site, across the 5′ UTRs of human, chimpanzee, macaque, mouse, rat and dog genes orthologous to NRAS (Table 1 and Supplementary Table 1 online).
Table 1.
Conservation of the NRAS 5′ UTR G-quadruplex
| Species | Sequencea | Positionb |
|---|---|---|
| Human | GGGAGGGGCGGG--U---CUGGG | -222 |
| Chimpanzee | GGGAGGGGCGGG--U---CUGGG | -222 |
| Macaque | GGGAGGGGCGGG--U---CUGGG | -222 |
| Mousec | GGGGGCGGGGCGGGGCUGGACUGGG | -220 |
| Rat | GGGUGGGGAGGGGCGGGG-UG----GGG | -221 |
| Dogc | GGGAGGGGCGGG-----AAUGGG | -730d |
| Consensus | GGGAGGGGCGGG--U---CUGGG |
Dashes represent gaps in the alignment. Nucleotides in bold are runs of guanines capable of forming G-quartets.
The position of the last G of the putative G-quadruplex sequence is given relative to the translation start site.
Mouse and dog each have a second G-quadruplex sequence motif in their 5′ UTRs (Supplementary Table 1).
The difference in position for dog is likely to be due to the incomplete annotation of introns, as all other NRAS ortholog sequences have an intron in the unspliced 5′ UTR (480 nucleotides in humans), whose removal reduces the distance to approximately 220 nucleotides in the spliced form.
To confirm that the putative RNA G-quadruplex NRQ folds into a stable quadruplex, we carried out biophysical experiments on the synthetic oligonucleotide 5′-UGUGGGAGGGGCGGGUCUGGG-3′. Circular dichroism (CD) spectroscopy has been widely used to characterize the structure of folded nucleic acid quadruplexes11. At pH 7.4, 100 mM KCl, the CD spectrum of NRQ showed a positive peak at 263 nm and a negative peak at 241 nm (Fig. 1a), which is the characteristic CD signature of a parallel quadruplex structure11,12. The thermal melting of quadruplex nucleic acids can be characterized by an inverse UV transition at 295 nm13, and has generally been found to have a significant and characteristic cation dependence14. At pH 7.4 and low K+ (1 mM), the UV-melting profile of NRQ at 295 nm showed a hypochromic shift (Fig. 1b), with a Tm value of 63 ± 1 °C (error here and below is s.e.m.); this result was corroborated by CD melting at 260 nm under comparable conditions, which also gave a Tm of 63 ± 1 °C (Supplementary Fig. 2 online). At 100 mM KCl, the folded NRQ G-quadruplex could not be unfolded, even at 95 °C, which is indicative of a very stable quadruplex. The NRQ RNA G-quadruplex was reasonably stable (Tm = 43 ± 1 °C) even in the absence of added stabilizing cations. Studies over a ten-fold strand concentration range (from 1 to 10 μM) showed no change in the Tm value (Supplementary Table 2 online), which is consistent with the melting of an intramolecular quadruplex. The monovalent ion dependence for stabilization of folded NRQ, as judged by Tm, was in the order K+ > Na+ > Li+ (Supplementary Table 3 online), which is characteristic of G-quadruplex nucleic acids14. Thus, NRQ folds into a very stable, parallel intramolecular G-quadruplex under near-physiological pH and salt conditions.
Figure 1.
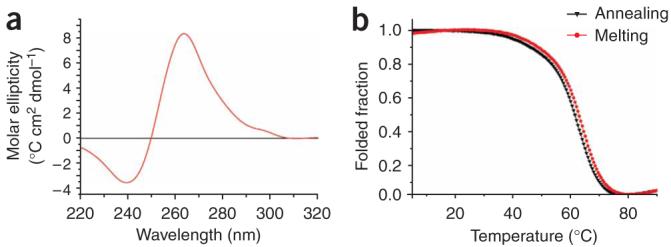
Biophysical analysis of the NRQ RNA G-quadruplex. (a) CD spectrum of NRQ at 4 μM strand concentration in 10 mM Tris-HCl, pH 7.4, and 100 mM KCl, at 20 °C. (b) Melting and annealing UV profiles of NRQ at 4 μM strand concentration in 10 mM Tris-HCl, pH 7.4, and 1 mM KCl.
