Electron transfer (ET) reactions in bioenergetics move electrons 10 Å and more between redox cofactors through insulating protein (1). Nowhere else does the wave nature of matter control a biological event in a more striking way: These shifting electrons tunnel quantum mechanically from donor to acceptor. Nature uses the physics of electron tunneling for good reason: Delocalized charge would dissipate energy and induce collateral redox damage. As described in a recent issue of PNAS (2), new studies of electron tunneling between hemes in the quinol-oxidizing cytochrome bo3, a heme–copper oxidase in the same family as cytochrome c oxidase, find that a specific tunneling conduit, or tunneling pathway, mediates the electron flow. This experiment, which shows that a dominant coupling pathway controls a physiological ET reaction, reveals the limitations of coarse-grained (average medium) models for protein electron tunneling and indicates why atomic-resolution descriptions can be essential. The perspective emerging from this study will help to determine how evolution acts on the molecular structure of electron tunneling pathways in the biological ET chains.
The Electron and the Protein
A jewel of 20th-century theoretical chemistry, ET theory (3–7), predicts that long-range ET rates are controlled by two factors: the propensity for thermally induced geometry fluctuations to bring the donor and acceptor into energetic degeneracy and the probability of electron tunneling between these degenerate states. The thermal fluctuations are well described by Marcus theory (3): these fluctuations depend on the coupling of the environment to the shifting electron charge. The electron tunneling probability is determined by the chemical structure of the tunneling barrier (3–7). Extensive theoretical and experimental work over the last 20 years has revealed fascinating symmetry, energy, and distance dependences of these tunneling propensities (7–10).
The invention of selective metallolabeling methods (8) paved the way for experimental (9) and theoretical (10) determinations of the protein medium dependence of distant donor–acceptor electronic couplings. Importantly, a structure-dependent pathway model was developed to describe how the protein structure affects the ET kinetics (10). The pathway model predicts rates proportional to the coupling strength along the dominant through-bond and through-space coupling routes for electrons from donor to acceptor. The key parameter in the pathway model is the relative strength of through-bond and through-space electronic propagation. The through-space exponential decay factor (≈2 Å−1) follows from the ≈5-eV binding energy for electrons in biological cofactors, and the softer through-bond decay exponent (≈1 Å−1) derives from interactions among covalent bonds (10, 11). The through-bond decay is associated with effective tunneling barriers of ≈1 eV, approximately the energy gap between the frontier orbitals of the protein and the redox-active orbitals of the cofactors. The pathway model accounts for anomalously slow ET rates in specific Ru-modified derivatives of cytochrome c, cytochrome b562, myoglobin, and high-potential iron protein (9, 12). Recent analysis of these rates finds that ET at long distances through heme edges favors average exponential distance dependence (here, simple exponential distance models and pathway models produce similar rate predictions), whereas coupling via axial heme ligands favors pathway-specific effects (12).
The question of how natural selection manipulates electronic couplings has remained open for some time.
Hopfield's (6) early semiclassical analysis of protein electron transfer predicted an average ET rate distance decay exponent of 1.4 Å−1. The approach of Dutton and coworkers (13) to biological ET initially adopted Hopfield's single decay exponent, based on kinetics in the photosynthetic reaction center. Dutton and coworkers (14) later modified the distance decay constant—interpolating between through-bond and through-space parameters—to capture the average physical features of the pathway model without using a fully atomistic view of the tunneling routes present in the pathway model (11) [the Dutton formulation is problematic in the endergonic regime (15)].
Atomistic Pathways and Average Packing Densities
The packing density and pathway models make similar predictions for many proteins (10). However, the packing density model makes many untested assumptions about how best to quantify a protein's packing density. Indeed, Jasaitis et al. (2) show that alternative plausible strategies can shift the computed packing density by 50% for cytochrome bo3 heme o3–heme b ET. Pathway and packing density methods will make qualitatively different ET rate predictions if the structure of the strongest pathway is not typical of the medium structure in which it is embedded (11). The density model only incorporates average characteristics of the protein medium between cofactors and it assigns a corresponding penalty factor for tunneling along the shortest line between the edges of the cofactors; the dominant tunneling pathway zigzags along the set of bonded and nonbonded interactions among specific atoms that establishes the strongest electronic route from donor to acceptor.
