Abstract
Certain plant varieties typically require prolonged exposure to the cold of winter to become competent to flower rapidly in the spring. This process is known as vernalization. In Arabidopsis thaliana, vernalization renders plants competent to flower by epigenetically silencing the strong floral repressor FLOWERING LOCUS C (FLC). As a result of vernalization, levels of lysine-9 and lysine-27 trimethylation on histone 3, modifications that are characteristic of facultative heterochromatin in plants, increase at FLC chromatin. We have identified a mutant, protein arginine methyltransferase 5 (atprmt5), that fails to flower rapidly after vernalization treatment. AtPRMT5 encodes a type II protein arginine methyltransferase (PRMT) that, in winter-annual strains, is required for epigenetic silencing of FLC and for the vernalization-mediated histone modifications characteristic of the vernalized state. Furthermore, the levels of arginine methylation of FLC chromatin increase after vernalization. Therefore, arginine methylation of FLC chromatin is part of the histone code that is required for mitotic stability of the vernalized state.
Plants have evolved a range of strategies to ensure that flowering occurs at the optimum time of the year for reproductive success. In some plants, a vernalization requirement is a key component of the reproductive strategy. Vernalization is the acquisition of the competence to flower resulting from exposure to the prolonged cold of the winter season (1). A vernalization requirement permits plants to become established during the fall season without the risk of flowering as winter sets in. During the cold of winter, these plants become vernalized, which enables them to flower during the favorable conditions of spring.
A study of natural variation in the vernalization requirement among Arabidopsis accessions led to some of the first examples of identifying genes that influence Arabidopsis life history traits. Napp-Zinn (2) identified FRIGIDA (FRI) as a locus that confers a vernalization requirement. FLOWERING LOCUS C (FLC) was later identified as cooperating with FRI to confer a vernalization requirement (3, 4). FRI encodes a plant-specific protein of unknown biochemical function (5), and FLC encodes a transcriptional regulator that is a repressor of the floral transition. FRI acts to maintain FLC transcription at levels sufficient to effectively repress flowering before vernalization (6, 7).
Vernalization can be considered an epigenetic phenomenon in the sense that the cold of winter induces a mitotically stable change in gene expression that persists well into the spring season after the inducing signal, cold, is no longer present. During winter, FLC chromatin undergoes a transition from an actively transcribed state to a heterochromatin-like state (8, 9). This change in chromatin structure is associated with a decrease in the level of histone modifications characteristic of active chromatin such as trimethylation of lysine-4 on histone 3 (H3K4) and H3 acetylation, and an increase in levels of the repressive modifications characteristic of inactive chromatin such as methylation of lysine-9 on histone 3 (H3K9) and lysine-27 on histone 3 (H3K27) (8–10). These repressive modifications are maintained in the spring and are likely to be part of the stable switch that “remembers” winter. Like many other epigenetic phenomena, the molecular nature of vernalization is consistent with the “histone code hypothesis,” which posits that the combination of covalent modifications to histone tails contributes to the creation of stable states of gene activity (11).
In Arabidopsis, a number of genes have been identified that are required for vernalization-induced epigenetic silencing of FLC. Of these, VIN3 is the only gene that is transcriptionally activated by prolonged exposure to cold (9). VIN3 can interact with the related protein VIN3-LIKE 1/VERNALIZATION 5 (12, 13) and can exist in an Arabidopsis PRC2 (Polycomb Repression Complex)-like complex, which includes VERNALIZATION 2, CURLY LEAF, SWINGER, and FERTILIZATION INDEPENDENT ENDOSPERM (14). In Drosophila and other animals, the PRC2 complex establishes methylation of H3K27, thus laying the foundation for the maintenance of stable transcriptional repression by the PRC1 complex (15).
Unlike the situation in animals, plants do not possess PRC1 components; instead, the repression of FLC is maintained by LHP1 (LIKE HETEROCHROMATIN PROTEIN 1) (10, 16). Furthermore, FLC repression is associated with H3K9 trimethylation as well as H3K27 trimethylation (13). In the lhp1 mutant, H3K9 methylation occurs at FLC chromatin during cold, but after a return to warm conditions H3K9 methylation levels are reduced, indicating that LHP1 is required to maintain vernalization-induced PRC2-like histone modifications at FLC (10). VERNALIZATION 1 is also required for vernalization-mediated H3K9 methylation at FLC chromatin (8, 9) and may be part of the system that renders the vernalized state mitotically stable.