To specifically evaluate the influence of the NRQ RNA G-quadruplex on the efficiency of translation, we cloned the 254-base-pair 5′ UTR of NRAS from human genomic DNA upstream of the firefly luciferase reporter gene and immediately downstream of the minimal T7 promoter. Whereas this plasmid (pSKC11) encodes the transcript UTRQ (Fig. 2a), which contains the native 5′ UTR of NRAS including the intact NRQ G-quadruplex motif, we also generated two controls. We obtained the plasmid pSKC12, which encodes the transcript DelQ (Fig. 2a), by deleting the first 29 base pairs of the NRAS 5′ UTR (that comprises the RNA G-quadruplex-forming sequence). The second control plasmid, pSKC13, which encodes the transcript MutQ (Fig. 2a), was derived from pSKC11 by a GGG-to-AAA substitution that was made at base pairs 15-17 of the NRAS 5′ UTR to disrupt RNA G-quadruplex formation but maintain the natural length of the 5′ UTR. For each system the corresponding 5′-capped RNA transcript was generated by in vitro transcription using T7 RNA polymerase, and the transcripts were subjected to in vitro translation using the rabbit reticulocyte lysate. We evaluated the efficiency of translation by the standard luminescence assay for luciferase catalytic activity15. Figure 2b describes the relative translation efficiency for each system. Deletion of the G-quadruplex motif (DelQ) resulted in a 3.7-fold increase in translation efficiency relative to the native sequence system, UTRQ. This indicates that the RNA G-quadruplex NRQ suppresses translation. The GGG-to-AAA quadruplex-destabilizing triple mutation (MutQ) caused a 3.6-fold increase in translation efficiency relative to UTRQ. Taken together, these data indicate that the natural 5′ UTR NRQ RNA G-quadruplex has an inhibitory effect on translation. Translational control of proto-oncogenes mediated via the 5′ UTR is generally complex6, and we postulate that this RNA G-quadruplex is one regulatory element within the 5′ UTR of NRAS. Factors that disrupt NRQ RNA G-quadruplex formation, such as mutations, may lead to oncogenesis.
Figure 2.
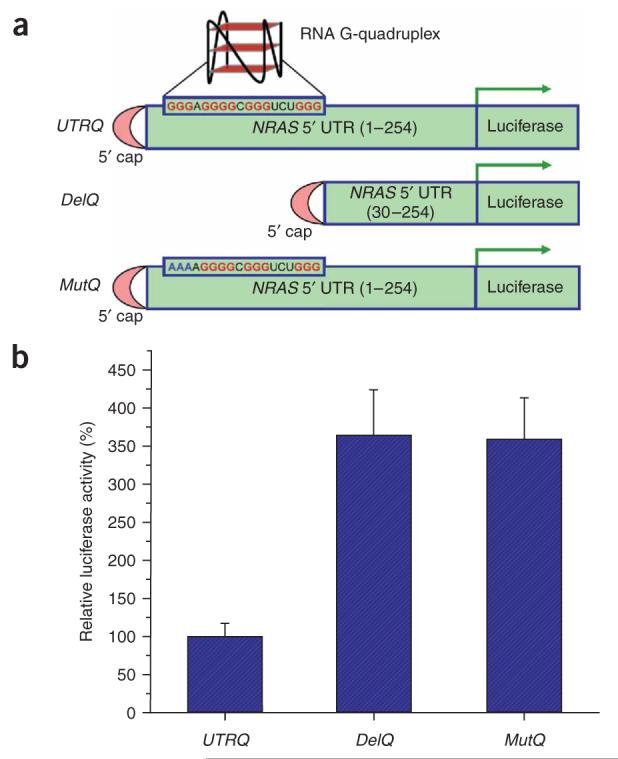
Effect of the NRAS 5′ UTR on the translational efficiency of chimeric RNA. (a) Schematic representation of mRNAs of luciferase reporter constructs: UTRQ, full-length (254 nucleotides) NRAS 5′ UTR; DelQ, in which the first 29 nucleotides of UTRQ, containing the G-quadruplex sequence, are deleted; and MutQ, full-length NRAS 5′ UTR with a GGG-to-AAA mutation (shown in blue) at nucleotides 15-17 to disrupt the RNA G-quadruplex formation. (b) Relative translation efficiency of the three constructs, as judged by quantitation of luciferase enzyme activity. Results were normalized relative to data for the UTRQ system. Error bars represent the s.d. of three independent experiments.
