Abstract
In mammalian cells, ceramide is synthesized in the endoplasmic reticulum and transferred to the Golgi apparatus for conversion to sphingomyelin. Ceramide transport occurs in a nonvesicular manner and is mediated by CERT, a cytosolic 68-kDa protein with a C-terminal steroidogenic acute regulatory protein-related lipid transfer (START) domain. The CERT START domain efficiently transfers natural d-erythro-C16-ceramide, but not lipids with longer (C20) amide-acyl chains. The molecular mechanisms of ceramide specificity, both stereo-specific recognition and length limit, are not well understood. Here we report the crystal structures of the CERT START domain in its apo-form and in complex with ceramides having different acyl chain lengths. In these complex structures, one ceramide molecule is buried in a long amphiphilic cavity. At the far end of the cavity, the amide and hydroxyl groups of ceramide form a hydrogen bond network with specific amino acid residues that play key roles in stereo-specific ceramide recognition. At the head of the ceramide molecule, there is no extra space to accommodate additional bulky groups. The two aliphatic chains of ceramide are surrounded by the hydrophobic wall of the cavity, whose size and shape dictate the length limit for cognate ceramides. Furthermore, local high-crystallographic B-factors suggest that the α-3 and the Ω1 loop might work as a gate to incorporate the ceramide into the cavity. Thus, the structures demonstrate the structural basis for the mechanism by which CERT can distinguish ceramide from other lipid types yet still recognize multiple species of ceramides.
Keywords: crystal structure, lipid transport, sphingomyelin, diacylglycerol
Ceramide is a common precursor for both sphingomyelin and glycosphingolipids, which are ubiquitous components of membranes in mammalian cells and play important roles in cell growth, differentiation, and apoptosis (1, 2). There are at least two pathways by which ceramide is transported from the endoplasmic reticulum (ER) to the Golgi apparatus: an ATP- and cytosol-dependent major pathway and an ATP- and cytosol-independent minor pathway (3–6). Recently, we have shown that the CERT protein mediates the major pathway of ceramide transport in a nonvesicular manner (7).
CERT, which is known as a splicing variant of Goodpasture-antigen-binding protein, GPBPΔ26 (8, 9), consists of three regions. The N-terminal region of ≈120 aa has a pleckstrin homology domain that binds to phosphatidylinositol 4-monophosphate. The middle region, of ≈250 aa, contains coiled-coil motifs and a FFAT motif that may participate in association with the ER (10). Finally, the C-terminal region, also of ≈250 aa, is homologous to the steroidogenic acute regulatory protein-related lipid transfer (START) domain (7, 11, 12). The START domain of CERT has the activity to extract ceramide from donor membranes and release the bound ceramide to acceptor membranes (7, 13). CERT specifically recognizes natural d-erythro ceramides, but not sphingosine, sphingomyelin, cholesterol, or phosphatidylcholine (PtdCho). CERT is able to efficiently transfer the natural ceramide isomers dihydroceramide and phytoceramide and also various ceramide molecular species having C14-C20 amide-acyl chains, but not longer amide-acyl chains (7, 13). CERT also transfers diacylglycerol, although with only ≈5% of its activity toward C16-ceramide (13). To elucidate the molecular mechanism of specific lipid recognition of CERT, we determined the crystal structures of the CERT START domain in its apo-form (2.2-Å resolution), in complex with C6-, C16-, and C18-ceramide (1.4- to 2.1-Å resolution), and with 1,2-didecanoyl-sn-glycerol (C10-diacylglycerol) (1.8-Å resolution). In each of the complex structures, one lipid molecule is buried in an amphiphilic cavity of the CERT START domain. The spatial arrangement of the ceramide recognizing residues in the cavity confers the specificities for hydrophilic parts and length of the amide-acyl groups of cargo lipids. Using a combination of mutational analysis of ceramide extraction and transfer activities based on these structures, we demonstrate the mechanism of ceramide recognition and subsequently discuss the uptake and release of ceramide by CERT.
Results
Overall Structures of the CERT START Domain.