Here we discuss the involvement of AtPRMT5, a type II protein arginine methyltransferase, in the vernalization-induced epigenetic silencing of FLC. AtPRMT5 was identified in a genetic screen for vernalization-insensitive mutants. In a winter-annual background, atprmt5 mutants fail to stably repress FLC after vernalization treatment, indicating that arginine methylation is required for maintenance of the silenced state. In atprmt5 mutants, global levels of symmetric dimethylation of arginine 3 of histone 4 (H4R3sme2) are greatly reduced, and this modification is reduced at FLC chromatin as well. Furthermore, in FRI;atprmt5 mutants, levels of H3K9 and H3K27 methylation at FLC chromatin do not increase after vernalization. Thus, AtPRMT5 is required for the repressive histone modifications that are thought to contribute to establishment and maintenance of the vernalized state in Arabidopsis winter annuals.
Results
atprmt5 Mutants Exhibit an Attenuated Vernalization Response.
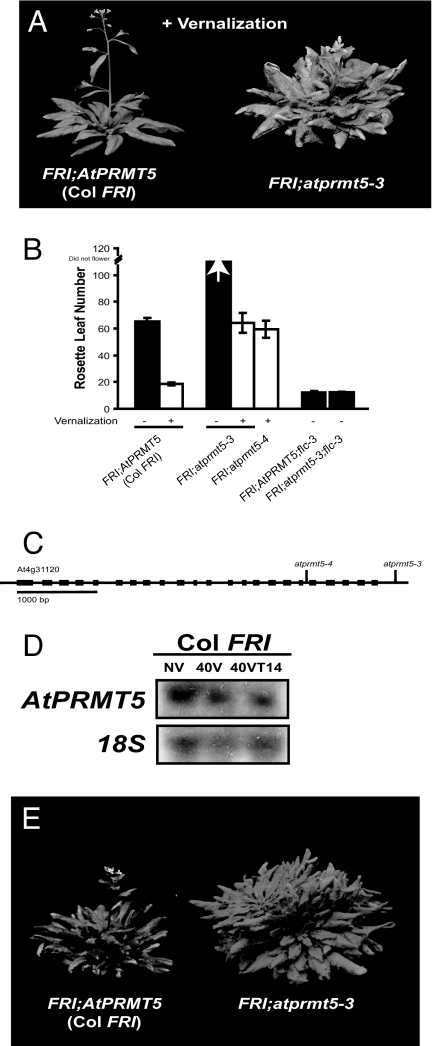
The Columbia (Col) accession of Arabidopsis thaliana contains a naturally occurring lesion in FRIGIDA and therefore does not have a strong vernalization requirement (5). Introgression of a functional allele of FRI into Col from the vernalization-requiring San Feliu-2 accession of Arabidopsis created a “nearly isogenic” Col line (Col FRI) that has a strong vernalization requirement and thus behaves as a winter annual (17). We identified a mutant from a genetic screen in Col FRI that fails to flower rapidly after a vernalization treatment (Fig. 1 A and B). The vernalization-insensitive phenotype is recessive, and a single T-DNA locus cosegregates with the mutant phenotype. Arabidopsis sequences that flanked the T-DNA insert aligned to the 3′ UTR of At4g31120, which is predicted to encode a protein arginine methyltransferase (Fig. 1C) (18). An alignment of the predicted primary amino acid sequence of At4g31120 to protein databases revealed high sequence identity to human PRMT5 (Protein Arginine Methyltransferase 5) (19). An additional At4g31120 mutant was isolated from the Salk T-DNA collection (Fig. 1C). This allele also displays a vernalization-insensitive phenotype when present in the Col FRI genetic background (Fig. 1B). Expression of At4g31120 during a vernalization time course revealed that, unlike VIN3, At4g31120 mRNA abundance is not affected by prolonged exposure to cold (Fig. 1D).
Fig. 1.
FRI;atprmt5 mutants display a vernalization-insensitive phenotype. (A) Image of FRI:AtPRMT5 versus FRI;atprmt5-3 after 40 days of cold treatment. (B) Flowering time represented as total rosette leaf number. (C) Gene structure of AtPRMT5 (At4g31120). Lines indicate location of T-DNA alleles [atprmt5-3 was isolated in our genetic screen, and atprmt5-4 was isolated from the Salk collection of T-DNA mutants (accession no. SALK_073624)]. (D) Northern blot analysis of AtPRMT5 during cold treatment. (E) Image of Col FRI and FRI;atprmt5-3 without vernalization treatment. RNA was isolated from seedlings without vernalization (NV), with 40 days of cold treatment (40VT0), and with 40 days of cold treatment followed by 14 days in the warm (40VT14).
