The repair of chromosome breaks is essential to maintain genome integrity. Both in mammalian cells and in budding yeast, repair in the G1 phase of the cell cycle is particularly dependent on nonhomologous end-joining (NHEJ) to repair broken chromosomes. NHEJ is important in repairing double-strand breaks (DSBs) that arise during both V(D)J recombination and class switch recombination (CSR) events in the immune system. End-joining also is responsible for generating chromosomal translocations that are associated with various cancers. Work by Guirouilh-Barbat et al. (1) published in a recent issue of PNAS and by several other laboratories (2–4) has revealed that–in addition to the “classical” NHEJ pathway–there is a robust alternative pathway that also contributes to the generation of translocations.
Our current understanding of the mechanisms of NHEJ results from a dynamic interplay between genetics and biochemistry both in mammals and in budding yeast (for reviews, see refs. 5–7). A major motivation to identify the genes responsible for NHEJ has been the study of end-joinings in both V(D)J recombination and Ig CSR in mammals, driven by the identification of immune-compromised humans. A second impetus has been the identification of genes conferring x-ray resistance. These studies led to the identification of DNA-protein kinase catalytic subunit (PKcs) and the Ku70 and Ku80 proteins, which play a central role both in immune cell recombination and in resistance to ionizing radiation. Further work led to the identification of a number of XRCC genes conferring radio-resistance. Subsequent studies in budding yeast confirmed that Ku proteins were also essential for NHEJ (there is no DNA-PKcs). Research using budding yeast identified other NHEJ components, notably DNA ligase 4, its partner Lif1 (whose mammalian homolog is XRCC4), and Nej1 (whose mammalian homolog is XLF/Cernunnos)†. Ku-deficient mice are viable but severely immunocompromised, whereas mice lacking DNA ligase 4 (LIG4) and XRCC4 die embryonically; however, they can be rescued by also knocking out the p53-mediated apoptotic pathway that eliminates cells with unrepaired DSBs (8–11). However, these immune-deficient mice almost always die of lymphomas characterized by gene fusions between the Ig heavy-chain IgH locus and c-Myc; they also show evidence of many other nonreciprocal translocations. Surprisingly, the translocation junctions still appear to be the result of nonhomologous end fusions rather than of homologous recombination between repeated sequences. Thus, there had to be an alternative NHEJ process not requiring LIG4 or XRCC4 and probably also not requiring the Ku proteins. A key feature of this pathway is that joinings occur preferentially between sequences near the ends that shared as much as 7 nt of homology (12). Similarly, end-joining of blunt-ended linearized plasmids in cells without KU80 involve the resection and annealing of short homologous sequences rather than simply joining the blunt ends (13). An analogous microhomology-mediated end-joining (MMEJ) pathway was seen in budding yeast in the case when the 4-bp protruding chromosome ends, created by a site-specific nuclease, were completely nonhomologous (14). Whereas NHEJ between ends that had even partial complementarity were all Ku-dependent, end-joinings between noncomplementary ends were efficient but Ku-independent. MMEJ must involve significant 5′–3′ resection of the ends to expose regions with 2- to 10-bp microhomologies. Yeast MMEJ also needs the Rad1-Rad10 endonuclease apparently to trim the 3′ ends extending beyond the annealed microhomologies.
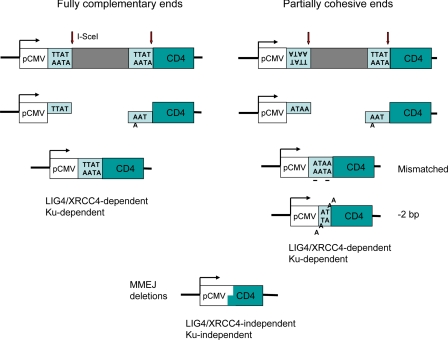
A systematic study of alternative pathways of NHEJ in mammalian cells was carried out by Lopez and coworkers (15) by using I-SceI site-specific endonuclease to create multiple chromosomal DSBs whose joinings could be scored by flow cytometry. They used I-SceI cleavage sites arranged so that the protruding 4′-bp 3′ ends were either fully complementary or (when one site was inverted) partially complementary. With fully complementary ends, most end-joinings were perfect ligations, restoring the cleavage site. Partially complementary ends also annealed despite mismatches within the junction, sometimes yielding small deletions or insertions caused by misalignments (Fig. 1). Deleting KU80 reduced NHEJ with complementary ends but surprisingly had little effect on the partially cohesive ends; however, many of the joints showed misaligned ends or larger deletions, with junctions containing short regions of microhomology, as in yeast MMEJ.
Fig. 1.
Induction of NHEJ by I-SceI endonuclease cleavage. I-SceI cleaves within an 18-bp nonsymmetric site to generate 4-bp, 3′ overhanging ends. Joining of the CMV promoter to the coding sequence of the CD4 gene creates CD4+ cells that can be scored by flow cytometry. Inversion of one of the cleavage sites creates partially complementary ends. Fully complementary ends are generally joined by perfect ligation when the “classical” NHEJ apparatus (including Ku proteins, LIG4, and XRCC4) is functional. With partially compatible ends, NHEJ still permits ligation even though two sites (underlined) are mispaired. Among other joinings of the overhanging ends are 2-bp deletions requiring removal of 2 nt from each end before they can be ligated. In the absence of Ku proteins or XRCC4, most ends are joined by the alternative NHEJ pathway by using MMEJ.