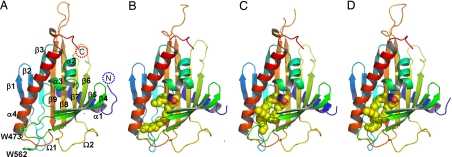
We first determined the crystal structures of the CERT START domain, in the apo-form (2.2-Å resolution) and in complex with C6-ceramide (1.40-Å resolution), by the multiwavelength anomalous dispersion (MAD) method with selenomethionine-labeled protein crystals [Fig. 1 A and B and supporting information (SI) Table 1]. Subsequently, structures in complex with ceramides with longer amide-acyl chains (C16- and C18-ceramide, at 1.40- and 1.65-Å resolution, respectively) and with C10-diacylglycerol (1.8-Å resolution) were solved by the molecular replacement method using the C6-ceramide-bound structure as a search model (Fig. 1 C and D; SI Fig. 8A and SI Table 2). The overall structure shares the same helix-grip fold of other START domains (14–16), with two α-helices, α1 and α4, at the N and C termini separated by nine β-strands and two shorter helices. Electron density for the first 12 or 13 residues was not observed in all cases, because of disorder. Two Ω-loops are inserted between β5-β6 (Ω1), and β7-β8 (Ω2) (Fig. 1A). There are two neighboring tryptophan residues exposed to the protein exterior: Trp-473 in the Ω1 loop and Trp-562 in the loop between β9 and α4 (Fig. 1A). In some structures, the electron density map was difficult to model in the loop regions of β5-β6 (Ω1) and β8-β9. Superposition of the apo- and lipid-bound CERT START domain structures reveals essentially no difference in the Cα chains (SI Figs. 7A and 8C). The values of rmsd are <1.0 Å for C6-, C16-, and C18- ceramides and C10-diacylglycerol.
Fig. 1.
Overall structure of the CERT START domain. (A) The CERT START domain (apo-form) in a ribbon representation. α-Helices (α1-α4), β-strands (β1-β9), and Ω loops (Ω1, Ω2) are numbered from the N to the C terminus. (B–D) Ribbon representation of the CERT START domain in complex with C6-, C16-, C18-ceramide, respectively. The ceramide molecules are represented as space-filling spheres in which yellow, blue, and red spheres represent C, N, and O atoms, respectively.
Amphiphilic Cavity of the CERT START Domain.
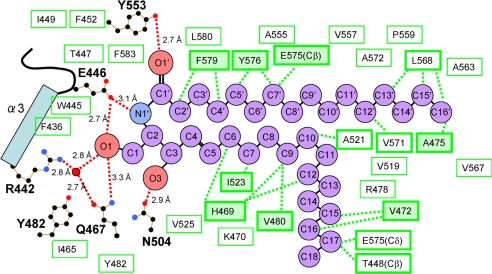
The CERT START domain has a long amphiphilic cavity in the center of the protein. This cavity is composed of curved β-sheets covered by three α-helices (α2, α3, and α4) and the Ω-loops. The cavity (≈12 × 21 × 8 Å) is lined by 26 hydrophobic and 10 polar and/or charged amino acid residues (Fig. 2 and SI Table 3). In the complex structures, the electron densities of the entire substrate molecule (C6-, C16-, and C18- ceramides or C10-diacylglycerol) can be clearly observed. One lipid molecule is buried in the amphiphilic cavity of the CERT START domain (Fig. 1), in agreement with our previous biochemical analysis showing a one-to-one binding stoichiometry of CERT and ceramide (13). Ceramide consists of two long aliphatic chains, the sphingosine backbone and the amide-acyl chain. Hydrophobic interactions of the two aliphatic chains of C16-ceramide with specific amino acid residues of the CERT START domain are summarized in Fig. 2 (in the case of complex structures with C6-, C18-ceramides and C10-diacylglycerol, the interactions are summarized in SI Fig. 9 A–C). The three complex structures with ceramides show a common set of eight amino acid residues interacting with ceramide molecules by hydrophobic interactions. Additional four to five amino acid residues contribute to the hydrophobic interaction in each structure.
Fig. 2.
Schematic representation of C16-ceramide recognition by the CERT START domain. Residues lining the amphiphilic cavity are shown. Red dashed lines, hydrogen bonds; red circles, water molecules; black, blue, and red dots, C, N, and O atoms, respectively, of the residues involved in the hydrogen network. Green boxes indicate residues contributing to the hydrophobicity of the cavity in general, whereas green boxes with thick borders indicate those with direct hydrophobic interactions, which are represented as green dashed lines. Among these, eight amino acid residues, which are common to all of the C6-, C16-, C18-ceramide complex structures, are indicated by thick-bordered green boxes filled with light green.