While our work was in progress, two recent articles reported the phenotype of mutations in AtPRMT5 in a Col (fri) background (20, 21). Those mutants were named atskb1-1/atprmt5-1 and atskb1-2/atprmt5-2. We hereafter refer to the additional alleles we present in this study as atprmt5-3 and atprmt5-4 (Fig. 1C).
AtPRMT5 Is Required to Repress FLC in the Absence of Vernalization.
In the absence of vernalization, atprmt5 mutants in Col FRI typically fail to flower after generating well over 120 rosette leaves from the primary meristem. In contrast, the Col FRI parental line flowers without vernalization after producing ≈60 rosette leaves in our growth conditions (Fig. 1 B and E).
atprmt5;flc-3 double mutants were generated to determine whether FLC was required for this enhancement of the delayed-flowering phenotype of atprmt5 in a Col FRI background. atprmt5;flc-3 double mutants flowered at the same time as flc-3 single mutants, indicating that enhanced FLC expression is responsible for the delay in flowering in Col FRI (Fig. 1B).
AtPRMT5 Is Required for Maintenance of Vernalization-Induced FLC Silencing in Col FRI, but Not for Repression of Other Members of the FLC Clade.
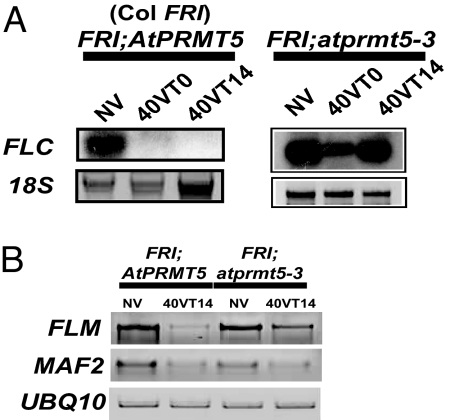
To determine whether the vernalization-insensitive phenotype of FRI;atprmt5 was due to a failure to repress FLC during and after a vernalizing cold exposure, we evaluated mRNA levels of FLC throughout a vernalization time course. In Col FRI, FLC is expressed at high levels before vernalization and is repressed by prolonged cold treatment (6) (Fig. 2A). In atprmt5 mutants in Col FRI, FLC is strongly expressed before vernalization and is only partially repressed during cold (Fig. 2A). Furthermore, FLC repression is not maintained after plants are removed from the cold (Fig. 2A). This failure to maintain FLC silencing likely contributes to the vernalization-insensitive phenotype of atprmt5.
Fig. 2.
atprmt5-3 flowering phenotypes are dependent on FLC. (A) Northern blot analysis of FLC mRNA levels in Col FRI and FRI;atprmt5-3. FLC expression resumes after plants are returned to warm conditions in the FRI;atprmt5-3 mutant, indicating that AtPRMT5 is required for vernalization-induced stable repression of FLC. (B) RT-PCR analysis of mRNA levels of members of the FLC clade.
Although, as noted above, FRI;atprmt5 mutants are later-flowering than the parental Col FRI line both with and without vernalization, FRI;atprmt5 mutant plants do flower more rapidly after a vernalizing cold exposure than nonvernalized mutants (Fig. 1B). This “partial” response to vernalization is not likely to be a result of FLC repression because stable repression of FLC fails to occur in FRI;atprmt5-3. Therefore, other floral repressors that are silenced by vernalization are likely contributing to the acceleration of FRI;atprmt5 flowering after vernalization (Fig. 1B).
FLC is a member of a clade of floral repressors that includes FLM/MAF1 and MAF2-5 (22–24). All members of this clade, except for MAF5, are subjected to repression by vernalization (22), and thus vernalization can accelerate flowering in flc mutants (25). The effect on flowering due to the vernalization-mediated repression of FLM/MAF1 and certain MAFs is most pronounced in short days (9, 25).
atprmt5;flc-3 double mutants and flc-3 mutants both flower more rapidly in short days after vernalization [supporting information (SI) Fig. 6], indicating that the partial vernalization response observed in atprmt5 might be due to repression of members of the FLC clade. Indeed, mRNA levels of two members of the FLC clade tested (FLM/MAF1 and MAF2) are reduced after vernalization in both FRI;atprmt5 and fri;atprmt5 when compared with their respective wild types (Fig. 2B; data shown for FRI backgrounds only). Thus, AtPRMT5 is required for the vernalization pathway to repress FLC but not for the vernalization pathway to repress some other members of the FLC clade.
AtPRMT5 Contributes to FLC Repression in the Absence of FRI.