In the aforementioned article in PNAS by Guirouilh-Barbat et al. (1) and in several recent papers (2–4), the characterization of an alternative, Ku-independent MMEJ mechanism in mammalian cells has been shown to be independent of LIG4 and XRCC4. In the article by Guirouilh-Barbat et al., joinings of I-SceI-induced DSBs were examined in XRCC4-deficient cells. Both with fully or partially cohesive ends, there was a >5-fold reduction in end-joinings, but the fact that a significant number of successful end-joinings remained again argued that there is a backup NHEJ system operating without XRCC4 (and presumably LIG4). Most of the junctions were deletions with microhomologies at the junction.
Yan et al. (3) examined the end-joinings associated with Ig IgH CSR in mouse B cells. The absence of LIG4 or XRCC4 reduces CSR substantially, but there were still many switches, most of which appear to have used microhomologies. Moreover, there were still many chromosomal translocations between the IgH locus and other chromosomes. Thus, there must be a “nonclassical” NHEJ mechanism that acts when LIG4/XRCC4 is absent. In a parallel study, de Villartay and coworkers (4) used a conditional XRCC4 knockout mutation to show that an alternative NHEJ pathway appeared to be used in CSR. Finally, Corneo et al. (2) made the surprising discovery that alternative NHEJ can be elicited in cells wild type for all general NHEJ factors. V(D)J recombination is initiated by RAG1/RAG2 proteins that not only create the DSBs but also hold and protect the ends. In Rag proteins deleted for certain regions, recombination is efficient, but the outcomes are skewed toward MMEJ events. Alternative NHEJ activity can be found even with wild-type Rag proteins. These studies make clear that MMEJ is an important process in preserving overall genome integrity.
Ku-deficient mice are viable but severely immunocompromised.
Little is known about the components of the alternative NHEJ (MMEJ) machinery or how the classical NHEJ machinery predominates when it is functional. DNA ligase 3, with its associated XRCC1 protein, is a candidate for the ligase that substitutes for LIG4 (16). It has also been suggested that PARP is important (17, 18). The filling-in of small gaps created during repair of noncompatible ends also requires DNA polymerases λ or μ (19, 20). Whether the Rad1–Rad10 homologs XPF-ERCC1 play a similar role in removing the extremities of the 3′ tails after microhomology mediated annealing, as in yeast, is not known; an intriguing clue comes from the fact that this complex is required in the fusions of telomere sequences that have partially cohesive ends (21). Yeast MMEJ also requires the Mre11–Rad50 complex (14); it will be interesting to see whether these proteins are also required in mammalian cells.
It has been known for a decade that Ku-defective mice are not as deficient in V(D)J recombination and are more viable than XRCC4- or LIG4-defective animals. This observation might lead one to the conclusion that there were XRCC4-dependent mechanisms that acted independent of Ku, but in fact it now looks as if this interpretation is incorrect. Instead it seems that Ku plays an XRCC4-independent role in regulating NHEJ, apparently in preventing the alternative MMEJ machinery from gaining access to the DSB ends when KU70/KU80 are associated with the ends. In budding yeast, deletion of Ku proteins allows more robust 5′–3′ resection of the ends, which is consistent with the idea that Ku acts to shelter ends from degradation. Resection of the ends is required for MMEJ. Indeed, the finding that Ku deficiency rescues the lethality of ligase 4-deficient cells (22) can be explained if the absence of Ku gives the alternative NHEJ pathway a chance to maintain genome stability.
Footnotes
The author declares no conflict of interest.
See companion article on page 20902 in issue 52 of volume 104.
The identification of yeast Nej1 and mammalian XLF/Cernunnos is a fascinating example of many possible strategies to identify the same gene. NEJ1 was found simultaneously by five groups using four different approaches, including gene-expression profiling, two-hybrid interactions, and screenings of libraries of yeast gene deletions. XLF/Cernunnos was identified both by complementation of a human immune-deficient mutation and by two-hybrid analysis.
References
- 1.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 3.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 4.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. J Exp Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daley JM, Palmbos PL, Wu D, Wilson TE. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 6.Burma S, Chen BP, Chen DJ. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR. J Biol Chem. 2007 in press. [Google Scholar]
- 8.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, et al. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 9.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 10.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 12.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Eur J Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Ma JL, Kim EM, Haber JE, Lee SE. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Audebert M, Calsou P. Biochem Biophys Res Commun. 2007 doi: 10.1016/j.bbrc.2007.11.132. in press. [DOI] [PubMed] [Google Scholar]
- 19.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Nucleic Acids Res. 2007;35:3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 22.Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR. DNA Repair (Amst) 2002;1:1017–1026. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]