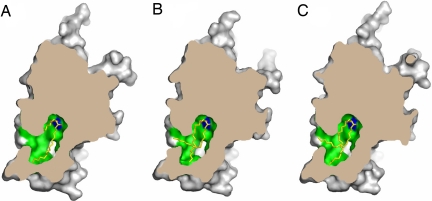
In the C6-ceramide bound structure, both aliphatic chains are enclosed completely in the hydrophobic part of the cavity, with some empty space remaining (Figs. 1B and 3A). In the C16- and C18-ceramide bound structures, however, both aliphatic chains completely fill the hydrophobic part of the cavity (Figs. 1 C and D and 3 B and C). Here, the sphingosine backbone bends at the C-11 position and is directed toward the outside of the protein and the ω methyl group of sphingosine backbone reaches the surface of the protein. Inspection of the space for the aliphatic group in the hydrophobic part of the cavity suggests that it might accept up to two more methylene groups, but not more. We also determined the structure of CERT START domain crystals grown in the presence of C24-ceramide, at 2.0 Å resolution (SI Table 2), but it does not show any electron density for the ceramide in the cavity (SI Fig. 7B). These results are consistent with our previous observation that CERT efficiently transfers ceramide molecules with C14-C20 chains, but not those with longer amide-acyl chains (13).
Fig. 3.
Molecular surface of the CERT START domain in complex with C6- (A), C16- (B), and C18- (C) ceramide cut at the level of the cavity, respectively. Ceramide molecules are drawn as sticks, in which yellow, blue, and red represent C, N, and O atoms, respectively. Hydrophobic and polar/charged amino acid residues inside the cavity are shown in green and blue, respectively. The outer surface and the cross-section of the CERT START domain are drawn in gray and in dark brown, respectively.
Ceramide Recognition Mechanism.
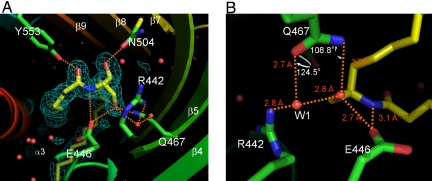
In addition to the hydrophobic interaction, the binding of ceramide to the CERT START domain is achieved through recognition of the amide- and hydroxyl- group of ceramide by a hydrogen bond network involving residues Arg-442, Glu-446, Gln-467, Asn-504 and Tyr-553, which are located at the far end of the amphiphilic cavity (Figs. 2, 3 and 4). The keto group of Glu-446 forms two hydrogen bonds with the amide-nitrogen (N1′) and the hydroxyl group (O1) of the ceramide. The O1 atom of the hydroxyl group of ceramide also forms water-molecule-mediated hydrogen bonds with the guanidino group of Arg-442 and with the amido-oxygen of Gln-467. In addition, the O1 atom has a direct hydrogen bond with the amido-nitrogen of Gln-467, although at an unusually long distance (3.3 Å). The oxygen atoms of Asn-504 and Tyr-553 also form hydrogen bonds with the O3 and O1′ atoms of ceramide, respectively. The interaction patterns described above are also observed in the complex structures with C6- and C18-ceramide (SI Fig. 9 A and B). Among these hydrogen bond interactions, Arg-442 may be less important than the others because of the indirect hydrogen bond mediated by the water molecule. In the apo-form structure, no solvent molecules are observed within the cavity except for two hydrogen-bonded water molecules mimicking the hydroxyl-groups of ceramide, near Asn-504 and Tyr-553.
Fig. 4.
Hydrogen bond network between the CERT START domain and C16-ceramide (A). Large white letters, amino acid residues interacting with the ceramide; small white letters, α-helix and β-sheet strands, numbered; green meshes, ceramide omit map contoured at 2.5 σ; orange dashed lines, hydrogen bonds; red circles, water molecules. In the wire model, N and O atoms are highlighted by blue and red, respectively. The ceramide molecule (yellow) and the side chains of the CERT START domain are shown by wire models. (B) Water-mediated stabilization of O1 oxygen of ceramide viewed in the direction opposite to A.