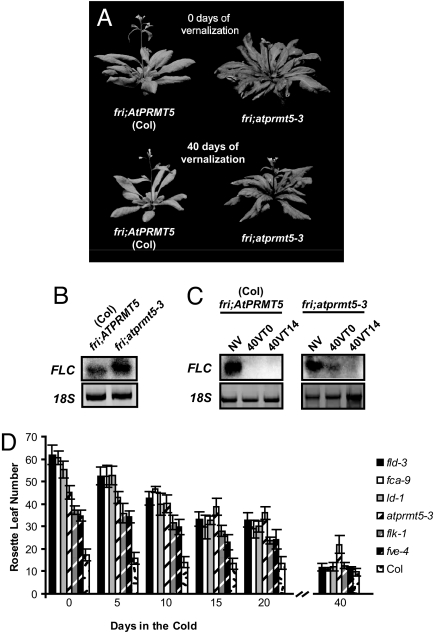
atprmt5 mutants also exhibit a delay in flowering in the wild-type Col genetic background, which lacks a functional FRI (Fig. 3 A and B; we refer to wild-type Col hereafter as Col fri). The delay in flowering in Col fri was also observed by Pei et al. (21) and Wang et al. (20). In Col fri, atprmt5-3 and atprmt5-4 mutants flower after producing ≈45 rosette leaves in contrast to Col, which flowers after generating ≈18 leaves in our growth conditions (Fig. 3B; data shown for atprmt5-3).
Fig. 3.
AtPRMT5 represses FLC before vernalization. (A) Image of Col fri and fri;atprmt5-3 before and after 40 days of vernalization. (B) Northern blot analysis of FLC mRNA expression in Col fri and fri;atprmt5-3. (C) Northern blot analysis of FLC mRNA expression in Col fri and fri;atprmt5-3 during a vernalization time course. (D) Flowering time, represented by total rosette leaf number, of known autonomous-pathway mutants and fri;atprmt5-3 with various days of cold treatment as indicated on the x axis.
Late flowering and vernalization responsiveness are defining characteristics of autonomous-pathway mutants (26). Autonomous-pathway genes are involved in FLC repression, and the delayed-flowering phenotype of autonomous-pathway mutants depends on FLC function; i.e., a lesion in flc suppresses the delayed flowering of autonomous-pathway mutants (25). fri;atprmt5 is similar to autonomous-pathway mutants because (i) its late-flowering phenotype is associated with elevated levels of FLC mRNA (Fig. 3B), (ii) its late flowering is suppressed by lesions in flc (Fig. 1B), and (iii) in fri;atprmt5 FLC is stably repressed by vernalization (Fig. 3C). Thus, in a fri, but not a FRI, background, vernalization is able to cause a stable suppression of FLC expression. Interestingly, the vernalization response of fri;atprmt5 (as measured by total leaf number after cold treatment) is attenuated relative to other autonomous-pathway mutants (Fig. 3D). The slightly attenuated vernalization response of fri;atprmt5 relative to other autonomous-pathway mutants may be to be due to “leaky” FLC repression in fri;atprmt5.
Global Levels of H4R3sme2 Are Reduced in atprmt5 Mutants.
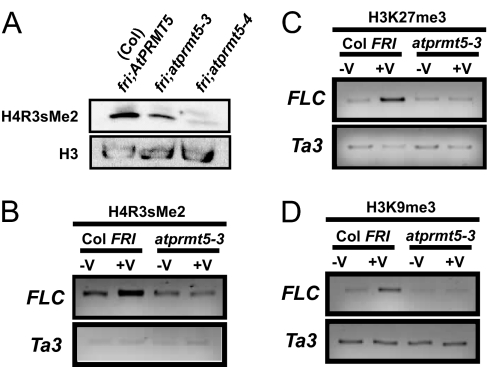
Arginine methylation is found in three forms: monomethylation, asymmetric dimethylation, and symmetric dimethylation. Of these forms, monomethylation and asymmetric dimethylation are generally associated with transcriptional activity whereas symmetric dimethylation is associated with transcriptional silencing (27). PRMT5 can symmetrically dimethylate arginine residues of proteins involved in RNA processing (28) and histones, specifically histone 4 (29). AtPRMT5 appears to be the only type II PRMT encoded in the Arabidopsis genome. Analysis of global levels of H4R3sme2 in fri;atprmt5-3 and fri;atprmt5-4 compared with Col fri revealed that loss of AtPRMT5 results in a genome-wide reduction in symmetric dimethylation of arginine-3 at histone 4 (H4R3sme2) (Fig. 4A; also reported in ref. 20). Furthermore, the difference between the two alleles of atprmt5 in the degree of H4R3sme2 level reduction correlates well with the strength of each allele predicted by the T-DNA position and the severity of the other associated mutant phenotypes (atprmt5-4 is more severe than atprmt5-3 as discussed below).