Coupling Routes and Tunneling Barriers in Cytochrome c Oxidase
The physics of tunneling in folded proteins is becoming clear, but the question of how natural selection manipulates electronic couplings has remained open for some time. The distance between cofactors certainly has an exponential effect on the ET rates, and this distance is a key control point for ET reactions. Yet, a critical question is whether specific protein structures are selected to accelerate or slow the rates at a given distance. Indeed, pathway theory suggests that such effects on the rates could be as large as 10- to 100-fold (10).
Cytochrome c oxidase, another heme–copper oxidase, is a significant target for the exploration of coupling pathway selection. Gray and coworkers (16, 17) identified a direct CuA-to-heme a coupling pathway that would support effective ET, as well as an axial-to-axial heme a-to-heme a3 pathway. Stuchebrukhov and coworkers (18) examined the axial coupling route and also identified a direct heme-to-heme route (via methyl groups on the D rings). Balabin, Onuchic, and coworker (19) showed that thermal fluctuations of the direct heme–heme route likely cause it to dominate the heme a-to-heme a3 interaction.
Cytochrome bo3
Jasaitis et al. (2) report ET kinetics between CO-dissociated ferrous heme o3 and low-spin ferric heme b on the nanosecond time scale. Indeed, the structure and heme-to-heme ET kinetics is very similar in cytochrome bo3 and in cytochrome c oxidase. The heme iron separations differ in the two proteins by less than 0.5 Å, and the nearby methyl groups distances differ by 0.8 Å, based on the x-ray structures [differences well within the range of thermal fluctuations (19)]; the coupling pathway structure is nearly the same in the two proteins, involving the bonded His-Phe-His path linking irons and the direct methyl-to-methyl through-space contact. Taking a minimum donor–acceptor distance of 7.8 Å, a (generic) packing density value (2, 14), and a typical ET reorganization energy (0.7 eV) places rates computed with packing analysis in agreement with experimental rates (20). However, recent studies have found a reorganization energy for cytochrome bo3 of <0.2 eV, so the kinetics cannot be accommodated with the packing density reported by Dutton and coworkers (20).
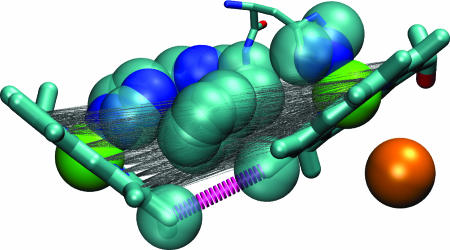
To rationalize the cytochrome bo3 ET kinetics by using average barrier height models, the packing density must be decreased by more than a factor of 2. This would make the tunneling barrier much more “vacuum-like.” Fig. 1 shows why the Dutton packing density prescription overestimates the tunneling medium density: It includes the axial–axial coupling residues, which are not relevant because direct heme–heme (methyl-to-methyl) interactions dominate (19). Moreover, small changes in the model parameters or medium sampling rules will shift atoms in or out of the packing density analysis. Jasaitis et al. (2) point to the “large local variation of the packing density” in the space between the hemes, meaning that the computed density is model dependent. Dominant single pathways are particularly likely to arise at short ET distances, so it is not surprising that difficulties with “coarse-grained” models occur for the cytochrome bo3 and cytochrome c oxidase heme-to-heme reactions.
Fig. 1.
The dominant ET coupling pathway (10, 19) is shown with a thick dashed line, and those atoms identified in packing density analysis are shown as spheres (14) for the two hemes in cytochrome bo3 [the position of the nearby Cu (orange) appears for reference]. The dominant coupling route involves a through-space jump between two porphyrin methyl groups. Thin gray lines connect all pairs of atoms of the porphyrin rings, and atoms with van der Waals surfaces intersecting these lines are drawn as spheres. The packing density model includes almost all atoms of the protein bridge, even those atoms that do not mediate ET to a substantial degree. The number of atoms captured in the packing density estimate is sensitive to small changes in the protein conformation, the parameterization of the van der Waals radii, and the rules for defining the tunneling volume.
When and Why Is a High-Resolution Theory of Tunneling Essential?