Ceramide Extraction and Transfer Activities of the CERT START Domain.
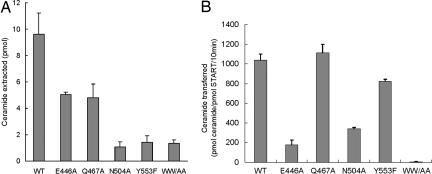
We next examined the importance of Glu-446, Gln-467, Asn-504 and Tyr-553, which are directly involved in the hydrogen-bond network with ceramide, for ceramide transfer. For this, we made alanine-replaced (E446A, Q467A, and N504A) and phenylalanine-replaced (Y553F) mutants. In addition, an alanine-replaced double mutant (W473A/W562A) was constructed to study the function of the exposed tryptophan residues. First, we assayed the extraction of ceramide from phospholipid vesicles. As shown in Fig. 5A, all of the mutants showed reduced ceramide extraction activities; these results indicate the importance of the residues. Next, we measured intermembrane ceramide transfer activity in a cell-free assay system (Fig. 5B). The transfer activities of the E446A and N504A mutants were <40% of the wild-type control, confirming the importance of these residues. The Y553F mutant showed 20% lower activity than the wild type, supporting the concept that the hydrogen bond between Tyr-553 and the O1′ atom of ceramide makes a contribution to the interaction. The W473A/W562A mutant has almost no ceramide transfer activity. No substantial difference in transfer activity was observed between the Q467A mutant and the wild type, although the Q467A mutant did have impaired extraction activity. This might be explained by formation of a water-mediated hydrogen bond between the O1 atom of ceramide and Arg-442, even when the hydrogen bonds of the O1 atom with Gln-467 are lost (Figs. 2 and 4). The transfer activity does not correlate one to one with the extraction activity, because the former measures the rate of the ceramide transfer from the donor to acceptor membranes whereas the latter detects the distribution of ceramide between the CERT START domain and the donor membranes.
Fig. 5.
Activities of the CERT START domain. (A) Ceramide extraction activities of the CERT START domain. (B) Ceramide transfer activities of the CERT START domain. The activities of the indicated constructs for C16-ceramide are shown as mean values with standard deviations calculated from triplicate experiments. WW/AA indicates W473A/W562A double mutant.
It has been reported that CERT is capable of transferring diacylglycerol, which structurally resembles ceramide, although at low efficiency (13). We measured and showed that the intermembrane diacylglycerol transfer activity of the CERT START domain is only 1% of the activity toward C16-ceramide (SI Fig. 10). In complex structure with C10-diacylglycerol, Asn-504 and Tyr-553 form hydrogen bonds with the carbonyl groups of sn-1 and sn-2 of the diacylglycerol, respectively (SI Figs. 9C and 11B). The amino group of Asn-504 also forms a hydrogen bond with the hydroxyl group of sn-3, which has another hydrogen bond with the backbone carbonyl oxygen of Phe-436. Glu-446 and Gln-467 are >3.7 Å away from the diacylglycerol. The low transfer activity for diacylglycerol is thus explained well, because six hydrogen bonds from every side participate in the binding of ceramide (Fig. 2), whereas fewer hydrogen bonds are formed unilaterally in the diacylglycerol complex (SI Fig. 9C). Interestingly, the N504A mutation slightly enhanced the activity of diacylglycerol transfer (SI Fig. 10), whereas this mutation reduced the activity of ceramide transfer (Fig. 5B). To elucidate this, we determined the crystal structure of the CERT START N504A mutant in complex with C10-diacylglycerol at 1.8-Å resolution (SI Table 2 and SI Fig. 8B). The diacylglycerol goes into a deeper position at the bottom of the cavity due to the reduced side-chain volume caused by the N504A mutation and forms a different set of hydrogen bonds compared with the wild type (SI Figs. 9 C and D and 11). The alternative bonds might change the specificity of the transfer activity.
Ceramide Transfer Activity of CERT in Vitro and in Vivo.
To assess the effect of the mutations on CERT, we made five full-length CERT mutants (E446A, Q467A, N504A, Y553F, and W473A/W562A) and measured ceramide transfer activities both in vitro and in vivo. First, we measured intermembrane ceramide transfer activity using a cell-free assay system (SI Fig. 12). All of the mutants except Q467A showed reduced activities, corroborating the results of the CERT START domain mutants (Fig. 5B).