Fig. 4.
AtPRMT5 is required for the epigenetic silencing of FLC. (A) Immunoblot analysis of symmetric dimethylation of arginine-3 on histone 4 (H4R3sme2) on protein extracts isolated from seedlings. Histone 3 (H3) was used as a loading control. (B–D) ChIP analysis of FLC chromatin before and after vernalization using antibodies against H4R3sme2 (B), H3K27me3 (C), and H3K9me3 (D). Ta3 was used to standardize our samples, and the FLC-P region was analyzed.
Levels of H4R3sme2 at FLC Chromatin Increase as a Result of Prolonged Exposure to Cold.
H4R3sme2 levels at FLC increase after a vernalizing cold treatment (Fig. 4B). As previously reported (8–10), methylation of H3K9 and H3K27 also increases as a result of prolonged cold treatment, and these two repressive histone modifications may be required, along with LIKE-HETEROCHROMATIN PROTEIN 1, for maintenance of the repressed state (10, 16). In atprmt5 mutants in Col FRI, no increase in H3K9 and H3K27 methylation occurs after cold treatment (Fig. 4 C and D), indicating that, in this winter-annual background, H4R3sme2 is required for repressive histone modifications such as trimethylation of H3K9 and H2K27 at FLC chromatin.
NDPK2 Is a FLC-Independent Target of AtPRMT5.
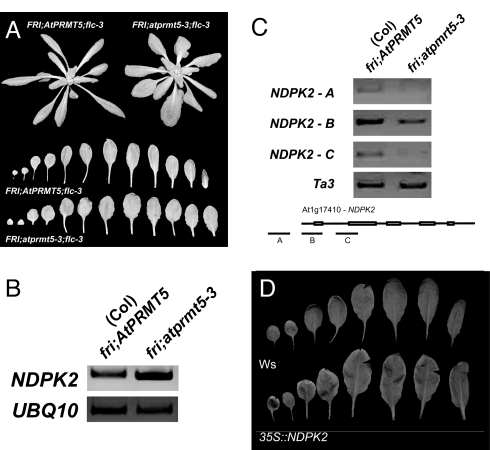
atprmt5 mutant plants have darker green leaves with shorter petioles and consequently form a compact rosette. These mutants also exhibit slight downward curling of the leaves, and mutant leaves are wider than wild type (Fig. 5A). These phenotypes are stronger in atprmt5-4 than atprmt5-3. Although the flowering phenotype of these alleles is identical, the nonflowering phenotypes are independent of FLC (data not shown). The pleiotropy associated with atprmt5 is not surprising given the global decrease in H4R3sme2 in atprmt5 mutants. We performed microarray analyses to identify genes that are derepressed in fri;atprmt5 mutants; such derepressed genes are potential targets of AtPRMT5 and are listed in SI Table 1. One candidate, NUCLEOTIDE DIPHOSPHATE KINASE 2 (NDPK2), exhibited a >3-fold increase of mRNA levels in the mutant compared with wild type. These data were confirmed by RT-PCR analysis of NDPK2 RNA levels (Fig. 5B). Furthermore, NDPK2 is indeed a locus that contains H4R3sme2; ChIP analysis of NDPK2 chromatin revealed the presence of H4R3sme2 in wild type compared with the fri;atprmt5-3 mutant, indicating that symmetric arginine methylation is required for proper levels of NDPK2 expression (Fig. 5C). Last, there is an expansion in leaf width in transgenic lines overexpressing NDPK2 when compared with the wild-type Wassilewskija accession (Fig. 5D). Because a similar expansion in leaf width is also observed in atprmt5 mutants (Fig. 5A), it is likely that the observed leaf expansion phenotype in atprmt5 mutants is due to the derepression of NPDK2. Interestingly, NM23 (the human homolog of NDPK2) is a target of PRMT5 (the human homolog of AtPRMT5) (30). It is intriguing that PRMT5 modifies NDPK2 chromatin with arginine methylation in both Arabidopsis and humans.
Fig. 5.
NDPK2 represents another target of AtPRMT5. (A) Image of pleiotropy present in FRI;atprmt5-3 compared with Col FRI (double mutants between FRI;atprmt5-3 and FRI;flc-3 still result in pleiotropy). atprmt5-3 mutants display shorter petioles, a compact rosette, and dark green leaves. (B) RT-PCR analysis of NDPK2 mRNA levels in Col fri and fri;atprmt5-3. (C) ChIP of NDPK2 chromatin from seedlings. Samples were immunoprecipitated with H4R3sme2 antibodies. (D) Overexpression of NDPK2 in the Wassilewskija (Ws) accession results in plants with some similarity to atprmt5-3.