On the basis of this study, we reasoned that analogous RNA G-quadruplexes may exist within the 5′ UTRs of other genes. To investigate this, we carried out a bioinformatics search for RNA G-quadruplexes in 5′ UTRs in the human genome. We examined 38,915 5′ UTRs from all known Homo sapiens genes in Ensembl (version 40, National Center for Biotechnology Information (NCBI) build 36). 2,922 of the 5′ UTRs contained one or more G-quadruplex motifs, giving a total of 3,321 5′ UTR G-quadruplex motifs. The density of G-quadruplex motifs in 5′ UTRs as a whole is 0.305 per kilobase, compared with a genome average of 0.063 per kilobase (for one strand only)2, which is a 4.8-fold enrichment of G-quadruplex motifs. It is noteworthy that the genes containing 5′ UTR G-quadruplex motifs include several other proto-oncogenes, including BCL2, JUN and FGR (Table 2).
Table 2.
Human proto-oncogenes containing 5′UTR G-quadruplex motifs
| Gene description | HGNC code | 5′ UTR length | Motif positiona | Motif sequence | Ensembl gene ID |
|---|---|---|---|---|---|
| GTPase NRAS | NRAS | 254 | -222 | GGGAGGGGCGGGUCUGGG | ENSG00000168638b |
| Proto-oncogene tyrosine-protein kinase | FGR | 186 | -156 | GGGUGAGAGGGGCAGGUGGGGCUAGGG | ENSG00000000938 |
| Thyroid hormone receptor-α | THRA | 483 | -85 | GGGCCUGGGUGGCAGGGGGUGGG | ENSG00000126351 |
| Transcription factor Maf | MAF | 812 | -170 |
GGGGGGAGGGAGGGCGGGCGCGGG GGGCGCGGGCAGGGCGGGGGGG |
ENSG00000178573 |
| Friend leukemia integration 1 transcription factor | FLI1 | 165 | -95 | GGGAGGGCCCAGGGCGCCAGGG | ENSG00000151702 |
| Proto-oncogene C-crk | CRK | 141 | -44 | GGGCGGCGGGCGCCGGGGGCCGGAGGGG | ENSG00000167193c |
| Apoptosis regulator | BCL2 | 493 | -42 | GGGGGCCGUGGGGUGGGAGCUGGGG | ENSG00000171791 |
| Mitogen-activated protein kinase kinase kinase 8 | MAP3K8 | 696 | -647 |
GGGCAAAUGAGGGGCGGCGGGGUGGCGGG UGGGGGGG |
ENSG00000107968d |
| Proto-oncogene tyrosine-protein kinase | ABL1 | 381 | -270 | GGGAACGCCAGGGGCCCCUGGGUGCGGACGGG | ENSG00000097007 |
| Proto-oncogene tyrosine-protein kinase | ABL1 | 381 | -212 | GGGCGGGGGCGGGCCUGGCGGG | ENSG00000097007 |
| Proto-oncogene tyrosine-protein kinase | ABL1 | 381 | -126 | GGGCGGGCGCGGGCGCGCGGGG | ENSG00000097007 |
| Proto-oncogene tyrosine-protein kinase | ABL1 | 381 | -66 |
GGGGCCGGGGGCGCCGGGGGGGCGCGCGGGCC GAGCCGGG |
ENSG00000097007 |
| Transcription factor AP1 | JUN | 1,257 | -1,231 | GGGGAGGGGACCGGGGAAGAGAGGG | ENSG00000177606 |
The position of the last G of the G-quadruplex motif is given relative to the translation start site.
Because of an annotation error in version 40 of Ensembl, whereby the NRAS gene was accidentally fused with the neighboring CSDE1 gene, we have used data from the original publication10, which is identical to that in previous versions of Ensembl and the manually curated vertebrate genome annotation (Vega) database.
There is also an alternative transcript (ENST00000300574) that has eight fewer bases in the 5′ UTR, upstream of the G-quadruplex motif.