Purified CERT protein can restore the ER-to-Golgi trafficking of ceramide in CERT deficient LY-A semiintact cells (5, 7). Next, we measured the amount of sphingomyelin synthesis, which reflects ceramide transfer, in the LY-A semiintact cell assay system (SI Fig. 13). The W473A/W562A mutant showed almost no sphingomyelin synthesis activity. Among the other mutants, the E446A mutant showed reduced activity, whereas the remaining three mutants had almost the same or higher activities. These results show the importance of Glu-446, and Trp-473/Trp-562 for ceramide transport (see below).
Discussion
We determined the structures of the CERT START domain with and without ceramides, all of which show basically the same amphiphilic cavity. The inner wall of the cavity is lined by many hydrophobic side chains and hydrophobic parts of amino acid residues, for instance, Cγ, Cδ, and Cε of lysine. The two aliphatic chains of ceramide interact with this hydrophobic wall. At the far end of the cavity, five polar and charged amino acid residues (Arg-442, Glu-446, Gln-467, Asn-504, and Tyr-553) make hydrogen bonds with the ceramide head group. Among them, Glu-446 is the most important for ceramide transfer activity, as shown by the lipid extraction and transfer experiments and semiintact cell assay described above. Other single point mutations in the cavity, H469D, A521D, and Y576D, also showed reduced ceramide transfer activity in LY-A cell lines (K.H. and K.K., unpublished results). These observations suggest that, for CERT to have activity, ceramide must be recognized not only by the hydrophobic inner wall but also by a specific hydrogen bond network.
To date, x-ray crystal structures of three other START domains have been reported: apo-forms of MLN64 (14) and StarD4 (15) and a lipid-bound form of phosphatidylcholine (PtdCho)-transfer protein (PCTP) (16). The structures resemble that of the CERT START domain reported in this work (rmsd values of the Cα atoms are 2.2, 2.7, and 3.6 Å, respectively) with similar tunnels or cavities with different degrees of amphiphilicity. In comparison with the amino acid sequences of the defined START family proteins, the START domain of CERT is most similar to that of PCTP (SI Fig. 14). Interestingly, the five residues at the far end of the amphiphilic cavity in the CERT START domain are not well conserved among different START family proteins. In the PtdCho-bound structure of PCTP, Arg-78 interacts directly with the phosphoryl group of PtdCho and forms a salt bridge with Asp-82. Such an Arg-Asp pair was suggested to be a motif for the recognition of a negative charge of cognate lipid head groups (16). Although the START domain of CERT also has Arg-442 and Glu-446 residues at the position corresponding to the Arg-78/Asp-82 pair of PCTP, the Arg-442/Glu-446 pair of CERT does not form a salt bridge, either in the apo-form or in the complexes with C6-, C16-, and C18-ceramides; instead, it interacts with ceramide by a hydrogen-bond network. PCTP interacts only with the trimethylammonium moiety and the phosphoryl group by a hydrogen-bond network, but ceramide and diacylglycerol do not have such moieties.
The amino acid residues making hydrophobic interactions with long aliphatic chains of lipid ligands are not conserved between the PCTP and the CERT START domain with the exception of Phe-579 (SI Fig. 14). Both of the acyl chains of diacylglycerol show some disorder toward their termini in the PCTP structure (16). In the CERT START domain, however, the whole molecule of ceramide is clearly observed, and the amide acyl chains stay inside the cavity. CERT may be able to distinguish the length of lipid amide-acyl chains that can be accepted in the hydrophobic cavity. It is conceivable that the cavity works as a molecular measuring device, termed “hydrocarbon rulers” as with the bacterial membrane acyltransferase PagP (17).