Dutton and coworkers (20) have argued that “[a]ny variance in the packing density of the tunnelling medium appears unrelated to function… .” This perspective seems to be at odds with what the heme–copper oxidases are teaching us. Evidence both from cytochrome bo3 and from the family of Ru-modified proteins points to three accessible regimes: (i) a single pathway limit, as in cytochrome bo3, where an average medium view is of little use (2); (ii) a regime of pathway-limited coupling through an electronic bottleneck, as was recently described for Ru-modified heme proteins with axial-coupled tunneling pathways (12); and (iii) a multipathway regime, common at longer distances, involving edge-coupled macrocyclic redox cofactors, where implementing either an average medium or pathway analysis produces similar results (12, 13).
Simple models hold great sway over the design and interpretation of biophysical experiments. The complexity of biology requires coarse graining in the models, but the resolution or graininess needs to match the scale of the phenomena under investigation. In the case of cytochrome bo3 and cytochrome c oxidase ET, average medium models seem to fail because the methyl groups that dominate the electron tunneling are much smaller than the dimensions over which the packing density model performs its averaging. In this regime, tunneling pathway analysis—or more detailed electronic structure theory—is essential to characterize the tunneling process. If the rate of heme-to-heme electron transfer in the heme copper oxidases is of consequence to evolution, evolution has confronted the atomic-scale features of the tunneling barrier structure in evolving the electron-tunneling kinetics. At least in electron-transfer proteins, “Darwin at the molecular scale” (20) cannot escape the rich and grainy atomic structure of matter.
ACKNOWLEDGMENTS.
Our research is supported by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20811 in issue 52 of volume 104.
References
- 1.Bertini I, Gray HB, Stiefel EI, Valentine JS. Biological Inorganic Chemistry. Sausalito, CA: University Science Books; 2007. [Google Scholar]
- 2.Jasaitis A, Johansson MP, Wikström M, Vos MH, Verkhovsky MI. Proc Natl Acad Sci USA. 2007;104:20811–20814. doi: 10.1073/pnas.0709876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus RA, Sutin N. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 4.Hush NS. Electrochim Acta. 1968;13:1005–1023. [Google Scholar]
- 5.Levich VG, Dogonadze RR. Dokl Akad Nauk SSSR. 1959;124:123–126. [Google Scholar]
- 6.Hopfield JJ. Proc Natl Acad Sci USA. 1974;71:3640–3644. doi: 10.1073/pnas.71.9.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bixon M, Jortner J, editors. Adv Chem Phys. 1999:106–107. [Google Scholar]
- 8.Winkler JR, Nocera DG, Yocom KM, Bordignon E, Gray HB. J Am Chem Soc. 1982;104:5798–5800. [Google Scholar]
- 9.Gray HB, Winkler JR. Q Rev Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 10.Beratan DN, Betts JN, Onuchic JN. Science. 1991;252:1285–1288. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 11.Jones ML, Kurnikov IV, Beratan DN. J Phys Chem A. 2002;106:2002–2006. [Google Scholar]
- 12.Prytkova TR, Kurnikov IV, Beratan DN. Science. 2007;315:622–625. doi: 10.1126/science.1134862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 14.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 15.Crofts AR, Rose S. Biochim Biophys Acta. 2007;1767:1228–1232. doi: 10.1016/j.bbabio.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez BE, Malmström BG, Winkler JR, Gray HB. Proc Natl Acad Sci USA. 1995;92:11949–11951. doi: 10.1073/pnas.92.26.11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regan JJ, Ramirez BE, Winkler JR, Gray HB, Malmström BG. J Bioenerg Biomembr. 1998;30:35–39. doi: 10.1023/a:1020551326307. [DOI] [PubMed] [Google Scholar]
- 18.Medvedev DM, Daizadeh I, Stuchebrukhov AA. J Am Chem Soc. 2000;122:6571–6582. [Google Scholar]
- 19.Tan ML, Balabin I, Onuchic JN. Biophys J. 2004;86:1813–1819. doi: 10.1016/S0006-3495(04)74248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser CC, Page CC, Dutton PL. Philos Trans R Soc London B. 2006;361:1295–1305. doi: 10.1098/rstb.2006.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]