There is also an alternative transcript (ENST00000375328) that has seven fewer bases in the 5′ UTR, downstream of the G-quadruplex motif. HGNC, Hugo Gene Nomenclature Committee.
Stable RNA hairpin secondary structures in the 5′ UTR of mRNAs inhibit the process of translation16. The intramolecular RNA G-quadruplex motif (NRQ) that we have identified in the 5′ UTR of NRAS is the first example to be elucidated of a quadruplex that inhibits translation. Detailed elucidation of the quadruplex structure and also of the quadruplex-mediated inhibitory mechanism will be the subject of future work. The presence of G-quadruplex motifs in the 5′ UTRs of 2,922 human genes suggests that the functional role of the NRAS RNA G-quadruplex may be a paradigm for other genes. Substituted flat polyaromatic heterocyclic molecular scaffolds that complement the surface of a G-tetrad have provided many small quadruplex-binding ligands17. Given the emerging interest in small molecules that target and stabilize nucleic acid quadruplexes with specificity18, this RNA G-quadruplex has obvious potential as a molecular target for small-molecule therapeutic agents that act by inhibiting mRNA translation.
METHODS
Bioinformatics
Genomic data was taken from Ensembl using version 40, corresponding to NCBI build 36, and was acquired using the EnsMart tool. It was then analyzed using quadparser2 and custom-written perl utilities.
Spectroscopy
For spectroscopic studies, we prepared RNA samples at 4 μM strand concentration in RNase-free water and degassed buffers containing 10 mM Tris-HCl, pH 7.4, and varying KCl concentrations from 1 mM to 100 mM. The samples were annealed by heating at 90 °C for 10 min and then slow cooling to 5 °C at a controlled rate of 0.2 °C min-1.
CD experiments were performed using a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller. Typically, a 200 μl sample was placed in a quartz cuvette with an optical path length of 1 mm, transferred to the spectropolarimeter and allowed to equilibrate at 20 °C for 10 min. Five CD scans, over the wavelength range of 220 to 320 nm, were performed at 50 nm min-1 with a 2-s response time, 1-nm pitch and 1-nm bandwidth, and their average was taken. For each experiment, a CD spectrum of the buffer was recorded and subtracted from the spectrum obtained for the RNA-containing solution. Data were zero-corrected at 320 nm.
UV-melting studies were carried out on a Varian Cary 1E UV-visible spectrophotometer equipped with a Peltier temperature controller. Typically, a 100 μl sample was transferred to a quartz cuvette with an optical path length of 1 cm and was covered with approximately 100 μl of mineral oil to prevent sample evaporation. This sample was transferred to the spectrophotometer and then heated to 90 °C and cooled to 5 °C, twice consecutively at 0.25 °C min-1 temperature gradient, and absorption data recorded at 295 nm were collected every 0.5 min on both annealing and melting steps. Tm values were determined using the van’t Hoff method19.
Plasmid construction
To construct the plasmid pSKC11, which encodes the 254-nucleotide UTRQ transcript, we PCR-amplified the 237 base pairs of exon-I of the NRAS 5′ UTR from human genomic DNA using Pfu DNA polymerase (Promega) and two sequence-specific primers: a forward primer tailed with HindIII (underlined) and a minimal T7 promoter (5′-AAGCTTTAATACGACTCACTATAGGGACGTCCCGTGTGGGAGGGGCG-3′; bold GG indicates positions that were mutated from AA in the natural 5′ UTR sequence to yield an efficient transcription template), and a restriction site SmlI-tailed (naturally present at the end of exon -I) reverse primer (5′-CTCAAGCTCAAGCTCCACTGCCTCTGC-3′). This amplified product (A) was digested with HindIII and SmlI, and purified by gel electrophoresis. We amplified the firefly luciferase gene from the pGL3 basic vector (Promega) using a forward primer tailed with SmlI (underlined) and the 17 base pairs of the 5′ UTR from exon I (5′-CTTGAGGTTCTTGCTGGTGTGAAATGGAAGACGCCAAAAACATAAAG-3′), and an EcoRI-tailed reverse primer (5′-GAATTCTTACACGGCGATCTTTCC-3′). This amplified product (B) was digested with SmlI and EcoRI and purified by gel electrophoresis. In a three-piece ligation, we ligated the two gel-purified fragments (A and B) at the SmlI site and inserted into the HindIII- and EcoRI-digested linear pUC18 vector (Invitrogen) to obtain the full-length (254 base pairs) NRAS 5′ UTR containing the G-quadruplex-forming sequence at its natural position (14 base pairs downstream of the 5′ cap). We confirmed positive clones for the full length of insert by DNA sequencing.