PCTP has enough space to accommodate the phosphocholine group in addition to the diacylglycerol moiety (16). The glycosylceramide transfer protein also has space to accommodate the sugar groups in addition to the ceramide moiety (18). In contrast, there is no extra space to accommodate any additional bulky group around the C-1 hydroxyl group of ceramide in the CERT START domain (Fig. 3). This structural feature clearly explains how CERT distinguishes ceramide from sphingolipids with large head groups. Among lipids, diacylglycerol seems to resemble ceramide stereochemically and thus is transferred by CERT at a low level. The transfer assay showed the E446A mutation of the CERT START domain impairs the activity of both ceramide and diacylglycerol, whereas Glu-446 is away from the diacylglycerol in the complex structure (SI Figs. 9C and 11). The residue may have another role in extracting lipid from the membranes, beyond merely holding the lipid by hydrogen bonds. The hydrogen bonds of Glu-446 with the amide nitrogen and the hydroxyl group of ceramide may be, in part, the reason for the preference of ceramide over diacylglycerol by CERT.
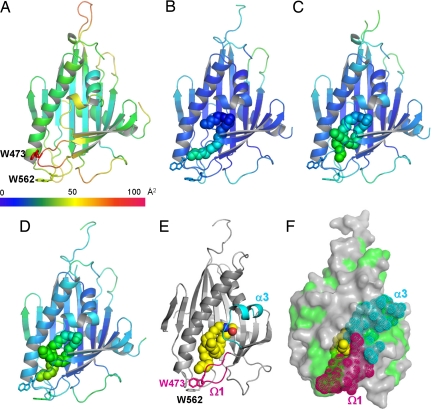
Comparison of the apo-form and ceramide-bound structures might give a clue regarding how the CERT START domain docks onto the target membranes to extract or release ceramides. As stated above, however, there are no drastic conformational changes between the apo-form and the ceramide-bound structures (Fig. 1 and SI Fig. 7A). This may be due to crystal packing that forces the ceramide bound CERT START domain structures to take a closed form similar to the apo-form structure. In both cases, the amphiphilic cavity is almost completely closed, except for one small opening surrounded by residues of α3, α4 and the Ω1 loop, which is too small for ceramide to enter (Figs. 3 and 6 E and F). However, higher-temperature factors found in the apo-form structure might explain the mechanism. Although α4 has rather low temperature factors, α3, Ω1, and Ω2 loops appear relatively mobile as compared with the rest of the apo-form CERT START domain (Fig. 6 A–D). The right-side wall of this entrance is formed by α3 and the Ω1 loop, which might act as the gate for ceramide to enter or leave the cavity (Fig. 6F). In addition, there are two tryptophan residues exposed to the protein exterior: Trp-473 in the Ω1 loop and Trp-562 in the loop between β9 and α4 (Fig. 6E). Because tryptophan residues have a high propensity for membrane interaction (19), we speculate these residues might interact with lipid bilayers when CERT forms contacts with ceramide in the membrane. Indeed, the W473A/W562A double mutant of the CERT START domain shows almost no ceramide extraction or transfer activities in cell-free assay systems (Fig. 5), and the mutant of CERT shows no ER-to-Golgi trafficking of ceramide in semiintact cells (SI Fig. 13). The W473A/W562A mutant of CERT retains the ability to localize at the Golgi region (SI Fig. 15), consistent with the idea that CERT can target the Golgi apparatus by recognizing phosphatidylinositol 4-monophosphate via the pleckstrin homology domain of CERT (7). In contrast, the WW motif of PITP-β is required for its localization to the Golgi region (20).
Fig. 6.
Temperature factors and a putative entrance to the amphiphilic cavity. (A–D) Ribbon diagrams of the CERT START domain colored according to the crystallographic B-factors for the apo-CERT START domain and in complex with C6-, C16-, and C18-ceramide, respectively. The color bar below A shows the crystallographic B-factor scale. (A–E) The side chains of Trp-473 and Trp-562 are drawn as stick models. The ceramide molecules are drawn as filled spheres. (E) Ribbon representation of the CERT START domain in complex with C18-ceramide. The structure is rotated by 45° around the y axis with respect to those shown in D. α3 and Ω1 loop are colored cyan and magenta, respectively. C18-ceramide is drawn as space-filling spheres, in which yellow, blue, and red spheres represent C, N, and O atoms, respectively. (F) Molecular surface of the CERT START domain in complex with C18-ceramide, drawn in the same orientation as in E. The hydrophobic surface is painted green, and the residues in α3 and Ω1 loop are highlighted as dotted spheres in cyan and magenta, respectively. C18-ceramide is drawn as in E.