Discussion
AtPRMT5 is required for repression of FLC in the absence of vernalization in both FRI and fri backgrounds, as well as for vernalization-mediated repression of FLC in a FRI background. In FRI;atprmt5 there is some degree of FLC repression during cold exposure, but FLC expression is restored to approximately pre-cold-treated levels after plants are removed from cold. Reduction of expression of Arabidopsis PRC2 components CLF, SWN, and FIE results in phenotypes similar to FRI;atprmt5 such as delayed flowering and an inability to maintain FLC repression after vernalization (14). Lesions in VRN1 and VRN2 also compromise the maintenance of FLC repression (31, 32). Thus, arginine methylation might function in conjunction with polycomb repression machinery to maintain stable repression of target genes.
An atprmt5 lesion causes a vernalization-insensitive phenotype in combination with the autonomous-pathway mutation luminidependens-1 (data not shown). Therefore, the vernalization insensitivity of atprmt5 is not due to a specific interaction of FRI with atprmt5, but rather is likely due to the combination of atprmt5 with situations in which FLC is highly transcribed.
The effect of AtPRMT5 on flowering appears to be specific to FLC. This is in contrast to VIN3, which affects FLC and several additional members of the FLC clade. One measure of FLC clade specificity is the effect of a mutation on the vernalization response in short days. For example, with vernalization, flc-3 single mutants and atprmt5;flc-3 double mutants both flower more rapidly in short days, whereas vin3;flc-3 double mutants do not (9). The “residual” short-day vernalization response of FRI:atprmt5 or flc-3 single mutants is likely due to vernalization-mediated repression of other members of the FLC clade. The lack of response in vin3 mutants likely results from a requirement for VIN3 for repression of most or all FLC clade members. The specificity of AtPRMT5 for FLC is not unprecedented: components of both the autonomous pathway and certain suppressors of FRI-mediated late flowering can also specifically affect FLC transcription without effect on other FLC clade members (22, 25, 33, 34).
In the absence of vernalization, loss of atprmt5 delays flowering in both the winter-annual Col FRI and the rapid-flowering Col fri strains. In this respect, AtPRMT5 behaves in a manner similar to genes in the autonomous pathway. Indeed, in a fri background, the only difference between atprmt5 and other autonomous-pathway mutants is the slight attenuation of the vernalization response (Fig. 3D). Interestingly, two autonomous-pathway mutants (fve and flowering locus d) also exhibited a mild late-flowering phenotype after vernalization when in the FRI background, similar to FRI;atprmt5 (Scott Michaels, personal communication, and our unpublished results). Other autonomous-pathway mutants present in a FRI background, such as fca, fpa, flowering locus k homology domain, and luminidependens, do not exhibit late-flowering phenotypes after vernalization. These results indicate that within the autonomous pathway there is specialization because a subset of autonomous-pathway genes are also required for vernalization in the presence of FRI.
We have shown that vernalization results in an increased level of H4R3sme2 at FLC chromatin. Moreover, the delayed-flowering phenotype of nonvernalized atprmt5 mutants is consistent with a model in which some degree of H4R3sme2 is required for “basal” repression of FLC. Indeed, Wang et al. (20) demonstrated, by immunoprecipitation, that AtPRMT5 interacts with FLC chromatin from nonvernalized plants. Thus, FLC chromatin appears to be a substrate of AtPRMT5 in both vernalized and nonvernalized plants. The increased level of H4R3sme2 at FLC after vernalization may result from increased AtPRMT5 activity or a reduction of activity that removes H4R3sme2.
Although the simplest model for the repression of FLC by AtPRMT5 is a direct methylation of H4R3 at FLC chromatin, it should be noted that mammalian PRMT5 associates with a large protein complex, referred to as the methylosome (35), which methylates components of a small nuclear ribonucleoprotein complex involved in splicing of target pre-mRNAs (28). Interestingly, many members of the Arabidopsis autonomous pathway contain RNA-binding domains (FCA, FPA, and FLK) that may have a function in splicing (36). Thus, it is possible that some portion of the effect of AtPRMT5 on FLC expression could be due to a failure to symmetrically dimethylate arginine residues of proteins involved in mRNA splicing.
AtPRMT5 appears to be a single-copy gene, and it is predicted to encode the only type II protein arginine methyltransferase in Arabidopsis. The viability of atprmt5 mutants may reflect that none of the alleles described in this or other studies (20, 21) are nulls, but a more likely possibility is that loss of AtPRMT5 activity does not result in a lethal disruption of gene expression. Indeed, lesions in other single-copy proteins involved in chromatin modification such as components of the PAF1 complex in both yeast and Arabidopsis are also viable (37, 38).