To construct pSKC12, the control plasmid encoding the transcript DelQ, we deleted the first 29 base pairs of the 5′ UTR, including the G-quadruplex-forming sequence, from the pSKC11 plasmid using the forward primer (5′-GGGTGCGGCCTGCCGCATGACTCG-3′) and an EcoRI-tailed reverse primer (5′-GAATTCTTACACGGCGATCTTTCC-3′). We gel-purified this amplified product and reamplified with a forward primer tailed with HindIII and a minimal T7 promoter (5′-AAGCTTTAATACGACTCACTATAGGGTGCGGCCTGCCGCATGACTCG-3′) and the above-mentioned EcoRI-tailed reverse primer. This reamplified product was inserted into the HindIII- and EcoRI-digested linear pUC18 vector. The second control plasmid, pSKC13, which encodes the MutQ transcript, was derived from the pSKC11 plasmid by a GGG-to-AAA substitution at base pairs 15-17 of the NRAS 5′ UTR using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. We sequenced all the constructs to confirm the presence of the intended changes.
In vitro transcription
The plasmids pSKC11, pSKC12 and pSKC13, which encode the transcripts UTRQ, DelQ and MutQ, respectively, were linearized at the 3′ end of the region to be transcribed using EcoRI. 5′-capped transcripts were synthesized in vitro using the mMessage mMachine T7 kit (Ambion). We purified all transcripts on 1% agarose gel, after DNase treatment. The RNA concentration was determined by UV spectroscopy. We confirmed the integrity and size of each transcript on 1% agarose gel.
In vitro translation and luciferase assay
We carried out in vitro translation of the mRNAs in a cell-free translation system consisting of extracts from nuclease-treated rabbit reticulocytes lysate (Promega) according to the manufacturer’s protocol. We measured firefly luciferase activity using luciferase assay reagent (Promega) according to the manufacturer’s protocol on a Varian Cary Eclipse fluorescence spectrophotometer working in bio/chemiluminescence mode.
ACKNOWLEDGMENTS
We thank Cancer Research UK, the Cambridge Commonwealth Trust and Trinity College, Cambridge for funding. S.B. is a Biotechnology and Biological Sciences Research Council career development fellow. J.L.H. is a research fellow at Trinity College, Cambridge. We thank Z. Jawad-Alami for useful discussions and S. Sewitz for critically reading this manuscript.
Footnotes
Supplementary Material
Note: Supplementary information is available on the Nature Chemical Biology website.
References
- 1.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Biomolecular Sciences; 2006. [Google Scholar]
- 2.Huppert JL, Balasubramanian S. Prevelance of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saccà B, Lacroix L, Mergny J-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis AE. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell Biol. 1999;31:73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Targeting Ras signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 8.Barbacid M. Ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 9.Eskandarpour M, et al. Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int. J. Cancer. 2005;115:65–73. doi: 10.1002/ijc.20873. [DOI] [PubMed] [Google Scholar]
- 10.Hall A, Brown R. Human N-ras: cDNA cloning and gene structure. Nucleic Acids Res. 1985;13:5255–5268. doi: 10.1093/nar/13.14.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structure for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang C-F, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 14.Hardin CC, Watson T, Corregan M, Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG) Biochemistry. 1992;31:833–841. doi: 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- 15.DiLella AG, et al. Utility of firefly luciferase as a reporter gene for promoter activity in transgenic mice. Nucleic Acids Res. 1988;16:4159. doi: 10.1093/nar/16.9.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 17.Searle MS, Balkwill GD. In: Quadruplex Nucleic Acids. Neidle S, Balasubramanian S, editors. Cambridge: RSC Biomolecular Sciences; 2006. pp. 131–153. [Google Scholar]
- 18.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 19.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]