Taken together, it is conceivable that the exposed tryptophan residues, Trp 473 and/or Trp-562, might orient the CERT START domain relative to the membrane, and the α3 and the Ω1 loop might act as the gate of the amphiphilic cavity for ceramide uptake and release. Glu-446, which is the most important residue in ceramide transfer, is located just after α3. In the case of MLN64, one of the cholesterol transfer proteins in the START domain family, molecular dynamics simulations indicated that the transient opening of the Ω1 loop is sufficient for uptake and release of cholesterols (21). Although the issue of the gating mechanisms needs further investigation, the present crystallographic study has revealed the atomic mechanisms underlying ceramide-specific recognition by the START domain of CERT, and showed that the amphiphilic cavity of the START domain is optimized for specific binding of natural ceramides.
Methods
Plasmid Construction.
Boundaries of the CERT START domain suitable for the crystallographic studies (residues 347–598) were identified by SDS/PAGE, mass spectroscopy, and N-terminal peptide sequencing of trypsin digested products of CERT. A cDNA encoding the START domain was subcloned into pGEX-5T1 (Amersham Biosciences) with a TEV protease recognition sequence, resulting in a plasmid with an N-terminal GST-tag and a protease cleavage site. Mutations were introduced with a QuikChange site-directed mutagenesis kit (Stratagene).
Purification of the CERT START Domain.
Transformed Escherichia coli DH5α cells were grown in Luria–Bertani medium. After addition of 0.1 mM isopropyl-d-thiogalactoside, the cells were cultured for 20 h at 30°C. The GST-fused CERT START domain was purified from soluble cell lysate by GS-4B affinity chromatography (Amersham Biosciences). After removal of the GST-tag by incubation with TEV protease (Invitrogen), the CERT START domain was purified by Ni-NTA affinity (Qiagen), GS-4B affinity, MonoQ ion-exchange (Amersham Biosciences), and Superdex-75 size-exclusion (Amersham Biosciences) chromatography columns. The selenomethionine (Se-Met)-labeled CERT START domain was purified with the same procedure from E. coli DL41 cells which were grown in minimal media containing Se-Met. Mutant proteins were expressed and purified using the same procedure described above. Circular dichroism spectra of the wild-type and mutants of the CERT START domain indicate that they have essentially identical secondary structures.
Crystallization.
Crystallization of the CERT START domain was performed by hanging-drop vapor diffusion. Protein samples (10–15 mg·ml−1) were mixed with an equal volume of precipitant solution [P1 crystal: 15–20% (wt/vol) PEG-3350, 0.1 M Mes-NaOH, pH 6.0–6.2; P212121 and P21 crystal: 10–15% (wt/vol) PEG-3350, 0.1 M Mes-NaOH, pH 6.0–6.2, 0.2 M potassium fluoride]. For cocrystallization, 100 mM ceramide (Avanti Polar Lipids) dissolved in 100% dimethyl sulfoxide was added to the protein solution at a final concentration of 1 mM, and the mixture was subjected to vapor diffusion. Single crystals with C6-ceramide appeared in 2–5 days at 20°C. Apo-form crystals and crystals with C16-, C18-, C24-ceramide, and C10-diacylglycerol were grown in the same condition after streak seeding, using C6-ceramide bound crystals as a seed. The crystals appeared in 2–5 days at 20°C.
X-Ray Data Collection and Structure Determination.
Crystals were cryoprotected in reservoir solutions supplemented with 10% glycerol and flash-frozen in nitrogen stream gas at 95 K. Data were collected at beamlines BL-5A, BL-6A, and AR-NW12A of the Photon Factory and processed with HKL2000 (22). The selenium positions were located and phases were calculated with SOLVE (23). Density modification and initial model building were carried out with RESOLVE (24) and ARP/wARP (25). Further manual model building was continued with XtalView (26), and refmac5 (27) was used for refinement of the models. Figs. 1, 3, 4, and 6 and SI Figs. 7, 8, and 11 were prepared with PyMol (http://www.pymol.org).
Ceramide Extraction Assay.
The activity of extraction of ceramide from phospholipid vesicles was determined as described (7), with minor modifications. The reaction mixture in the present study contained purified CERT (30 pmol), bovine albumin (5 μg), and phospholipid vesicles (composed of 40 nmol of phosphatidylcholine, 10 nmol of phosphatidylethanolamine, and 0.1 nmol of 14C-ceramide) in 50 μl of 20 mM Hepes-NaOH buffer (pH 7.4) containing 50 mM NaCl and 1 mM EDTA.