A definitive role for histone arginine methylation has not yet been established. There is a correlation between active gene expression and H4R3asMe2 (asymmetric dimethylation) and between H4R3sme2 and repressed loci (39, 40), although only a few loci that contain histone arginine methylation have been studied and there are examples to the contrary. Our study is consistent with a role for H4R3sme2 in the epigenetic silencing of FLC. Indeed, the vernalization-mediated increase in H4R3sme2 at FLC chromatin is the first example of a dynamic change in H4R3sme2 at a specific locus in response to an environmental signal. H4R3sme2 may be necessary to provide a proper chromatin substrate to enable levels of other vernalization-mediated histone modifications such as H3K9 and H3K27 methylation to increase. Alternatively, the increase in H4R3sme2 may occur concurrently with both H3K9 and H3K27 methylation. In either scenario it is clear that H4R3sme2 is required for both methylation of H3K9 and H3K27 and the subsequent epigenetic silencing of FLC.
Very few targets of PRMT5 have been reported in any organism. We identified NDPK2 as a possible target because it was derepressed in fri;atprmt5 mutants relative to wild-type Col fri. ChIP analysis of the NDPK2 locus revealed the presence of H4R3sme2. Interestingly, in human cell cultures NM23 (the human homolog of NDPK2) is a direct transcriptional target of PRMT5 (the human homolog of AtPRMT5) (28). In the absence of human prmt5, NM23 is derepressed, and, conversely, in cell lines overexpressing human PRMT5, NM23 expression levels are reduced. It is intriguing that NDPK2 is a target of PRMT5-mediated arginine methylation in both Arabidopsis and humans. That this PRMT5 target is conserved may be coincidence (i.e., convergent evolution) or an ancient conserved relationship.
Methods
Plant Material.
All genotypes used in this work are in the Columbia (Col) background with the exception of the 35S::NDPK2, which is in the Wassilewskija accession. The atprmt5-4 insertion line in the Col background was isolated from the Salk collection (http://signal.salk.edu; accession no. SALK_073624) (41).
Growth Conditions.
Plants were grown in long days (16 h light/8 h dark) or short days (8 h light/16 h dark) at 22°C under cool-white fluorescent lights (Sylvania) at 100 μmol·m−2·s−1. For experiments involving vernalization, seeds were incubated on agar-solidified medium containing 0.65 g/liter Peters Excel 15-5-15 fertilizer (Grace Sierra) at 4°C for 2 days, transferred to short days at 22°C, allowed to germinate for 5 days, and then transferred to 4°C in short days for the number of days noted in the figure legends.
T-DNA-Flanking Sequence Analysis.
The sequence flanking the T-DNA of atprmt5-3 was obtained by thermal asymmetric interlaced PCR (18). T-DNA borders were defined by sequencing PCR products obtained by using a T-DNA border primer and a gene-specific primer.
RNA Isolation and Blotting.
In all cases, total RNA was isolated from 7-day-old seedlings by using TRIzol (Invitrogen) following the manufacturer's protocol. RNA was subsequently purified and concentrated by using an RNeasy column (Qiagen). For RNA blots, 15 μg of total RNA was used per lane except for the fri;AtPRMT5 (Col) samples, in which 20 μg of total RNA was used (Fig. 2D). Samples were separated on a denaturing gel and transferred by capillary blotting to Nytron N membranes (Sigma–Aldrich). RNA was fixed to the membrane by using a Stratalinker UV cross-linker (Stratagene). Hybridization using a 32P-labeled AtPRMT5 probe corresponding to the first 300 bp of the coding sequence was performed in the QuickHyb solution from Stratagene according to their protocol. Membranes were exposed to film for 3 days at −80°C (Fuji Film Super Rx).
RT-PCR.
First-strand cDNA synthesis was performed on 5 μg of RNA with the MLV-RT First-Strand Synthesis System (Promega) for RT-PCR and a primer containing the M13 primer sequence with an oligo dT extension (5′-GTAAAACGACGGCCAGTCCCT15-3′). PCR amplification of the cDNA was performed with Ex-Taq (Takara). FLM (5′-CCTCCGGAAAACTCTATGACTCT-3′ and 5′-CGGTATTTTGTTGCCGGAGCTACTTC-3′), MAF2 (5′-GGCTCCGGAAAACTCTA-CAAGTC-3′ and 5′-TGATGGTGATTACTTGAGCAGCGGA-3′), and UBQ (5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′ and 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′) were amplified by using the indicated primers and the following cycles: 95°C for 4 min followed by 22 cycles for UBQ, 38 cycles for FLM, and 34 cycles for MAF2 of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s. Amplified fragments were separated on a 1.2% agarose gel.
Western Blot Analysis.
Nuclear protein extracts were isolated from a chromatin preparation as described in the ChIP protocol in ref. 29. Proteins were separated on a 12.5% SDS/PAGE and transferred to a nitrocellulose membrane (Amersham Biosciences). Total H4R3 symmetric dimethylation was determined by using anti-H4R3 symmetric dimethyl antibodies (Abcam, catalog no. 5823-200) using the recommended 1:2,000 dilution.
ChIP Assays.
Chromatin samples were prepared as described (31). Immunoprecipitations were performed by using a ChIP assay kit (Millipore, catalog no. 17-295) following the manufacturer's suggested protocol modified as previously described (42). Antibodies against trimethyl H3K9 and trimethyl H3K27 were obtained from Upstate Biotechnology. Primers used to detect Ta3 and the FLC-P region were previously described (10). Primers used to detect NDPK2 chromatin are as follows: region A forward, 5′-TGGTATTAAAGCAGCAAATTGTGCGA-3′; region A reverse, 5′-AAGAACAAAGTTAATTGACATAAATA-3′; region B forward, 5′-ATGGTGGGAGCGACTGTAGTTAGTAA-3′; region B reverse, 5′-CAAACCATAGAAGCTACAAGGTGAGGAA-3′; region C forward, 5′-GAGCATTATAAGGATCTTAGTGCTA-3′; region C reverse, 5′-CCAACACCTTCCCAAGCCTTTACCA-3′. All ChIP assays were performed at least three times from at least two chromatin samples prepared from biological replicates.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Dae-Jin Yun (Gyeongsang National University, Chinju, South Korea) for the 35S::NDPK2 seeds and the University of Wisconsin Gene Expression Center for assistance with our microarray experiments. Work in R.M.A.'s laboratory was supported by the College of Agricultural and Life Sciences and the Graduate School of the University of Wisconsin, National Institutes of Health Grant 1R01GM079525, and National Science Foundation Grant 0209786.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710423104/DC1.
References
- 1.Chouard P. Annu Rev Plant Physiol. 1960;11:191–238. [Google Scholar]
- 2.Napp-Zinn K. In: La Physiologie de la Floraison. Champagnat P, Jaques R, editors. Paris: Collogues Internationaux Centre National de la Recherche Scientifique; 1979. pp. 217–220. [Google Scholar]
- 3.Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T. Plant J. 1994;6:911–919. [Google Scholar]
- 4.Lee I, Michaels SD, Masshardt AS, Amasino R. Plant J. 1994;6:903–909. [Google Scholar]
- 5.Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 6.Michaels SD, Amasino RM. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 9.Sung S, Amasino RM. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 10.Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Nat Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. Curr Biol. 2007;17:73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 13.Sung S, Schmitz RJ, Amasino RM. Genes Dev. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. Proc Natl Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringrose L, Paro R. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 16.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. Proc Natl Acad Sci USA. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee I, Amasino RM. Plant Physiol. 1995;108:157–162. doi: 10.1104/pp.108.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 19.Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zeng W, Kim MS, Allen PB, Greengard P, Muallem S. EMBO J. 2007;26:2768–2776. doi: 10.1038/sj.emboj.7601701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei Y, Niu L, Lu F, Liu C, Zhai J, Kong X, Cao X. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scortecci KC, Michaels SD, Amasino RM. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- 25.Michaels SD, Amasino RM. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koornneef M, Hanhart CJ, van der Veen JH. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 27.Wysocka J, Allis CD, Coonrod S. Front Biosci. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 28.Meister G, Eggert C, Bühler D, Brahms H, Kambach C, Fischer U. Curr Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 29.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendall AR, Levy YY, Wilson A, Dean C. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 32.Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- 33.Michaels SD, Bezerra IC, Amasino RM. Proc Natl Acad Sci USA. 2004;101:3281–3285. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz RJ, Hong L, Michaels S, Amasino RM. Development. 2005;132:5471–5478. doi: 10.1242/dev.02170. [DOI] [PubMed] [Google Scholar]
- 35.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson GG, Quesada V, Henderson IR, Dijkwel PP, Macknight R, Dean C. Biochem Soc Trans. 2004;32:565–566. doi: 10.1042/BST0320565. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Doyle MR, Amasino RM. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh S, Zhang H, Ludwig P, van Nocker S. Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. Mol Cell Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 41.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 42.Johnson L, Cao X, Jacobsen S. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.