Lipid Transfer Assay.
Lipid transfer between artificial phospholipid membranes was assayed by using radioactive C16-ceramide or dioleoylglycerol (American Radiolabeled Chemicals) as transfer substrates, as described (13), with several minor modifications. Because ceramide transfer activity of the CERT START domain is far higher than that of CERT (7), the amount of the CERT START domain per assay of ceramide transfer was reduced to 50 fmol (1.45 ng) in the ceramide transfer assay. Protein [2.5 pmol (72.5 ng)] was used for diacylglycerol. In addition, to prevent adsorption of the CERT START domain on the tube wall, polypropylene 1.5-ml assay tubes were coated with BSA by soaking with 250 μl of 0.2 mg/ml BSA (fatty-acid free, Sigma) for 5 min before use. The reaction mixtures of lipid transfer assay were supplied with 5 μg of BSA. BSA alone catalyzed no appreciable transfer of ceramide or diacylglycerol under the experimental conditions used.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Ms. Tamie Aoki-Katsuta for technical assistance in protein purification and crystallization. This work is supported by the Protein 3000 Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), a MEXT Grant-in-Aid for Scientific Research (B), the Japan Health Sciences Foundation, and the TORAY Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2E3M, 2E3N, 2E3O, 2E3P, 2E3Q, 2E3R, 2E3S, 2Z9Y, and 2Z9Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709191105/DC1.
References
- 1.Hanada K, Kumagai K, Tomishige N, Kawano M. Biochim Biophys Acta. 2007;1771:644–653. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Futerman AH, Hannun YA. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, Nishijima M. J Biol Chem. 1998;273:33787–33794. doi: 10.1074/jbc.273.50.33787. [DOI] [PubMed] [Google Scholar]
- 4.Fukasawa M, Nishijima M, Hanada K. J Cell Biol. 1999;144:673–685. doi: 10.1083/jcb.144.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funakoshi T, Yasuda S, Fukasawa M, Nishijima M, Hanada K. J Biol Chem. 2000;275:29938–29945. doi: 10.1074/jbc.M004470200. [DOI] [PubMed] [Google Scholar]
- 6.Yasuda S, Kitagawa H, Ueno M, Ishitani H, Fukasawa M, Nishijima M, Kobayashi S, Hanada K. J Biol Chem. 2001;276:43994–44002. doi: 10.1074/jbc.M104884200. [DOI] [PubMed] [Google Scholar]
- 7.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 8.Raya A, Revert F, Navarro S, Saus J. J Biol Chem. 1999;274:12642–12649. doi: 10.1074/jbc.274.18.12642. [DOI] [PubMed] [Google Scholar]
- 9.Raya A, Revert-Ros F, Martinez-Martinez P, Navarro S, Rosello E, Vieites B, Granero F, Forteza J, Saus J. J Biol Chem. 2000;275:40392–40399. doi: 10.1074/jbc.M002769200. [DOI] [PubMed] [Google Scholar]
- 10.Kawano M, Kumagai K, Nishijima M, Hanada K. J Biol Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 11.Loewen CJ, Roy A, Levine TP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soccio RE, Breslow JL. J Biol Chem. 2003;278:22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- 13.Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K. J Biol Chem. 2005;280:6488–6495. doi: 10.1074/jbc.M409290200. [DOI] [PubMed] [Google Scholar]
- 14.Tsujishita Y, Hurley JH. Nat Struct Biol. 2000;5:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 15.Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Proc Natl Acad Sci USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Nat Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 17.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yau W, Wimley WC, Gawrisch K, White SH. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 20.Phillips SE, Ile KE, Boukhelifa M, Huijbregts RPH, Bankaitis VA. Mol Biol Cell. 2006;17:2498–2512. doi: 10.1091/mbc.E06-01-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murcia M, Faráldo-Gómez JD, Maxfield FR, Roux B. J Lipid Res. 2006;47:2614–2630. doi: 10.1194/jlr.M600232-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terwilliger TC. Acta Crystallogr D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrakis A, Morris RM, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 26.McRee DE. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 27.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.